Abstract
Secreted aspartic proteases (Saps) are extracellular proteolytic enzymes that enhance the virulence of Candida pathogens. These enzymes therefore represent possible targets for therapeutic drug design. Saps are inhibited by nanomolar concentrations of the classical inhibitor of aspartic proteases pepstatin A and also by the inhibitors of the HIV protease, but with the Ki of micromolar values or higher. To contribute to the discussion regarding whether HIV protease inhibitors can act against opportunistic mycoses by the inhibition of Saps, we determined the structure of Sapp1p from Candida parapsilosis in complex with ritonavir (RTV), a clinically used inhibitor of the HIV protease. The crystal structure refined at resolution 2.4 Å proved binding of RTV into the active site of Sapp1p and provided the structural information necessary to evaluate the stability and specificity of the protein-inhibitor interaction.
Introduction
The treatment strategy used against HIV/AIDS relies on a combination of at least three compounds: two nucleoside reverse transcriptase inhibitors, and one non-nucleoside reverse transcriptase inhibitor or one protease inhibitor (PI). This regimen, known as Highly Active Antiretroviral Therapy (HAART) leads to a dramatic decrease in viral loads and an improvement in CD4 countsCitation1. The marked reduction of oropharyngeal candidiasis (OPC), which is one of the common HIV-related diseases, has been observed particularly in the patients receiving treatment with protease inhibitors. It has been hypothesized that HIV PR inhibitors are active also against aspartic proteases secreted by Candida species, and can thus alleviate opportunistic candidiasisCitation2.
Most of the pathogenic yeasts of the genus Candida possess a gene family encoding secreted aspartic proteases (Saps) that are important for the virulence of these microorganisms. Saps belong to the same family as HIV PR, sharing the catalytic mechanism and some of the structural features. They participate in the establishment of infection by facilitating adhesion to host surfaces. Saps also degrade host defense molecules such as the Fc portion of immunoglobulin molecules and contribute to host tissue damageCitation3,Citation4. Pepstatin A, a specific inhibitor of aspartic proteases, was found to exert a protective effect in the early stages of a Candida attack, and comparable results were obtained also with the HIV PR inhibitors ritonavir, amprenavir, saquinavir, nelfinavir, indinavirCitation5–9. These studies also indicate that protease inhibitors may be active at least against surface mycoses including OPC, although in deep-seated systemic infection models the efficacy of protease inhibitors has not been unequivocally provenCitation10.
A doubt concerning the anti-fungal effect of HIV PR inhibitors arises from high Ki values that were obtained for these compounds and Saps from several pathogenic Candida speciesCitation10,Citation11,12. While pepstatin A and its analogs inhibit Saps in nanomolar concentrations, inhibition constants for Saps and HIV PR inhibitors ritonavir, saquinavir, indinavir and nelfinavir are within the micromolar range or higher. It is therefore questionable, whether OPC reduction in AIDS patients should be attributed to the inhibition of Saps, or rather to the overall recovery of host anti-microbial immunity. On the other hand, micromolar concentration of HIV PR inhibitors can be regularly found in the plasma of patients under the HAART treatmentCitation13.
To prove the binding of HIV PR inhibitors to Candida secreted aspartic proteases on a molecular level, we co-crystallized Sapp1p, the secreted protease of Candida parapsilosis, in complex with HIV-1 PR inhibitor ritonavir (RTV).
OPC is most frequently caused by C. albicans, however, other Candida species are being increasingly isolated from oropharyngeal lesions, and C. parapsilosis often ranges as the second most common OPC causative agentCitation14,Citation15. Ritonavir (RTV) was designed by Abbot laboratories and gained FDA approval in 1996 under the name NorvirCitation16. Due to the frequent occurrence of side effects and a high cross-resistance with other PIs, its use as a HIV protease inhibitor was gradually abandoned. Unexpectedly, RTV has proven to be a potent inhibitor of cytochrome P-450 3A4, the microsomal enzyme responsible for the bulk of the metabolism of other HIV PR inhibitors. Therefore, RTV is currently used almost exclusively as a pharmacokinetic boosting agent.
The crystal structure of Sapp1p from Candida parapsilosis in complex with ritonavir was determined at a resolution of 2.4 Å. The interactions of RTV in the active site of Sapp1p were analyzed and compared to the available structure of HIV-1 PR in complex with RTV and the structure of Sapp1p complex with pepstatin A, which is the classical inhibitor of aspartic proteases.
Materials and methods
Protein preparation
Sapp1p was purified from its natural source. C. parapsilosis strain P-69 was obtained from the mycological collection of the Faculty of Medicine, Palacky University, Olomouc, Czech Republic. The yeasts were cultivated in a YCB-BSA medium (1.2% yeast carbon base, 0.2% BSA, 15 mM sodium citrate, pH 4.0). Sapp1p was purified from the cell-free supernatants obtained from C. parapsilosis cultivation for 72 h at 30°C in a rotation shaker. The cells were harvested by centrifugation (5000g for 15 min). The supernatant was filtered using Stericup (Milipore, Billerica, MA, USA). The cell-free supernatant containing secreted proteases was applied to an SP Sepharose fast flow (Sigma-Aldrich, St. Louis, MO, USA) equilibrated with 15 mM of sodium citrate pH 4.0. The fractions containing Sapp1p/Sapp2p were eluted with gradient 0–300 mM NaCl, collected and concentrated using an Amicon Ultra – 30k (Millipore) to a volume of 5 ml. The concentrate was loaded on the HiLoad Superdex™ 75 26/60 (GE Healthcare, Piscataway, NJ, USA) column equilibrated in 25 mM Bis-Tris HCl, pH 6.3. After gel chromatography, the active fractions containing the mixture of Sapp1p and Sapp2p were collected. Sapp1p and Sapp2p were separated by chromatofocusing on Mono P 5/200 GL (GE Healthcare) column equilibrated with 25 mM Bis-Tris, pH 6.3. Elution of Sapp1p and Sapp2p was performed using a gradient of 10% polybuffer 74, pH 4.0 (GE Healthcare). The efficiency of purification steps was analyzed using SDS-PAGE, Western blotting, and activity assays. Protein analyses and proteolytic activity assays were previously describedCitation17.
Protein crystallization
The Sapp1p-ritonavir (RTV) complex was prepared by mixing the enzyme with a 100-fold molar excess of RTV (dissolved in dimethyl sulfoxide) and concentrated by ultrafiltration to a concentration of 25 mg·ml−1 using Amicon Ultra− 0.5 ml 30k (Millipore). Initial crystallization trials were performed with the help of the Gryphon crystallization workstation (Art Robbins, Sunnyvale, CA, USA) by the vapor diffusion method in sitting drop mode at 19°C in 96-well plates; 0.2 μl protein solution was mixed with a 0.2 μl well solution and the mixture was equilibrated over a 50 μl reservoir solution. The PEGs Suite and JSCG Core I Suite (QIAGEN, Germantown, MD, USA) were used for the initial crystallization condition screen. Initial microcrystals appeared after several days in various conditions containing 100 mM MES pH 6.5 and PEG 200–400 as the precipitant. Further optimization was done manually, and involved changing to the hanging drop mode in the 24-well crystallization plates (EasyXtal DG-Tool, QIAGEN). Crystals were obtained by mixing 3 μl of the protein-RTV complex solution with 1 μl of reservoir solution composed of 100 mM MES pH 6.5, and 30% PEG 400. Crystals appeared after one day in the form of crystal clusters (Supplementary Figure S1) and reached a full size of 200 × 150 × 100 µm within 2 days. These crystals were unstable and during several days started to disintegrate. For data collection, one crystal was removed from the cluster and cryocooled in liquid nitrogen after 2 days of growth.
Data collection and structure determination
Diffraction data for the 2.4 Å resolution set were collected at 100 K using the MX14.2 beamline at BESSY, Berlin, Germany. Data were integrated and reduced using MOSFLMCitation18 and scaled using SCALACitation19 from the CCP4 suiteCitation20. Crystal parameters and data collection statistics are given in .
Table 1. Crystal data and diffraction data collection and refinement statistics.
Structure was determined by molecular replacement using the program MolrepCitation21. The search model was derived from the structure Sapp1p-pepstatin A inhibitor complex structure (PDB code 3FV3Citation17). Model refinement was carried out using the program REFMAC 5.2Citation22 from the CCP4 packageCitation20. Manual building was performed using CootCitation23. Medium NCS (non-crystallographic symmetry) restraints were applied during the refinement. The final refinement steps included TLS (translation/libration/screw) refinementCitation24. The quality of the final model was validated with MolprobityCitation25. The shape complementary valueCitation26 was calculated by the SC program implemented in CCP4Citation20. The reported value represents an average from the individual protein-inhibitor complexes present in the asymmetric unit. Refinement statistics are given in . Figures showing structural representations were prepared with the program PyMOLCitation27. Atomic coordinates and experimental structure factors have been deposited with the Protein Data Bank with the code 3TNE.
Results and discussion
Crystal structure
The crystal structure of the Candida parapsilosis Sapp1p, produced from its natural source, in complex with ritonavir (RTV) was determined by molecular replacement and refined using data to 2.4 Å resolution ().
Figure 1. (A) Overall three dimensional structure and secondary structure elements of Sapp1p complexed to RTV. The protein in the ribbon representation is colored by secondary structure, the RTV molecule is represented by sticks. the transparent protein solvent accessible surface area is also shown. (B) The 2Fo-Fc electron density map for RTV is contoured at 0.8σ. The carbon atoms of RTV are shown in green, oxygen, nitrogen and sulfur atoms are colored red, blue and golden, respectively. Two catalytic aspartates are depicted with carbon atoms colored yellow. (C) The detail of the RTV interaction with residues in the Sapp1p active site. Hydrogen bonds are shown as dashed lines; the number represents distance between hydrogen bond donor and acceptor in Å.
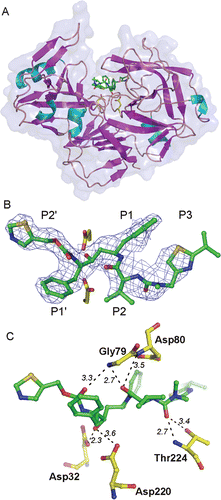
Obtaining a well-shaped single crystal for the diffraction analysis of the Sapp1p-RTV complex represented a major obstacle. Due to the low affinity of RTV toward Sapp1p, a mixture of different protein species (Sapp1p in free form and complexed with RTV) was present in the crystallization drops. This heterogeneity resulted in a suboptimal crystal nucleation and growth, and resulted in the formation of crystal clusters (Supplementary Figure S1).
As revealed by diffraction analysis, the individual parts of the clusters belonged to two different crystal forms with diverse compositions. Crystal form I belonged to the P212121 space group with cell dimensions a = 87.45 Å, b = 87.69 Å, and c = 160.5 Å. Crystal form I contained four protein subunits in the unit cell with no inhibitor present in active site. Crystal form II belonged to space group P212121 with cell dimensions a = 61.6 Å, b = 61.8 Å, and c = 158.0 Å. The crystal form II contained two protein subunits in the unit cell and the continuous map for RTV was found in the active sites of both protein molecules. The optimization of crystallization conditions to obtain single crystals of crystal form II failed even at an extremely high excess of inhibitor over protein and thus a part of the crystal cluster was used to collect a complete dataset. The diffraction pattern consisted of diffraction from several crystal lattices (Supplementary Figure S1); only the major lattice was selected during the data processing and we ascertained that the error models for data integration and scaling were properly chosen. Data collection statistics are shown in .
The orthorhombic crystal form contains two protein-inhibitor complexes in the asymmetric unit (AU) with a solvent content of 36.7%. The final crystallographic model consists of two molecules of the Sapp1p-RTV complexes, each molecule containing 339 protein residues and one RTV molecule.
The two protein chains in the AU are quite similar: the root mean square deviation (RMSD) for superposition of 339 Cα atoms is 0.281 Å; a value within the range observed for different crystal structures of identical proteinsCitation28. The refined Sapp1p-RTV structure shows no significant conformational changes in the protein chain compared to the structure of the same protein in complex with pepstatin A (PDB code 3FV3Citation17). The RMSD for superposition of 339 Cα atoms of different protein chains was 0.445–0.502 Å.
All atoms of RTV were modeled into an electron density map with a well-defined central region and rather disordered parts for substituents in positions P2 and P3 (). Due to a suboptimal crystal and consequently the diffraction data quality, the quality of the resulting crystal structure is also suboptimal. Nevertheless, all crystallographic statistical indicators of structure quality are acceptable (see refinement statistics in ) and the crystallographic model can evidently be used to analyze the structural features of RTV binding to Sapp1p.
Inhibitor binding
Sapp1p structure is composed of two domains forming a central binding cleft that accommodates a substrate (). Each domain provides one catalytic aspartate positioned at the bottom of the active site. The active site is covered by an anti-parallel β-sheet flap (residues 71–89), a common feature of aspartic proteases.
RTV binds to the Sapp1p active site in an extended conformation and occupies the S2-S3′ enzyme substrate binding subsites. The hydroxyl group of the central RTV moiety forms two hydrogen bonds with the side-chains of the two catalytic aspartates Asp32 and Asp220 (). Additional polar interactions involve hydrogen bonds with main-chain atoms of Ser35 (N), Gly79 (N), and Thr224 (N), and also a side-chain of Thr224 (Oγ1) (). Upon the complex formation, RTV buries 313 ÅCitation2 of solvent accessible surface area, which represents 92.3% of its total solvent accessible surface area.
Superposition of our structure with pepstatin A (PepA), a typical inhibitor of aspartic proteases, bound to the active site of Sapp1p is shown in . The Ki values for inhibition of Sapp1p by PepA and RTV are 0.3 nMCitation11 and 1.9 μMCitation11, respectively, a difference of four orders of magnitude. A comparison of inhibitor interactions in the Sapp1p active site showed suboptimal interactions of RTV over PepA. The central hydroxyl group of RTV and PepA interacts with catalytic aspartates in a similar way, however the interactions with individual substrate binding pockets differ substantially. The only exception is the P2 substituent, which is structurally similar in RTV and PepA. Bulky aromatic substituents in positions P1 and P1′ of RTV form very few interactions (P1′ phenyl) or even some unfavorable interactions (e.g. P1 phenyl with Ile30) with their corresponding Sapp1p substrate binding pockets. Thiazolyl substituents in P3 and P2′ positions do not fit into corresponding S3 and S2′ sites. Suboptimal filling of the enzyme subsite binding pockets by RTV can be documented by a comparison of the shape complementarity of the Sapp1p active site with RTV and PepA, respectively. The shape complementarity (SCCitation26) value for PepA is 0.80 ± 0.02, while the SC value for RTV is 0.68 ± 0.02. Such a low value points to a limited stability and specificity of the protein-ligand interaction.
Figure 2. (A) Comparison of RTV and PepA binding modes in the Sapp1p active site. Carbon atoms of RTV and PepA are shown in green and yellow, respectively. Oxygen, nitrogen and sulfur atoms are colored red, blue and golden, respectively. Protein is shown by a solvent accessible surface area with underlying catalytic aspartates shown as sticks. Substrate binding sites S4-S3′ are labeled. Flap region (residues 71–89) is omitted from the figure for clarity. (B) Superposition of Sapp1p and HIV-1 PR complexes with RTV. Structure of HIV-1 PR in complex with RTV (PDB code 1HXWCitation16) is colored pink; the structure of Sapp1p in complex with RTV is colored green. Superposition was done by matching the catalytic triads (catalytic aspartates are highlighted as sticks at the bottom of the active sites). (C) Comparison of RTV binding modes in the active sites of Sapp1p (carbon atoms colored green) and HIV-1 PR (carbon atoms colored pink).
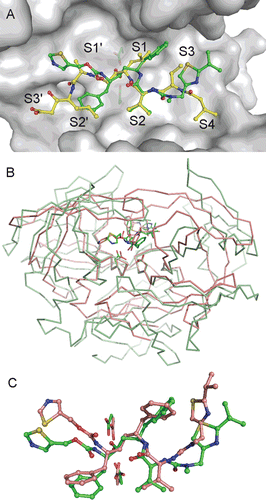
RTV was designed and selected to fit the active site of HIV-1 PR and the reported Ki value for inhibition of HIV-1 PR with RTV is 0.12–0.17 nMCitation29,Citation30. The superposition of the Sapp1p structure with the structure of HIV-1 protease is shown in and the superposition of the binding modes of RTV in the Sapp1p and HIV-1 PR binding sites, respectively, is depicted in . The central part of RTV binds similarly to HIV-1 PR and Sapp1p as a result of the sequence and structural conservation of the catalytic triads in the aspartic proteases. Position of P1 and P1′ substituents are also quite similar, although the conformation of the aromatic rings differs. The substituent in positions distal from the central hydroxyl of RTV differ in structure, especially the position of thiazolyl groups in P2 and P3′, which were specifically designed to form an optimal interaction with hydrophobic residues Pro81 and Val82 of HIV-1 PR in the S3 subsite, which is not obviously present in Sapp1p.
In conclusion, the structural analysis showed that RTV can specifically bind to the Sapp1p active site and forms specific polar interactions with catalytic aspartates and additional hydrogen bonds with the residues in S1 and S1′ subsites. The interaction of the central part of RTV with the enzyme catalytic site is similar to the interaction of PepA with Sapp1p as well as to interaction of RTV with HIV-1 PR. Non-polar interactions of RTV with Sapp1p are quite limited. The low shape complementarity of RTV with individual substrate binding subsites results in a low inhibition efficiency of RTV toward Sapp1p in the micromolar range. Although the structure of the Sapp1p-RTV complex by itself does not explain the reported anti-mycotic activity of the HIV protease inhibitors, it sheds light on the concept of a multi-target effect of ritonavir.
Supplementary Material
Download PDF (457.5 KB)Acknowledgements
This work was supported by research projects AV0Z40550506, and AV0Z50520514 awarded by the Academy of Sciences of the Czech Republic. Additional support was awarded also by Ministry of Education of the Czech Republic (grant LC 531), and by the Czech Science Foundation (grants 310/09/1945 and 203/09/0820). Diffraction data were collected on beamline MX14.2 at BESSY, Berlin, Germany. The authors wish to thank Devon Maloy for critical proofreading of the manuscript.
Declaration of interest
The authors declare no conflicts of interest.
References
- Piacenti FJ. An update and review of antiretroviral therapy. Pharmacotherapy 2006;26:1111–1133.
- Munro CA, Hube B. Anti-fungal therapy at the HAART of viral therapy. Trends Microbiol 2002;10:173–177.
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 2003;67:400–428.
- Hruskova-Heidingsfeldova O. Secreted proteins of Candida albicans. Front Biosci 2008;13:7227–7242.
- Cassone A, De Bernardis F, Torosantucci A, Tacconelli E, Tumbarello M, Cauda R. In vitro and in vivo anticandidal activity of human immunodeficiency virus protease inhibitors. J Infect Dis 1999;180:448–453.
- Wurzner R, Bektic J, Lell CP, Fuchs A, Stoiber H, Speth C, Lass-Florl C, Borg-von Zepelin M, Dierich MP. HIV protease inhibitors attenuate adherence of Candida albicans to epithelial cells in vitro. FEMS Immunol Med Microbiol 2001;31:65–71.
- Schaller M, Bein M, Korting HC, Baur S, Hamm G, Monod M et al. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect Immun 2003;71:3227–3234.
- Monod M, Borg-von Zepelin M, Meyer I, Thomssen R, Wurzner R, Sanglard D, Telenti A. HIV-Protease inhibitors reduce cell adherence of Candida albicans strains by inhibition of yeast secreted aspartic proteases. J Invest Dermatol 1999;113:747–751.
- Fallon K, Bausch K, Noonan J, Huguenel E, Tamburini P. Role of aspartic proteases in disseminated Candida albicans infection in mice. Infect Immun 1997;65:551–556.
- Falkensammer B, Pilz G, Bektic J, Imwidthaya P, Jöhrer K, Speth C et al. Absent reduction by HIV protease inhibitors of Candida albicans adhesion to endothelial cells. Mycoses 2007;50:172–177.
- Pichová I, Pavlícková L, Dostál J, Dolejsí E, Hrusková-Heidingsfeldová O, Weber J et al. Secreted aspartic proteases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae. Inhibition with peptidomimetic inhibitors. Eur J Biochem 2001;268:2669–2677.
- Dostál J, Hamal P, Pavlícková L, Soucek M, Ruml T, Pichová I et al. Simple method for screening Candida species isolates for the presence of secreted proteinases: a tool for the prediction of successful inhibitory treatment. J Clin Microbiol 2003;41:712–716.
- Danner SA, Carr A, Leonard JM, Lehman LM, Gudiol F, Gonzales J et al. A short-term study of the safety, pharmacokinetics, and efficacy of ritonavir, an inhibitor of HIV-1 protease. European-Australian Collaborative Ritonavir Study Group. N Engl J Med 1995;333:1528–1533.
- Baradkar VP, Mathur M, Kumar S. Hichrom candida agar for identification of Candida species. Indian J Pathol Microbiol 2010;53:93–95.
- Back-Brito GN, Mota AJ, Vasconcellos TC, Querido SM, Jorge AO, Reis AS et al. Frequency of Candida spp. in the oral cavity of Brazilian HIV-positive patients and correlation with CD4 cell counts and viral load. Mycopathologia 2009;167:81–87.
- Kempf DJ, Marsh KC, Denissen JF, Mcdonald E, Vasavanonda S, Flentge CA, Green BE, Fino L, Park CH, Kong XP, Wideburg NE, Saldivar A, Ruiz L, Kati WM, Sham HL, Robins T, Stewart KD, Hsu A, Plattner JJ, Leonard JM, Norbeck DW. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. PNAS 1995;92:2484–2488.
- Dostál J, Brynda J, Hrusková-Heidingsfeldová O, Sieglová I, Pichová I, Rezácová P. The crystal structure of the secreted aspartic protease 1 from Candida parapsilosis in complex with pepstatin A. J Struct Biol 2009;167:145–152.
- Leslie AG. Integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 1999;55:1696–1702.
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 2006;62:72–82.
- The CCP4 suite: programs for protein crystallography.Acta Crystallogr D Biol Crystallogr 1994;50:760–763.
- Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr 2000;56:1622–1624.
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 2011;67:355–367.
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126–2132.
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 2001;57:122–133.
- Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 2003;50:437–450.
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol 1993;234:946–950.
- DeLano WL. 2002. The PyMOL Molecular Graphics System. DeLano Scientific LLC, San Carlos, CA, USA. Available from <http://www.pymol.org>.
- Betts MJ, Sternberg MJ. An analysis of conformational changes on protein-protein association: implications for predictive docking. Protein Eng 1999;12:271–283.
- Rinnová M, Hradilek M, Barinka C, Weber J, Soucek M, Vondrásek J et al. A picomolar inhibitor of resistant strains of human immunodeficiency virus protease identified by a combinatorial approach. Arch Biochem Biophys 2000;382:22–30.
- Klabe RM, Bacheler LT, Ala PJ, Erickson-Viitanen S, Meek JL. Resistance to HIV protease inhibitors: a comparison of enzyme inhibition and antiviral potency. Biochemistry 1998;37:8735–8742.
- Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 1992;355:472–475.
