Abstract
Context: Isolation and characterization of OXA-162, a novel variant of OXA-48.
Objectives: Klebsiella pneumoniae isolate with decreased susceptibility to carbapenems was recovered from a Turkish patient. This study aimed at characterizing the carbapenem resistance determinants of this isolate.
Materials and methods: Antibiotic susceptibility tests, analytic isoelectric focusing (IEF), cloning and sequencing were performed. Cloned β-lactamase was purified by means of preparative gel electrophoresis and the kinetic constants were determined under initial rate conditions.
Results: The identified blaOXA-162 gene was located on a ca. 45-kb plasmid carrying a transposon consisted of two IS1999-2 elements. OXA-162 differed from OXA-48 by a single amino acid substitution (Thr213Ala) which increased the catalytic efficiency (kcat/KM) of OXA-162 towards imipenem and meropenem. Also this substitution caused a gain of hydrolysis ability towards doripenem. Analysis of OXA-162 model implied that the amino acid change might generate an extension in the opening of the substrate entry site and might cause extended hydrolytic activity towards imipenem, meropenem and doripenem.
Discussion and conclusion: OXA-162, a derivative of OXA-48 has enhanced catalytic properties.
Introduction
The spread of carbapenem-hydrolyzing β-lactamases among Enterobacteriaceae is an emerging health threatCitation1. Carbapenemases inactivate carbapenem antibiotics and increase the minimum inhibitory concentration (MIC) in most cases proportional to their hydrolytic activity and expression level. In some instances, MICs do not exceed the resistant breakpointsCitation2. However, available data imply that carbapenemase production is clinically significant, even in infections caused by low-level carbapenemase-producersCitation3.
Carbapenemases belonging to Ambler class A, class B and class D are widespread. The first cloned and sequenced Ambler class A carbapenemase is NMC-A, identified in 1990 in a clinical isolate of Enterobacter cloacae (NOR-1), which hydrolyses imipenem and aztreonamCitation4,Citation5. Due to the intercontinental spread, Klebsiella pneumoniae carbapanemase (KPC) variants are more popular among all other class A carbapenemases, of which the first member was identified in a K. pneumoniae isolate before 1997 in USACitation6. Class B carbapenemases are metallo-enzymes, typically hydrolyze carbapenems but not aztreonam, are detected mainly among Pseudomonas and Acinetobacter species and are sporadic among the members of EnterobacteriaceaeCitation7. Class D carbapenemases (OXA-carbapenemases) are highly versatile; they disseminate on various mobile elements among nosocomial species and some members are natural in certain genomic species of AcinetobacterCitation8,Citation9.
According to sequence homology, OXA-carbapenemases are clustered in eight subgroups, most of which are weak enzymes and detected in Acinetobacter spp.Citation10. In 2001, a novel OXA-carbapenemase, OXA-48, was identified in a clinical K. pneumoniae isolate in Istanbul, TurkeyCitation11. Subsequent studies documented that blaOXA-48 spread on at least two distinct plasmids in Turkey as well as in Belgium, Egypt, France and LebanonCitation12,Citation13. One of these plasmids was a 70-kb plasmid carrying blaOXA-48.
We identified a variant of OXA-48 in a clinical isolate of K. pneumoniae from Kocaeli University Hospital. The genetic environment and enzymatic properties of this novel variant, assigned as OXA-162, were studied in detail.
Materials and methods
Bacterial strains, plasmids and antimicrobial resistance studies
K. pneumoniae strain KIRK1 was isolated from a patient (2 years of age) with nosocomial pneumonia. The patient was treated in the pediatric intensive care unit (ICU) of Kocaeli University Hospital in 2009. This was the first detection of Klebsiella with unusual MICs to carbapenem antibiotics in this hospital. During the following months, there were three more cases who were suffering from nosocomial infections caused by K. pneumoniae with carbapenem-resistant phenotypes. The outbreak ceased after the application of strict infection control measures in the ICU. The strain was initially identified by VITEK 2 (bioMérieux Inc., Marcy l’Etoile, France) and later confirmed in our laboratory using conventional techniques such as colony morphology, sugar fermentation properties, oxidase reaction and gram-staining. For cloning and transformation experiments, DH10B (Invitrogen, CA, USA) was the host cell and pBK-CMV (Stratagene, NJ, USA) was the plasmid used. The clone producing OXA-48 in E. coli (pVT-1) that was used in this study was a giftCitation11.
As recommended by the Clinical and Laboratory Standards Institute (formerly NCCLS), antibiotic MICs were studied by broth microdilution assay with a final inoculum of 5 × 104 CFU/wellCitation14. The MICs of imipenem, meropenem and doripenem were confirmed by E-test (bioMérieux Inc., France). Endpoints of the resistance tests were read after 18 h of incubation at 35°C.
Antimicrobial agents and their sources were as follows: ampicillin (Mustafa Nevzat, Istanbul, Turkey), cefepime (Bristol-Myers Squibb, NJ, USA), ceftazidime (GlaxoSmithKline, Middlesex, UK), doripenem (Janssen-Cilag, NJ, USA), ertapenem (Merck Sharp & Dohme, NJ, USA), imipenem (Merck, NJ, USA), meropenem (AstraZeneca, DE, USA), piperacillin and piperacillin/tazobactam (Wyeth-Ayerst, CA, USA).
DNA purification, restriction analysis and transformation
Plasmid DNA was isolated using the alkaline lysis method, purified via spin columns (NucleoSpin Plasmid Columns, MACHEREY-NAGEL GmbH & Co. KG, Germany) and run on 0.8% agarose gel. The size estimation was done by comparison of the relative mobility of restriction fragments against GeneRuler™ 1-kb DNA Ladder (Fermentas, Lithuania). Genomic DNA, for polymerase chain reaction (PCR) amplification experiments, was isolated by heating a dense bacterial suspension at 95°C for 10 min and sedimenting the debris with high speed centrifugation.
Plasmids were transformed by electroporation. Transformants were selected on 100 µg/mL ampicillin and the existence of a resistance determinant confirmed by the susceptibility phenotype, PCR and isoelectric focusing (IEF)Citation15.
Gene amplification, cloning and sequence analysis
PCR was accomplished in a final volume of 50 µL composed of 1.5 mM MgCl2, 0.8 mM dNTPs, 50 pmol primers each, 2.5 U of Taq polymerase (Fermentas, Lithuania) and 5 µL DNA extract. To amplify fragments longer than 1 kb, long PCR Enzyme Mix (Fermentas, Lithuania) replaced Taq polymerase and the elongation step was adjusted to 1 min for every 1-kb target. To amplify the gene blaOXA-48-type, forward (5′-TATGCGTGTATTAGCCTTATCGGCTG-3′) and reverse primers (5′-CGCGCTAACCACTTCTAGGGAATAAT-3′) were used with 55°C annealing temperature.
To clone the resistance determinants, the plasmid (pKRU-1) was isolated as mentioned above and digested with either BamHI, EcoRI or PstI. The fragments were gel purified and ligated to the corresponding arms of the gel purified and dephosphorylated pBK-CMV at 16°C overnight. Ligation mixtures were electroporated to DH10B at 1700 V and β-lactamase-positive clones were selected on LB-agar supplemented with 100 mg/L ampicillinCitation15. Disk diffusion phenotypes obtained by imipenem, cefepime, meropenem, ampicillin, amoxicillin/clavulanate disks and the IEF profiles of clones displayed the existence and types of resistance determinants.
Dye terminator cycle sequencing with the ABI Prism BigDye Terminator kit (Applied Biosystems, Foster City, CA, USA) was used for sequencing. The assay was carried out according to standard protocol. Data were collected on an ABI 377 automated fluorescence sequencer (CA, USA)Citation9. Sequence data were read and edited by free-ware programs ChromasPro (http://www.technelysium.com.au) and BioEdit (version 7.0.5.2).
Enzyme purification, IEF and kinetic studies
Cells were lysed by sonication in 50 mM Tris–HCl (pH 8.8), then treated with DNase and RNase. Crude cell extracts were prepared by centrifugation of the cell lysate at 25,000×g for 30 min at 4°C. For conventional enzyme purification, a published protocol was followed and crude cell extracts were sequentially applied to Q-Sepharose (HiTrap Q HP, GE Healthcare, NJ, USA) and S-Sepharose (HiTrap SP, GE Healthcare, NJ, US) columnsCitation11. Preparative gel electrophoresis (Prep Cell Model 491 from BioRad, CA, USA) was used for purification of OXA-48 and OXA-162. Gel running and elution buffers for preparative gel electrophoresis contained 25 mM Tris–HCl (pH = 8.3), 192 mM glycine and 10% glycerol. The acrylamide concentration was optimized to 8% and a gel length of 5-cm with 2.5 cm column dimension was used for purification of both enzymes. A total of 3.0 mL crude cell extract which contained approx. 30 mg of protein was mixed with 1 mL of 3× loading buffer (0.0625 M Tris–HCl, pH 6.8, 10% glycerol, 0.025% Bromophenol Blue) and loaded onto the Prep Cell (Model 491; BioRad, CA, USA) and run for 12 h at 8 W and fractions were collected with an elution rate of 1.5 mL/min. After 12 h of run, the collection rate was increased to 3 mL/min and 400 fractions (3 mL each) were collected. To locate the activity containing fractions, 40 µL from every tenth fraction was analyzed by nitrocefin activity assay. Once the enzyme was located, every fraction near the activity containing fractions was analyzed. High activity containing fractions were combined, concentrated to a final volume of 3 mL and stored in liquid nitrogen as droplets until use.
Protein concentration was measured by Lowry assay and SDS-PAGE/BSA titration method and fold of purification was estimated. In SDS-PAGE/BSA titration method, different concentrations of commercially available pure BSA (BioRad, USA) ranging from 10 to 600 ng were run on a 12% SDS-PAGE. The gel was fixed and stained in SYPRO ruby and the intensities of each band was measured by using PD-Quest analysis software (BioRad, USA) to generate a standard curve. Similarly run and stained SDS-PAGE gels containing different volumes of the purified enzymes were then used to estimate protein concentration of OXA-48 and OXA-162. Purity and the relative molecular weight of OXA-162/OXA-48 were assessed by SDS-PAGE. The kcat and KM values were determined by analyzing β-lactam hydrolysis under initial rate conditions with a UV spectrophotometer (UV 1601; Schimadzu, Japan). Enzyme kinetics was studied in 50 mM phosphate buffered saline (pH = 7.4) containing 50 mM NaHCO3 with relevant antibiotics, namely cefepime, ceftazidime, doripenem, ertapenem, imipenem, meropenem and ampicillin.
Analytical IEF was performed on 5% polyacrylamide gels containing ampholytes (pH range = 3–10; Fluka, Switzerland) with a Model 111 Mini IEF Cell (Bio-Rad Laboratories, USA). Enzymes were focused at constant 1 W for 45 min and detected by overlaying the gel with 1 mM nitrocefin solution. Known enzymes SHV-1 (pI = 7.6), OXA-14 (pI = 6.2) and TEM-1 (pI = 5.4) were used as referencesCitation16.
Protein modeling and antibiotic docking
A comparative homology model was built in the Swiss-Model server (http://swissmodel.expasy.org/) using the OXA-48 tertiary structure as the templateCitation17. The crystal structure for OXA-48 is available in PDB entry 3HBR. The modeled structure was analyzed with three different visualization programs: Swiss-Pdb Viewer (http://spdbv.vital-it.ch/download.html), MolMol (http://www.mol.biol.ethz.ch/groups/wuthrich_group/software) and PyMol (http://www.pymol.org/rel/). Molecules were superimposed in Swiss-Pdb Viewer with the iterative magic fit tool or in MolMol with the fit tool. When using Swiss-Pdb Viewer, specific amino acid residues were selected from the control panel whenever needed; when using MolMol and PyMol, selection dialog boxes were used. Amino acid sequences were aligned using ClustalW at EBI (http://www.ebi.ac.uk/emboss/align/index.html). Rigid docking models were created using PatchDock via a web-based server (http://bioinfo3d.cs.tau.ac.il/PatchDock/)Citation18,Citation19. Antibiotic structures for docking were obtained from DrugBank (http://www.drugbank.ca/).
GenBank accession number
The sequence of blaOXA-162 was submitted to GenBank and assigned the accession number “bla HM015773”.
Results and discussion
Characteristics of the K. pneumoniae isolate; origin, resistance phenotype and plasmid profile
The K. pneumoniae strain KIRK1 was the first Klebsiella isolate in Kocaeli University with unusually high MICs for carbapenems such as 8 mg/L for ertapenem, 4 mg/L for imipenem and 1 mg/L for meropenem. The strain was resistant to levofloxacin and gentamicin while intermediate to amikacin and susceptible to tigecycline. The patient was treated with amikacin and tigecycline after detection of the resistant Klebsiella, but unfortunately did not survive.
Crude extract from K. pneumoniae KIRK1 showed considerable activity over carbapenems and harbored a single plasmid (pKRU-1). Transformation of the naturally occurring plasmid carried by Klebsiella to E. coli DH10B enabled us to recover a carbapenemase producing E. coli (pKRU-1). Restriction fragment analysis of pKRU-1 by various enzymes and especially by PstI documented that it is approximately 45-kb. IEF with crude extracts from both KIRK1 and the transformant showed two activities at pI points 5.4 and 7.2 (relative to control enzymes TEM-1, OXA-14 and SHV-1; data not shown).
Cloning plasmidic bla genes, sequencing the recombinant plasmid possessing OXA-162
Two β-lactamases were cloned by PstI fragments; one is focused at pI 5.4 and the other is focused at pI 7.2. Both were narrow spectrum enzymes, not hydrolyzing expanded-spectrum cephalosporins or carbapenems. However, a BamHI-clone exhibited apparent carbapenemase activity. The enzyme responsible for this activity was focused at pI 7.2 which superimposed the narrow spectrum enzyme of pI 7.2 on the IEF gel.
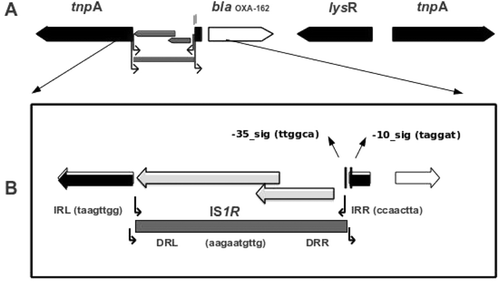
Sequence analysis showed that the recombinant plasmid (pMSH-3) from the BamHI-clone was carrying a 5656-bp insert, of which 5645-bp was similar to the composite transposon Tn1999.2. This transposon was typically characterized by the existence of two inverted copies of the insertion sequence IS1999 on the left and right extremes. The left insertion sequence IS1999 located immediately upstream to blaOXA-48 was disrupted by IS1RCitation13. The sequence mapping of the construct from the recombinant plasmid pMSH-3 was almost identical with Tn1999.2, except that blaOXA-162 replaced blaOXA-48. OXA-162 is a novel variant of OXA-48 and is defined by one amino acid substitution at position 213 (Thr replaced by Ala) of OXA-48. The schematic map of the recombinant plasmid pMSH-3 is shown in .
Figure 1. (A) Schematic map of the construct on the recombinant plasmid pMSH-3-(OXA-162). The coding regions are shown as arrow-head boxes indicating the direction of open reading frame. The insertion sequence IS1R was shown as gray-shaded box. (B) The left insertion sequence IS1999 is disrupted by IS1R between the dedicated promoter signal sequences while leaving the -10 signal intact.
Biochemical properties of OXA-162 β-lactamase and antibiotic resistance phenotypes
Despite of repeated trials and various optimization efforts, by using conventional column chromatography, we failed to purify both enzymes, OXA-48 and OXA-162. The highest purity obtained was only five-fold with a low percent of recovery indicating that in our hands conventional column chromatography failed with these enzymes. By using a preparative gel electrophoresis system, however, we purified both enzymes to an acceptable level (>90% according to gel analysis). Fold of purification was calculated to be 155-fold for OXA-48 and 45-fold for OXA-162. However, fold of purification increased to 357 for OXA-162 with increased acrylamide concentration. Based on the comparisons of catalytic efficiency constants (kcat/KM values), OXA-162 exhibited close hydrolytic activities over the studied cephalosporins and carbapenems except doripenem. For doripenem, OXA-162 displayed activity with a catalytic efficiency constant of 3.55E+04 M−1·s−1 distinguishing OXA-162 from OXA-48 ().
Table 1. Kinetic parameters for OXA-48 and OXA-162.
In a previous study, OXA-48 was reported to hydrolyze ceftazidimeCitation11 In a more recent study, however, the authors could not detect ceftazidime hydrolysis with OXA-48Citation17. We also could not detect ceftazidime hydrolysis activity in crude extracts or with purified OXA-162 and OXA-48. The increase in MIC values of OXA-162 possessing strains for ceftazidime indicated that OXA-162 might have a weak catalytic activity that is below our detection limit but works in vivo due to enzyme abundance.
OXA-162 producing clone exhibited four- to eight-fold higher MICs for carbapenems, except meropenem, than OXA-48 producing clone (). MICs for cefepime and piperacillin were found to be high with OXA-162 producers as well. The most likely explanation for these high MIC values observed with OXA-162 producing clones might be the presence of relatively higher concentration and enhanced activity of OXA-162 in cells in comparison to the OXA-48 producing clone.
Table 2. Minimum inhibitory concentration (MIC) of OXA-162 producing K. pneumoniae strain KIRK1, its transformant, OXA-162 producing clone and OXA-48 producing clone.
In the recent literature, contrasting kinetic data have been presented on OXA enzymesCitation20,Citation21. We also faced difficulties in comparing antibiotic hydrolyzing enzyme variants, such as OXA-48 versus OXA-162. Several reasons may be counted for explanationCitation1: difficulty in obtaining sufficiently pure enzymes. Our experience with OXA-48 and OXA-162 showed that these enzymes are hard to purify to homogeneity (at least in our hands) by using the reported protocolsCitation11,Citation17. This is perhaps, to some extent due to the lack of details provided by the authors who report the purification. In this study, we reported a novel and simple approach for purification of OXA-type enzymes to homogeneityCitation2. Another factor that affects the reported kinetic parameters significantly is the method of measuring concentration of the purified enzymes. Any deviation from true concentration values will cause generation of incorrect and incomparable data. This is especially important when data produced by different laboratories are compared. In this study, we estimated the pure enzyme concentrations by using SDS-PAGE/BSA titration method and Lowry assay. The former one although is more difficult to perform, produced a much more reliable protein concentration estimation. Another important point to emphasize in terms of comparing kinetic data produced among different laboratories is the assay reaction conditions. It has been now recognized that the carbamylation of the lysine residue near the catalytic serine is essential for catalysisCitation21. Therefore, as we did in this study, the kinetic measurements for OXA-type enzymes should be performed in alkaline buffers saturated with sodium bicarbonateCitation20.
Protein modeling and antibiotic docking
Analysis of kinetic data indicated that Ala substitution at position 213 (221 according to Ambler class D β-lactamases [DBL] position)Citation22–24 enhanced the activity of OXA-48 (). The question of how significantly a single amino acid change (Thr → Ala) affects the enzyme structure is a matter of discussion. In this regard, we modeled OXA-162 using OXA-48 as the template. The model structure differed from the template structure with a little deviation (RMSD 0.260) which indicated that the overall structures were quite similar. Closer examination and superimposition of the two structures revealed several dissimilar regions located on the loops. Similar observations were reported elsewhereCitation17.
The presence of three active site motifs was agreed upon in Ambler class D β-lactamases. In OXA-48, these motifs are formed by the residues Ser70-Thr71-Phe72-Lys73 (Motif I), Ser118-Val119-Val120 (Motif II) and Lys208-Thr209-Gly210 (Motif IIICitation17). These active site motifs are also present in OXA-162. Their presence indicated that the active site cavities of both enzymes are similar in shape, dimensions and charge distribution. Despite the presence of the same active site residues, the functional properties of both enzymes differ. This implies the direct or indirect involvement of surrounding residues in catalysis. Based on molecular dynamics simulations of OXA-48, a recent report predicted eight amino acid residues involved in catalysis, at least in meropenem hydrolysisCitation17. Among the residues, Lys73 and Val 120 are located in motifs I and II, respectively. However, rest of the residues are in the surrounding residues of the active site motifs (Trp105, Trp157, Leu158, Thr213, Arg214 and Arg250). These residues were also conserved in OXA-1, OXA-10, OXA-13 and OXA-24 for which crystal structures are available in Protein Data Bank under the accession numbers of 1M6K, 1K55, 1H5X and 3G4P, respectively. In general, the active site surrounding residues are known to be important in restricting or allowing the entrance of the substrates to the active sites. For OXA-48, two residues (Gln124 and Arg214) were especially pointed out in narrowing the entrance to the active siteCitation17. In this study, we demonstrated that Thr213Ala change in OXA-162 generated 1 Å extension in substrate entry site (). Therefore, although open to the discussion, it is possible to attribute the enhanced kinetic properties of OXA-162 to the widening of active site entry relative to OXA-48.
Figure 2. Examination of the active site regions of OXA-48 and modeled OXA-162. (A) OXA-48 homodimer (active site entry regions were pointed out with arrow heads and active site residues were colored). (B) Depiction of OXA-48 entry site. Dark orange colored amino acid (Thr213) is where the mutation occurred. Asp101 was located across the mutated amino acid. The measured distance between Thr213 and Asp101 was 9.12 Å. This was predicted to be the putative antibiotics entry site. (C) Depiction of the modeled OXA-162 entry site. Dark orange colored amino acid is the mutated amino acid (Ala188). Asp77 was located across the mutated amino acid. The measured distance between Ala188 and Asp77 was 10.07 Å which indicated the presence of 1 Å widening of the predicted entry site. Note that the corresponding amino acids for Ala188 and Asp77 in OXA-48 were Thr213 and Asp101, respectively.
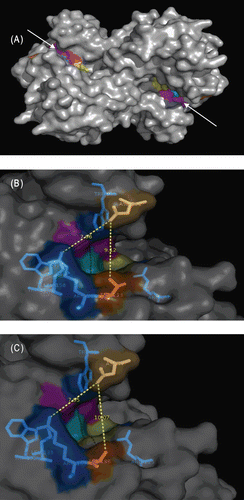
To gain a better understanding of the hydrolysis of antibiotics, we docked selected antibiotics with both enzymes and searched for any explicit differences. We were especially interested in doripenem and cefepime. However, there seem to be no apparent differences between the two enzymes. Doripenem docked to both structures similarly. The docking results of cefepime to both structures implied that cefepime failed to fully enter to the active sites of both enzymes and part of active site motifs II and III could not possibly involve in hydrolysis. However, knowing that a conformational change in the active site of OXA-48 and OXA-162 is expected as it occurs in the active site of OXA-13Citation17, and knowing that the docking results represent rigid conformations, it would be difficult to arrive at a reliable conclusion. It is perhaps best to examine solution structures in order to reliably evaluate the rigid docking models.
Conclusion
The transposon associated with dissemination of blaOXA-48 and blaOXA-162 is worrisome. This study demonstrated that the hydrolytic activity of OXA-48 may enhance even by a single amino acid change even if the amino acid is located outside the catalytic site. When this is coupled with an increased expression, evolved variants may confer resistance to almost all clinically relevant antibiotics.
Acknowledgment
We are grateful to Patrice Nordmann for providing us the strain E. coli DH10B-pVT-1 (OXA-48).
Declaration of interest
Authors do not have a relationship with pharmaceutical companies that might pose conflict of interest.
References
- Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 2009;9:228–236.
- Anderson KF, Lonsway DR, Rasheed JK, Biddle J, Jensen B, McDougal LK et al. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol 2007;45:2723–2725.
- Weisenberg SA, Morgan DJ, Espinal-Witter R, Larone DH. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn Microbiol Infect Dis 2009;64:233–235.
- Nordmann P, Mariotte S, Naas T, Labia R, Nicolas MH. Biochemical properties of a carbapenem-hydrolyzing beta-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob Agents Chemother 1993;37:939–946.
- Naas T, Vandel L, Sougakoff W, Livermore DM, Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob Agents Chemother 1994;38:1262–1270.
- Yigit H, Queenan AM, Rasheed JK, Biddle JW, Domenech-Sanchez A, Alberti S et al. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing beta-lactamase KPC-2. Antimicrob Agents Chemother 2003;47:3881–3889.
- Fritsche TR, Sader HS, Toleman MA, Walsh TR, Jones RN. Emerging metallo-beta-lactamase-mediated resistances: a summary report from the worldwide SENTRY antimicrobial surveillance program. Clin Infect Dis 2005;41 Suppl 4:S276–S278.
- Evans BA, Hamouda A, Towner KJ, Amyes SG. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin Microbiol Infect 2008;14:268–275.
- Vahaboglu H, Budak F, Kasap M, Gacar G, Torol S, Karadenizli A et al. High prevalence of OXA-51-type class D beta-lactamases among ceftazidime-resistant clinical isolates of Acinetobacter spp.: co-existence with OXA-58 in multiple centres. J Antimicrob Chemother 2006;58:537–542.
- Walther-Rasmussen J, Høiby N. OXA-type carbapenemases. J Antimicrob Chemother 2006;57:373–383.
- Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 2004;48:15–22.
- Carrër A, Poirel L, Eraksoy H, Cagatay AA, Badur S, Nordmann P. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob Agents Chemother 2008;52:2950–2954.
- Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G et al. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 2010;54:1369–1373.
- National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard. Wayne, Pennsylvania, USA; 2003.
- Meric M, Kasap M, Gacar G, Budak F, Dundar D, Kolayli F et al. Emergence and spread of carbapenem-resistant Acinetobacter baumannii in a tertiary care hospital in Turkey. FEMS Microbiol Lett 2008;282:214–218.
- Gacar GG, Midilli K, Kolayli F, Ergen K, Gundes S, Hosoglu S et al. Genetic and enzymatic properties of metallo-beta-lactamase VIM-5 from a clinical isolate of Enterobacter cloacae. Antimicrob Agents Chemother 2005;49:4400–4403.
- Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L et al. Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol 2009;16:540–547.
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 2005;33:W363–W367.
- Duhovny D, Nussinov R, Wolfson HJ. Efficient unbound docking of rigid molecules. In Gusfield et al., Ed. Proceedings of the 2′nd Workshop on Algorithms in Bioinformatics(WABI), Lecture Notes in Computer Science 2452. Rome, Italy: Springer Verlag, 2002. pp 185–200.
- Bou G, Oliver A, Martínez-Beltrán J. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother 2000;44:1556–1561.
- Santillana E, Beceiro A, Bou G, Romero A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc Natl Acad Sci USA 2007;104:5354–5359.
- Ledent P, Raquet X, Joris B, Van Beeumen J, Frère JM. A comparative study of class-D beta-lactamases. Biochem J 1993;292 (Pt 2):555–562.
- Naas T, Nordmann P. OXA-type beta-lactamases. Curr Pharm Des 1999;5:865–879.
- Couture F, Lachapelle J, Levesque RC. Phylogeny of LCR-1 and OXA-5 with class A and class D beta-lactamases. Mol Microbiol 1992;6:1693–1705.
