Abstract
Three glutamic acid derivatives, two boron-containing and one imide-containing compound, were synthesized and tested for antimicrobial activity targeting glutamate-racemase. Antimicrobial effect was evaluated over Bacillus spp. Docking analysis shown that the test compounds bind near the active site of racemase isoforms, suggesting an allosteric effect. The boron derivatives had greater affinity than the imide derivative. In vitro assays shown good antimicrobial activity for the boron-containing compounds, and no effectiveness for the imide-containing compounds. The minimum inhibitory concentration of tetracycline, used as standard, was lower than that of the boron-containing derivatives. However, it seems that the boron-containing derivatives are more selective for bacteria. Experimental evidence suggests that the boron-containing derivatives act by inhibiting the racemase enzyme. Therefore, these test compounds probably impede the formation of the bacterial cell wall. Thus, the boron-containing glutamic acid derivatives should certainly be of interest for future studies as antimicrobial agents for Bacillus spp.
Introduction
Antimicrobial agents, which represent an important advance in modern medicine for the treatment of infectious diseases, currently interfere with one of four cellular processes: cell wall biosynthesis, protein synthesis, DNA replication and repair, or folic acid coenzyme biosynthesis inhibition (this coenzyme is necessary for the biosynthesis of timineCitation1). Although heralded in the 1940′s as the beginning of the end of infectious diseases, during the last 20 years, more of these agents have lost their efficiency due to their abuse in medicine and agriculture. The design and development of new drugs has not been able to keep pace with these losses in the repertoire of treatment alternatives, leading to a troublesome situation that poses a threat to public health. Consequently, efforts to discover and design novel antimicrobial agents have been intensifiedCitation2–4.
The design of new agents is based on modifying the chemical structure of natural bacterial substrates in such a way that they damage the microbes. Several substrates have been modified in the last few years by adding boron and imides. The imides have shown antimicrobial, antispasmodic and analgesic effectsCitation5. On the other hand, boron-containing compounds are antimicrobial and anticancer agents as well as inhibitors of several different enzymesCitation6,Citation7.
Amino acids constitute one type of natural substrate that has been used for many years as the starting material for the development of antimicrobial agentsCitation8,Citation9. Whereas l-amino acids are used by eukaryotes, d-amino acids are important for the bacterial cell wall formation. One of the most important amino acids for Gram negative and Gram positive bacteria is d-glutamic acid, which is produced by the catalytic activity of the racemase enzyme (RacE) using l-glutamic acid as a substrate, and is part of the structure of peptidoglycans (mureine), a vital component of bacterial cell wallsCitation8,Citation10.
Due to the search for drugs with greater selectivity for RacE that could impede bacterial cell wall development, in the present study both d-glutamic acid and l-glutamic acid were modified and tested as RacE inhibitors. An in silico study was carried out to design these antibacterial agents, based on the theory that a certain moiety with similarity in structure to an endogenous compound, but with key modifications, could interact with greater selectivity and potency on enzyme targets. Hence, three glutamic acid derivatives were synthesized as tested for antimicrobial activity: two boron-containing glutamic acid compounds, (+)-(S)-4-(2,2-diphenyl-1,3,2-oxazaborolidin-5-oxo) propionic acid (l-DFB) and (-)-(R)-4-(2,2-diphenyl-1,3,2-oxazaborolidin-5-oxo) propionic acid (d-DFB), as well as one imide-containing glutamic acid compound, (S)-2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl) pentanedioate (l-GM).
These two boron-containing analogues have already been reported as having potent activity in globus pallidus neuronsCitation11. The aim of the current contribution was to evaluate the efficiency of these three test compounds as antimicrobial agents against Bacillus spp strains, the latter of which were chosen based on their pathogenicity and their lack of sensitivity to antibiotics currently used. Hence, three compounds with higher affinity for the RacE (according to docking studies) were synthesized. In vitro assays were carried out to assess their antimicrobial effectiveness, which was expressed as their minimum inhibitory concentration (MIC expressed in μg/mL), on cells of Bacillus spp strains. Additionally, a determination was made of the inhibitory concentration 50 (IC50 expressed in μg/mL) of the test compounds on RacE extracts from bacterial cells of the aforementioned strains.
Materials and methods
Maleic anhydride, diphenylboric acid 2-aminoethyl ester, d-glutamic acid, l-glutamic acid, dimethyl sulfoxide (DMSO), and other reagents were acquired from Sigma-Aldrich (Quimica SA, Toluca, México). All the solvents used were of reactive grade and purified before their use. All the melting points were determined in open glass capillary tubes in an Electrothermal 9300 device, and are uncorrected. All the IR spectra were recorded in KBr pellets on a Midac spectrometer. 1H-NMR (300 MHz), 13C-NMR (78.7 MHz) and 11B-NMR (90 MHz) spectra were recorded on a Bruker Avance 300-DPX spectrometer. Luria-Bertani (LB) and Mueller-Hinton (MH) culture mediums were purchased from BD Diagnostics. The bacterial strains, Bacillus cereus (B. cereus) ATCC 11778, Bacillus subtilis (B. subtilis) ATCC 6633 and Bacillus anthracis (B. anthracis) QHI were kindly supplied by Dr. Graciela Castro Escarpulli of the Bacteriology laboratory of the Microbiology Department, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, México.
Molecular modeling
The RacE 1 (RacE1, PDB ID: 2DWU) and RacE 2 (RacE2, PDB ID: 2GZM) structures employed in this study were described by May, et al.Citation10, which were taken from the Protein Data Bank (PDB). Before doing docking studies, the water molecules co-crystallized in the structures of RacE1 and RacE2 were eliminated. Through docking simulations, using the AutoDock 4.0.1 (http://autodock.scripps.edu/resources/adt/) softwareCitation12 in the Linux platform, the free energy (ΔG) values were obtained and the type of binding interactions between ligand and enzyme was determined. In this way, we could establish whether or not the ligand arrived to the active site of the RacE1 and RacE2 enzymes. To evaluate the atomic partial charge contribution, we calculated the Kollman partial charges after adding polar hydrogen atoms to these enzymes, using the AutoDocktools (ADT) programCitation12. The method utilized was the Genetic-Lamarckian algorithm implemented in the ADT program, with the following parameters: 100 runs, 100 individuals in the population, 1 × 107 evaluations of energy and 27,000 generations. The rest of the values were left at their default settingCitation12. Then, a grid box (60 × 60 × 60 Å) with a spacing of 0.375 Å was used to consider the recognition site reportedCitation10. A calculation of the affinity of each ligand on these receptors was made by calculating the sum of the partial contributionsCitation12. The structures of the compounds were optimized at their lowest energy by utilizing Density Function Theory B3LYP/6-31G* in the Gaussian 03 programCitation13, which was then converted to *.pdb format with Molekel 4.3Citation14. Then, the hydrogen atoms and Gasteiger charges of the ligand were assigned using the ADT programCitation12. The receptor-ligand complexes were visualized and analyzed by employing the VMD 1.8 programCitation15.
Synthesis of glutamic acid derivatives
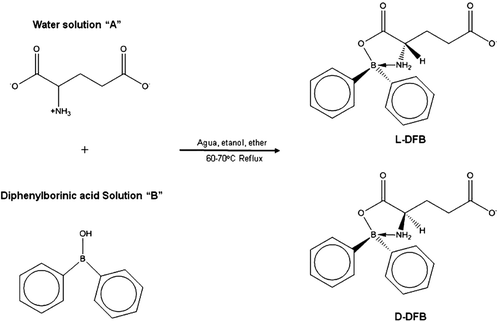
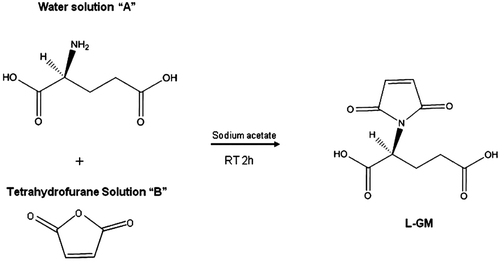
The method used to prepare compounds l-DFB and d-DFB () was previously reported by our work group, published elsewhereCitation11. The structures of compounds were determined with IR and NMR (1H and 13C). The spectroscopic data was in accordance with reports in the literatureCitation11,Citation16. The new imide compound, l-GM, was synthesized according to previous reports on the synthesis of arylimidesCitation5,Citation17,Citation18, with some modifications in order to obtain the compound in a single step. Consequently, a new methodology was established in our laboratory () for the synthesis of the derivative of glutamic acid with maleic anhydride. Briefly, solution A was prepared by dissolving (+)-(S) glutamic acid (1.0 g) in a minimum amount of distilled water. Then NaOH 0.5 N was added dropwise until reaching the desired pH of 9. Solution B was prepared by dissolving maleic anhydride (0.66 g) in 10 mL of tetrahydrofurane (solution B). Solution A and B were then mixed with constant stirring for 2 h at room temperature. When the target product was precipitated, it was filtered, washed with hexane, dried and weighed to give (S)-2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl) pentanedioate compound in a 90% yield as a white powder, mp 150–152 °C, IR (KBr) υ (cm-1): 1539 (C=Oas), 3060 (C=Cas), 3446 (NHas); 1H NMR (300 MHz, DMSO-d6, δ ppm): 6.15 (s, 2H), 3.650 (t, 1H), 2.383 (m, br 2H), 1.983 (m, br 2H); 13C NMR (68.7 MHz, DMSO-d6, δ ppm):177.17, 173.81, 170.78, 134.22, 53.78, 30.09, 25.49.
Determination of the minimum inhibitory concentration
The Minimum inhibitory concentration (MIC) was determined for the strains of B. cereus, B. subtilis and B. anthracis by the microdilution technique in Mueller Hilton broth, according to the Clinical and Laboratory Standards Institute (CLSI, formerly NCCLS) criteria for Staphylococcus aureusCitation19. Prior to testing, each strain was sub-cultured twice on Trypticase soya agar (TSA) plates containing 5% sheep blood (Difco, BD) and incubated at 37°C overnight. Starting from the seeded bacterial strains on cultivation plates (LB agar), a suspension was prepared with a density equivalent to 0.5 on the McFarland scale (about 1 to 5 × 106 cell/mL). Dilutions were carried out with this solution in order to obtain a final concentration of 5 × 102 to 2.5 × 103 cell/mL in Mueller Hinton (MH) broth (Difco, BD). A stock solution of 1 mg/mL was prepared from d-DFB, l-DFB and l-GM. Afterwards; this was employed to prepare a dilution of 256 to 0.5 µg/mL in DMSO. Beginning with well number one (100 µL/well), triple serial dilutions were carried out with the first ten wells. Then, 100 µL/well of the suspension were added to the bacterial suspension in wells one to eleven. Well 11 was the control for bacterial growth (100 µL DMSO), while well 12 was the test control for sterility of the medium. The microplates were incubated at 37°C for 24 h, at which time the corresponding readings were made at 541 nm on a Multiskan EX photometer for microplates. The concentration that inhibited the growth of 50% of each of the Bacillus spp strains (MIC50) was established, so as that which inhibited 90% of the strains (MIC90). The MIC range of each antimicrobial agent for each strain was determined using tetracycline as a positive control.
Preparation of cellular extracts
The cell extracts containing the RacE were obtained as previously reportedCitation3,Citation10,Citation20 from three Bacillus strains: B. cereus, B. subtilis and B. anthracis. Briefly, these three strains were each separately cultivated in 50 mL of LB culture medium at 37°C for 16 h. The cell cultures were harvested at 5000 rpm for 5 min, eliminating the supernatant. Afterwards, the cells were resuspended with 10 mL of buffer, 0.1 M of Tris-HCl (pH 8) and 0.1 g of glass pearls, achieving the breakage of the cell at 4°C with pulses every 30 s. The lysate was centrifuged at 13000 rpm for 10 min. The supernatant was dialyzed with the same buffer during 24 h at 4oC and employed as the cell extract.
Cellular extract assays
Extracts of the RacE from cells of three bacterial strains, B. cereus, B. subtilis and B. anthracis, were assayed by employing a previously reported method (Ashiuchi, et. alCitation20.).
Results and discussion
Docking studies
Glutamic acid, a moiety which is the natural substrate of RacE, has several groups that can be chemically modified, giving rise to inhibitors with much better affinity for the active site or some allosteric site than the natural substrate. Various studies have reported that some glutamic acid derivatives are very selective for RacE and have potent antimicrobial activityCitation21,Citation22. In the current contribution, three glutamic acid derivatives, two analogues of l-glutamic acid and one of d-glutamic acid, were synthesized and tested as RacE inhibitors.
First, the docking procedure employed, was tested by using glutamic acid with the RacE (), and compared to previously reported dataCitation10. Then, docking and structural studies were carried out in order to explore which modification in the target moiety groups of glutamic acid would probably lead to a compound with the greatest affinity for the active site of the RacE. It was found that what most influenced affinity for this enzyme was the presence of a 5-membered heterocycle moiety in the three test compounds, two of them containing a boron atom (l-DFB and d-DFB) and the other an imide (l-GM).
Figure 1. Binding site of: (a) d-glutamic acid and (b) l-glutamic acid on the RacE1 isoform with two principal amino acid residues, (c) d-glutamic acid and (d) l-glutamic acid on the RacE2 isoform with two principal amino acid residues. Carbon atoms are colored cyan; oxygen atoms, red; nitrogen atoms, blue; sulfur atoms of the protein, yellow for enzyme active site.
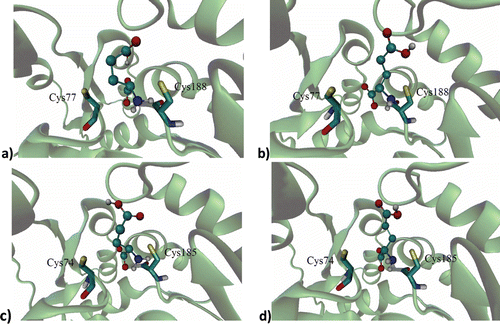
Previous docking studies have used crystallized RacE in order to explore the active site of the RacE in detailCitation23, identifying the amino acid residues that participate in the ligand-receptor interactionCitation3,Citation8. In the active site of RacE1, the natural substrate binds to Cys77 and Cys188, while in the active site of RacE2 this ligand binds to Cys74 and Cys185. Thus, residues change the configuration of glutamic acid from l to d or S to R by abstracting or adding a hydrogen atomCitation10,Citation23.
In each of the enzyme isoforms evaluated, the three test compounds were relatively distant from the two principal residues involved in the catalytic activity by the natural substrates. Regarding RacE1 (specifically Cys77 and Cys188), the imide group of the l-GM compound was found to be at a distance of 8.39 Å, whereas d-DFB was at 9.03 Å and l-DBF at 14.11 Å (). In relation to RacE2, the three test compounds were even more distant from the active site and therefore did not show any interaction with residues Cys74 and Cys185. Although the test compounds were relatively distant from the active site of both isoforms of RacE, their high affinity for an allosteric site of the enzyme suggests that they make conformational changes affecting the catalytic site, which would diminish the affinity for the natural substrate of the enzyme. This result is due to the fact that both RacE isoforms have a narrow pocket that forms the active site, which can be easily blocked.
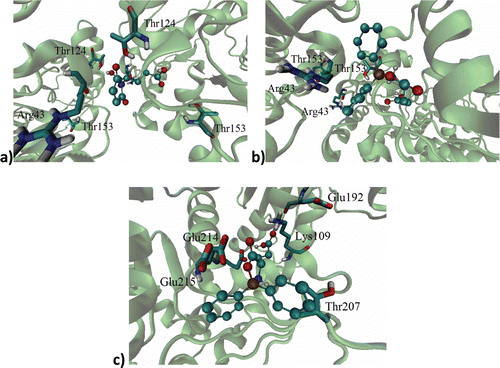
Figure 2. Binding site of: (a) l-GM, (b) d-DFB, (c) l-DFB on the RacE1 isoform, with two principal amino acid residues. Carbon atoms are colored cyan; oxygen atoms, red; nitrogen atoms, blue; sulfur atoms, yellow for enzyme active site.
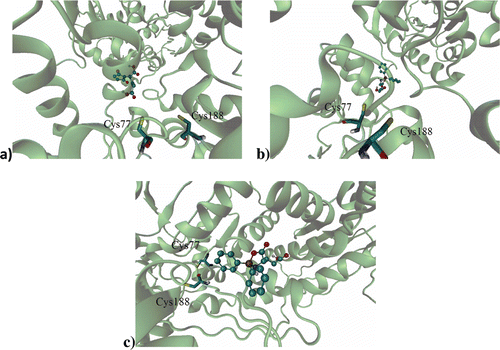
Apart from the principal residues, the natural substrate was close to the following residues of RacE1: Thr121, Gly47, Asn78, His190, Thr189, Tyr46, Ser15, Pro192 and Ala152. Likewise, the natural substrate was close to the following residues of RacE2: Ser12, Tyr43, Asp11, Thr186, Thr76, Asn75, Gly44, Pro42 and Val149. These results are in agreement with reports in the literatureCitation3,Citation10,Citation21.
Regarding l-GM, in the interaction with RacE1 it was close to Arg43, Thr124 and Thr153. With this same isoform, d-DFB was close to Arg43 and Thr153, while l-DFB was close to Glu192, Glu214, Glu215, Thr207 and Lys109 (). In the interaction with RacE2, l-GM was close to Lys262, Ile243, Gly244 and Lys242, while both d-DFB and l-DFB were close to Gly244, Ile243, Lys242 and Lys262. By comparing the residues for which the natural substrates have affinity with this data, it seems that the test compounds have affinity for an allosteric site.
Figure 3. Ligand on racemase enzyme allosteric site of: (a) l-GM, (b) d-DFB, (c) l-DFB on the RacE1 isoform, with some amino acid residues. Carbon atoms are colored cyan; oxygen atoms, red; nitrogen atoms, blue.
To compare affinity, the free energies (δG) of the test compounds and the natural substrates were determined in relation to RacE1 and RacE2 (). According to the δG, the affinity of the imide derivative is similar to that of l- and d-glutamic acid for both RacE1 and RacE2. However, the affinity of the boron-containing derivative is notably higher for both enzyme isoforms. This suggests that the boron atom could play an important role in protein recognition and could induce the conformational changes.
Table 1. Free energies values and Kd of the natural substrates of l and d-glutamic acid and all three test compounds in relation to RacE1 and RacE2, according to docking studies.
The bonding interaction of the test compounds and the natural substrates with the two enzyme isoforms is congruent with the comparative δG values. In a previously reported study, it is mentioned that the affinity of l- and d-glutamic acid for both enzyme isoforms is based on hydrogen bondsCitation10.
The current docking study found that hydrogen bonds are also responsible for the affinity of the imide derivative for both enzyme isoforms. A hydrogen bond is formed between the ω-carbonyl oxygen of this compound and the backbone amide of Thr124, between the carbonyl oxygen of the compound and Arg43, and between the imide of the compound and the hydroxyl side chain of Thr153. Thus the slight affinity of the imide derivative for both isoforms can be explained by the well-known weak nature of hydrogen bonds. Contrarily, the higher affinity of the boron-containing compounds for both isoforms can be attributed to the fact that not only hydrogen bond formation was found (between the ω-carbonyl oxygen of compounds and amine group on the side chain of Lys262), but also a much stronger type of bond based on π-cation interactions of the aromatic ring of the derivatives with the amine group of Lys242.
Synthesis of glutamic acid derivatives
The compounds were synthesized as reported and all compounds were obtained with good yields. The structures of the l-GM, d-DFB and l-DFB derivatives were established by their corresponding spectroscopic data (IR, 1H-NMR and 13C-NMR). For l-DFB the phenyl groups bonded to boron (the borinic ester) and the carboxyl group, evidenced by the presence of bands in the IR spectrum at 1724 cm-1 and 1602 cm-1, respectively, which arises from the asymmetrical and symmetrical stretching modes.
Assays with the MIC method
Antimicrobial activity of the test compounds and tetracycline was evaluated in vitro against three bacterial strains: B. cereus ATCC 11778, B. subtilis ATCC 6633 and B. anthracis QHI. The inhibitory effect of all compounds was compared to the growth of bacteria in wells without any compound added. The MICs of the test compounds and of the standard drug are presented in .
Table 2. MIC50 and MIC90 for the Bacillus spp. strains tested, by the microdilution technique in Mueller Hilton broth.
Tetracycline was selected as a positive control for inhibition of growth of the Bacillus strains used in this study because clinically it is the most frequently used for Bacillus spp. This drug, with a MIC ranging from 16 to 0.03125 µg/mL, was inhibitory for B. subtilis and B. anthracis. A concentration of 0.25 µg/mL effected 50% inhibition, while a concentration of 0.5 µg/mL caused 90% inhibition. A concentration of 8 µg/mL of this standard compound inhibited B. cereus at 50%, while 16 µg/mL resulted in 90% inhibition. As can be seen, tetracycline has a relatively low MIC, very close to that reported in strains of B. anthracis isolated in clinical studies. The two test compounds that gave the best results against the three bacterial strains were d-DFB and l-DFB, with a MIC ranging from 256 to 0.5 µg/mL. For the B. cereus strain, a concentration of 64 µg/mL of d-DFB showed a 50 % inhibition, while a concentration of 128 µg/mL inhibited this strain at 90%. d-DFB at a concentration of 32 µg/mL inhibited the strains of B. subtilis and B. anthracis by 50%, while a concentration of 64 µg/mL inhibited these strains by 90%. Curiously, these same concentrations of l-DFB inhibited the B. cereus and B. anthracis strains by 50% and 90%. For the B. subtilis strain, a concentration of 16 µg/mL of l-DFB showed 50% inhibition, while a concentration of 32 µg/mL resulted in 90% inhibition.
Hence, the MIC of the boron-containing compounds was relatively higher compared to tetracycline. However, tetracycline inhibits protein synthesis in a non-selective form. Contrarily, the test compounds probably inhibit cell wall synthesis, which would make them selective for bacterial versus human cells. The MIC of the test compounds is close to that of other antibiotics, such as ceftriaxone (MIC90 32 µg/mLCitation24), which also inhibit cell wall synthesis.
Cellular extract assays
Since the aforementioned inhibitory values are in relation to the bacterial cell, they do not necessarily reflect the activity of the compounds on the enzyme. To confirm that these values express inhibition of the enzyme, we obtained cellular extracts containing the RacE from the three bacterial strains, according to a reported methodCitation20. A determination was made of the formation of 2, 4-dinitrophenylhydrazone of α-ketoglutarate by measuring the increase in the absorbance at 550 nm (read on a spectrophotometer of visible UV). One unit of enzyme was considered as the quantity necessary to catalyze the formation of 1 µmol of l-glutamate per minute. Consequently, we could test the compounds on cellular extracts and obtain the inhibitory concentration 50 (IC50).
The results of the IC50 and the MIC with these assays were similar for d-DFB and l-DFB in relation to all three bacterial strains as those found with the whole bacterial cell. Once again, l-DFB showed greater affinity than d-DFB. The greater in vitro inhibitory effect of l-DFB on bacterial growth is not surprising, because the natural function of RacE is the change from the l- to d-configuration. Therefore, the recognition site is chiral, and consequently can distinguish between the two chiral ligands. For l-GM, no inhibition of the enzyme was found when using the same concentration as that used to inhibit the bacterial cell. It was necessary to double the concentration to find the IC50 for the imide derivative on the extract ().
Table 3. Determination of IC50 of compound on Bacillus spp.
The IC50 for the boron-containing compounds was higher in relation to B. cereus (<64 µg/mL for d-DFB and <32 µg/mL for l-DFB), with a greater sensitivity shown to B. anthracis (32 µg/mL for both boron-containing derivatives), and the greatest sensitivity to B. subtilis (<32 µg/mL for d-DFB and <16 µg/mL for l-DFB).
Judging by the X-ray of the structure for l-DFB, this compound has a 5-membered ring. Therefore, the d-boron-containing enantiomer must have this same structural geometry. The 11B-NMR analysis showed a chemical shift of 4.5 ppm for both compounds, confirming the tetravalent structureCitation16,Citation23. Consequently, these two compounds can pass through the lipidic membrane of the bacteria relatively quickly, compared to tetracycline, in order to approach the active site. Contrarily, tetracycline needs to pass through this lipidic membrane by passive diffusion through hydrophilic pores in order to inhibit protein synthesisCitation25. Similarly, it is likely that the hydrophilicity of l-GM makes it less lipophilic than d-DFB and l-DFB, which could explain the fact that at the assayed concentrations, l-GM did not inhibit the growth of the three Bacillus spp. strains.
Conclusions
Two boron-containing glutamic acid compounds, l-DFB and d-DFB, as well as one imide-containing glutamic acid compound, l-GM, were tested in silico for their affinity to two RacE isoforms. The in silico analysis suggested that all three test compounds disrupt enzyme function by allosteric binding to RacE1 and RacE2. On the other hand, the in vitro results shown that only two of the three test compounds, d-DFB and l-DFB, actually inhibit growth of the three bacterial strains employed in the present study. Although tetracycline shown a lower MIC than these two boron-containing compounds, it acts on bacteria by inhibiting protein synthesis. The fact that the two boron-containing glutamic acid derivatives inhibiting the RacE means that they probably act by inhibiting bacterial cell wall formation. This property together with their greater lipophilicity could confer these two test compounds the capacity of acting more quickly and selectively than tetracycline. These results suggest that boron-containing compounds could represent an important focus for future research.
Acknowledgments
The authors are grateful to CONACyT (132353), ICyTDF (PIRIVE 09-9), SIP, COFAA/IPN EDI/IPN and the scholarship from CONACyT to Feliciano Tamay-Cach. We thank Bruce Allan Larsen for reviewing the use of English in the manuscript.
Conflict of interest
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article.
References
- Fisher SL. Glutamate racemase as a target for drug discovery. Microb Biotechnol 2008;1:345–360.
- Taal MA, Sedelnikova SE, Ruzheinikov SN, Baker PJ, Rice DW. Expression, purification and preliminary X-ray analysis of crystal of Bacillus subtilis glutamate racemase. Acta Cryst 2004; 60:2031–2034.
- Dodd D, Reese JG, Louer CR, Ballard JD, Spies MA, Blanke SR. Functional comparison of the two Bacillus anthracis glutamate racemases. J Bacteriol 2007;189:5265–5275.
- Lewis K. Antibiotics: recover the lost art of drug discovery. Nature 2012;485:439–440.
- Cechinel-Filho V, Campos F, Corrêa R, Yunes RA, Nunes RJ. Chemical aspects and therapeutic potential of cyclic imides: a review. Quim Nova 2003; 26:230–241.
- Soriano-Ursúa MA, Mancilla-Percino T, Correa-Basurto J, Querejeta E, Trujillo-Ferrara JG. Give boron a chance: boron containing compounds reach ionotropic and metabotropic transmembrane receptors. Mini Rev Med Chem 2011;11:1031–1038.
- Petasis-Nicos A. Expanding roles for organoboron compounds-versatile and valuable molecules for synthetic biological and medicinal chemistry. Aust J Chem 2007; 60:795–798.
- Yoshimura T, Esak N. Amino acid racemases: functions and mechanisms. J Biosci Bioeng 2003;96:103–109.
- Kim KH, Bong YJ, Park JK, Shin KJ, Hwang KY, Kim EE. Structural basis for glutamate racemase inhibition. J Mol Biol 2007;372:434–443.
- May M, Mehboob S, Mulhearn DC, Wang Z, Yu H, Thatcher GR et al. Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design. J Mol Biol 2007;371:1219–1237.
- Araujo Alvarez JM, Querejeta E, Oviedo A, Trujillo Ferrara JG. Stereospecific activity of two glutamate analogs. Chirality 2004;16:586–591.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem 1998; 19:1639–1662.
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 03, Revision C.02. Wallingford CT: Gaussian, Inc., 2004.
- Flükiger PF. Development of the molecular graphics package MOLEKEL and its application to selected problems in organic and organometallic chemistry, 1992. Thèse No 2561, Département de chimie physique, Université de Genève, Genève.
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996;14:33–8, 27.
- Trujillo J, Höpfl H, Castillo D, Santillan R, Farfán N. X-ray crystallographic study of boroxazolidones obtained from L-ornithine, L-methionine, kainic acid and 2,6-pyridinedicarboxilic acid. J Organomet Chem 1998;571:21–29.
- Cava MP, Deana AA, Muth K, Mitchell MJ. N-Phenylmaleamida. Organic Syntheses Coll 1973;5:944–946.
- Andricopulo AD, Yunes RA, Nunes RJ. Synthesis and Antibacterial Activity of Cyclic Imides: 3,4 dicloromaleimides and 3-cloro-4-substituted-maleimides. Quim Nova 1998; 21:573–577.
- National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically-Sixth Edition: Approved Standard M7-A6. Wayne, PA:NCCLS, 2003.
- Ashiuchi M, Soda K, Misono H. Characterization of yrpC gene product of Bacillus subtilis IFO 3336 as glutamate racemase isozyme. Biosci Biotechnol Biochem 1999;63:792–798.
- Lundqvist T, Fisher SL, Kern G, Folmer RH, Xue Y, Newton DT et al. Exploitation of structural and regulatory diversity in glutamate racemases. Nature 2007;447:817–822.
- de Dios A, Prieto L, Martín JA, Rubio A, Ezquerra J, Tebbe M et al. 4-Substituted D-glutamic acid analogues: the first potent inhibitors of glutamate racemase (MurI) enzyme with antibacterial activity. J Med Chem 2002;45:4559–4570.
- Puig E, Mixcoha E, Garcia-Viloca M, González-Lafont A, Lluch JM. How the substrate D-glutamate drives the catalytic action of Bacillus subtilis glutamate racemase. J Am Chem Soc 2009;131:3509–3521.
- Mohammed MJ, Marston CK, Popovic T, Weyant RS, Tenover FC. Antimicrobial susceptibility testing of Bacillus anthracis: comparison of results obtained by using the National Committee for Clinical Laboratory Standards broth microdilution reference and Etest agar gradient diffusion methods. J Clin Microbiol 2002;40:1902–1907.
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 2001;65:232–60.