Abstract
Kinetic characterization of lipase inhibition was performed by activity measurement and mass spectrometry (MS), for the first time with serine-protease inhibitor 3,4-dichloroisocoumarin (DCI). Inhibition of Streptomyces rimosus extracellular lipase (SrLip), a member of the SGNH superfamily, by means of DCI follows the mechanism of two-step irreversible inhibition. The dissociation constant of the noncovalent E•I complex and first-order rate constant for inactivation were determined by incubation (Ki* = 26.6 ± 2.8 µM, k2 = 12.2 ± 0.6 min–1) or progress curve (Ki* = 6.5 ± 1.5 µM, k2 = 0.11 ± 0.01 min–1) method. Half-times of reactivation for lipase inhibited with 10-fold molar excess of DCI were determined by activity measurement (t1/2 = 11.3 ± 0.2 h), matrix-assisted laser desorption/ionization (MALDI, t1/2 = 13.5 ± 0.4 h), and electro-spray ionization (ESI, t1/2 = 12.2 ± 0.5 h) MS. The active SrLip concentration was determined by incubating the enzyme with near equimolar concentrations of DCI, followed by activity and MS measurement.
Introduction
Regulation of enzyme activity is very important for biochemical research and for various applications in medicine, physiology, pharmacology, toxicology, agriculture, and industry. Inhibitors bound in the active site of an enzyme (especially analogs of the transition state of the catalyzed reaction) can serve as extremely useful probes to reveal the mechanism of enzyme catalysisCitation1.
In most general definition, irreversible inhibitor of an enzyme is a substance with effects that cannot be reversed by removing the inhibitor by dialysis or dilutionCitation2. Nevertheless, irreversible inhibitors are, in most cases, directed to the active site of the enzyme because of their structural similarities to substrates, and show competitive kinetic behaviorCitation2. The binding of an inhibitor to an enzyme can proceed through one or two steps, and can be even more complicated by the presence of substrate in an inhibition mixtureCitation3 (). It is usually implied that the covalent bond formed in these particular cases is stable indefinitely; however, enzyme activity can be regained due to chemical degradation of the bound inhibitor or some other reasons. This process can be referred to as “reactivation” rather than “inhibition reversal”Citation4.
Scheme 1. Reaction schemes for irreversible inhibition (according to Ref. Citation3).

Preparation or selection of an efficient inhibitor for a particular enzyme implies knowledge about mechanism of enzyme–substrate interactions. In the study of new enzymes, their catalytic mechanisms are, in most cases, unknown. Therefore, described and tested inhibitors of a certain enzyme class are often used to probe the activity of a new enzyme. 3,4-dichloroisocoumarin (DCI) was discovered in 1985 as a general mechanism-based serine-protease inhibitorCitation5 but it turned out that it inhibits other serine hydrolases as well. The proposed mechanism of its action involves opening of the isocoumarin ring by a serine protease, followed by enzyme acylation. Formed acid chloride reactive group can either react with an active-site nucleophile (e.g. histidine) to form a stable diacylated structure (alkylated acyl-enzyme derivative, doubly covalent enzyme–inhibitor complexCitation6) or can be hydrolyzed by water ().
Scheme 2. Proposed mechanism of inhibition of serine hydrolases by 3,4-dichloroisocoumarin. Adapted from Ref. Citation5.
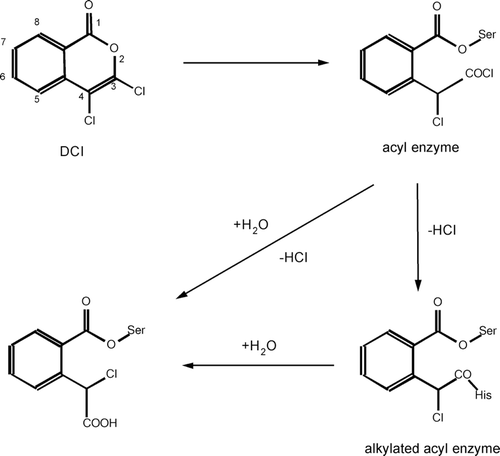
DCI is readily available, stable in dimethyl sulfoxide (DMSO) and fairly stable in aqueous media. It is specific toward serine proteases and analogous enzymes. Its reactivity is more pronounced than that of phenylmethylsulfonyl fluoride (PMSF), and DCI is less toxic than di-isopropylfluoridate. It could be applied in prevention of undesirable proteolysis due to serine proteases and elucidation of the physiological role of serine proteasesCitation5,Citation7. Even more, it has been found to induce apoptosisCitation8, to inhibit enzymes responsible for tumor growthCitation9,Citation10, to participate in prevention of transcriptional factors activationCitation11, to block priming of monocytesCitation12, to prevent infection of cells with Toxoplasma gondii parasiteCitation13, to inhibit growth of Mycobacterium tuberculosisCitation14, and to inhibit coagulant proteases from snake venomCitation15.
Lipase inhibition is of great significance in understanding catalytic mechanisms of these enzymes, and in treatment of obesity and several lipase-associated diseasesCitation16–20. Various compounds of natural and synthetic origin are constantly tested with respect to lipase inhibitionCitation21–28.
Extracellular lipase from Streptomyces rimosus (SrLip), a member of GDSL-family of lipolytic enzymes, also classified as SGNH-hydrolase, has been in the focus of our research. This small enzyme (24 kDa) exhibits pronounced stability at high temperatures and pH values and also in various organic solventsCitation29,Citation30. Recently, its substrate promiscuity, i.e. multifunctionality was demonstratedCitation31. These properties make SrLip very attractive to study the mechanism of ester hydrolysis catalyzed by lipases. To this purpose, we performed inhibition experiments with several serine-blocking agents. Out of compounds investigated, PMSF had low effect on SrLip activity, only when added during substrate hydrolysisCitation29. Tetrahydrolipstatin (THL) inhibits SrLip significantly in excess of organic solvent (50% 2-propanol) only. It binds covalently to the catalytic residue Ser-10 of the enzyme, exclusively. Mass spectrometric and capillary gel electrophoresis-on-a-chip experiments revealed that approximately 50% of SrLip molecules are covalently bound to THLCitation32. The partial binding of the inhibitor can be understood as a partially reactivation process of the enzyme as also observed in the inhibition study of human pancreatic lipase with THLCitation33. The inhibition of SrLip with DCI, on the other hand, was fast and complete, and was used to identify the active-site Ser-10Citation34. However, the inhibition effect was reversed after prolonged time of incubation of enzyme with inhibitor. To get close insight into the inhibition mechanism of SrLip, we decided to further inspect the DCI inhibition of SrLip. Here we describe the kinetic characterization of inhibition of S. rimosus extracellular lipase by means of DCI.
Materials and methods
Materials
Lipase substrates, p-nitrophenyl caprylate (pNPC) and p-nitrophenyl palmitate (pNPP), and inhibitor, DCI, were obtained from Sigma (St. Louis, MO). Buffer chemicals were purchased from Kemika (Zagreb, Croatia). Chromatographic media used for lipase purification were supplied by GE Healthcare (Piscataway, NJ) and ZipTip-C4 pipette tips by Millipore (Billerica, MA). All organic solvents were analytical grade (Sigma) and water used was of analytical grade quality too. All kit chemicals used for capillary gel electrophoresis-on-a-chip experiments were obtained from Agilent Technologies (Waldbronn, Germany). The vacuum matrix-assisted laser desorption/ionization (MALDI) matrix compound 2,6-dihydroxyacetophenone (2,6-DHAP) was obtained from Fluka (Buchs, Switzerland) and used without further purification. Trifluoroacetic acid (TFA; Sequenal™ quality) was purchased from Pierce (Rockford, IL).
Enzyme purification
Overexpressed lipase was purified from culture filtrate of S. rimosus strain R6-ZGL3(pDJ7) that contains a high-copy-number plasmid harboring S. rimosus lipase geneCitation35, as already describedCitation36. The purity of prepared enzyme solution was confirmed by capillary gel electrophoresis using lab-on-a-chip technology (Agilent Technologies).
Protein determination
Protein concentration was routinely assayed according to BradfordCitation37. In active-site titration experiments, protein concentration was also determined by measuring absorbance at 280 nm, on a NanoDrop1000 instrument (ThermoFisher Scientific, Waltham, MA), using molar extinction coefficient ε = 26,275 M−1 cm−1 calculated by ProtParam tool on ExPasy server (www.expasy.ch) for mature lipase assuming all cysteines form disulfide bridges.
Lipase activity assay
Lipase activity was determined according to Abramić et al.Citation29. In short, 1 mM emulsion of pNPP (or pNPC) was prepared by vigorously mixing a 40mM stock solution of substrate in dioxane with 50 mM sodium phosphate buffer solution pH = 8 (buffer 1), so that final amount of dioxane was 2.4%, followed by a 2 min sonication. The reaction was started by addition of small amount of enzyme or enzyme–inhibitor solution to 1 mL of substrate emulsion and was followed by increase in absorbance at 410 nm. All experiments were performed at room temperature, on Camspec M-501 spectrophotometer (Spectronic Camspec, Garforth/Leeds, UK).
Enzyme inhibition – incubation method
An inhibition was initiated by adding 1.5 µL of inhibitor solution in DMSO to 148.5 µL of buffered enzyme solution and the final concentrations were 0.14 µM lipase, 1.1–100 µM DCI, 1% DMSO, 10 mM sodium phosphate buffer with 0.2 M NaCl, and 10 mM ethylenediaminetetraacetic acid (EDTA) pH = 6.8 (buffer 2). Aliquots were removed at different times and residual activity was measured toward pNPP. In parallel, a control experiment with all compounds except inhibitor was performed to assess lipase stability in DMSO.
Second-order inhibition rate constants (kobs/[I]) were obtained for each inhibitor concentration by plotting ln([E]t/[E]o) vs. time and dividing the slope (kobs) by the inhibitor concentration. The t1/2 values were calculated from kobs according to the equation: t1/2 = −ln(1/2)/kobs, for inhibitor concentrations up to 13.8 µM. Dissociation constant of the noncovalent E•I complex, Ki*, and first-order rate constant for inhibition, k2, were calculated from following equation: ![]() , according to Powers and KamCitation6. Data were fitted to this equation by program GraphPad Prism 4.0 (GraphPad Software, La Jolla, CA).
, according to Powers and KamCitation6. Data were fitted to this equation by program GraphPad Prism 4.0 (GraphPad Software, La Jolla, CA).
Determination of inhibition rate in the presence of substrate – progress curve method
The inhibition rate constant was also determined in the presence of substrate (pNPP) as described by Tian and TsouCitation38. One microliter of enzyme solution was added to a substrate emulsion (prepared as described above, in buffer 1), which contained inhibitor and 5 % DMSO (final lipase concentration in the reaction mixture: 0.4 nM). The increase in absorbance was monitored at 410 nm until no further release of p-nitrophenol was observed. Control experiments containing all compounds excluding inhibitor or lipase were performed to assess lipase activity in the presence of DMSO or spontaneous inhibitor hydrolysis, respectively.
In this series of experiments, the substrate concentration was fixed at 1 mM (2.4% dioxane) and DCI concentration varied from 0.015 to 20 µM. In program GraphPad Prism 4.0, kobs was determined from the equation: log([P]∞ − [P]t) = log[P]t – 0.43 × kobs × t, where [P]∞ and [P]t are the concentrations of a product (p-nitrophenol) formed at infinite time (total inactivation) and at time t, respectively. When kobs was plotted against [I]o, ![]() and k2 were calculated as in the case of incubation method.
and k2 were calculated as in the case of incubation method.
Lipase active-site titration
In the active-site titration experiments, the lipase was incubated with the inhibitor in buffer 2 for 15 min at room temperature and the residual activity was measured by pNPP assay. An enzyme concentration in an incubation mixture was 3.0 µM, as determined on NanoDrop1000 instrument. The inhibitor concentration ranged from 0.5 to 10 µM. One aliquot of each incubation mixture was also prepared for mass spectrometric analysis, using ZipTip-C4 technology (10 µL of sample eluted into 10 µL of acetonitrile/1% aqueous formic acid 1:1 (v/v)).
Percentage of lipase activity after 15-min incubation with inhibitor was plotted against inhibitor concentration. The active enzyme concentration was calculated from this plot, as intercept on X-axisCitation39.
Reactivation kinetics
Lipolytic activity (measured toward pNPC) was followed for longer times of incubation at room temperature without removal of any residual inhibitor, under conditions described under Enzyme inhibition – incubation method. In addition, aliquots of incubation mixtures were taken and prepared for mass spectrometric measurements, using ZipTip-C4 technology (10 µL of sample eluted into 10 µL of acetonitrile/1% aqueous formic acid 1:1 (v/v)).
The reactivation half-time was calculated from plots of the percent of active enzyme vs. time by curve fitting performed in GraphPad Prism 4.0 program.
Determination of spontaneous hydrolysis rates of the inhibitor
An aliquot of DCI (final concentration 50–200 µM) in DMSO was added to the buffer solution (buffer 1 or buffer 2), so that the final DMSO concentration was 5 or 1%, respectively. Spontaneous hydrolysis was monitored by following the decrease in absorbance at 325 nm.
Since the decrease in absorbance at 325 nm was linear with time, zero-order hydrolysis rate constants and half-lives were calculated from line slopes.
MALDI mass spectrometry
All sample preparations were performed on stainless-steel target slides using the dried-droplet technique. One microliter of the sample solution and 1.0 µL of a saturated solution of 2,6-DHAP in acetonitrile/0.1% TFA 1:3 (v/v) were mixed on the target in the given order and dried at room temperature in a gentle stream of air.
The MALDI measurements were performed with a curved field reflectron time-of-flight mass spectrometer (AXIMA CFR+, Shimadzu Biotech–Kratos Analytical, Manchester, UK) equipped with a pulsed nitrogen laser. The instrument was operated in the positive ion linear mode applying an accelerating voltage of 20 kV. Delayed extraction was used and the delay time was set according to the molecular weight of the analyte in order to optimize resolution. Typically, mass spectra were acquired by averaging 441 single laser shots. External calibration was performed for molecular mass assignment of the intact protein and the lipase–inhibitor complex using myoglobin from equine skeletal muscle and carbonic anhydrase II from human erythrocytes (both from Sigma–Aldrich, St. Louis, MO). All mass spectra were smoothed using the company-supplied Savitzky–Golay algorithm.
Electro-spray ionization mass spectrometry
For molecular weight determination of the SrLip and the SrLip–DCI complex, nano-LC separation was performed on an Ultimate 3000 Titanium LC system using an Acclaim PepMap nano column (75 µm I.D. × 150 mm length, particle diameter 3 µm) (LC Packings/Dionex, Amsterdam, NL). The column temperature was kept at 20°C and the flow rate at 300 nL/min. Nano-LC conditions: solvent A, 0.1% formic acid; solvent B, 0.1% formic acid/acetonitrile (95/5, v/v); gradient: 0–5 min 5% B, 5–20 min increasing B to 95%, 20–30 min 95% B constant, 30–32 min decreasing B to 5%, 32–40 min 5% B constant; injection volume, 1 µL.
The nano-LC system was coupled on-line to an HCTplus 3D-ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany) under the following experimental nano electro-spray ionization (nanoESI) conditions: spray voltage, 4.0 kV; gas flow, temperature and ion transmission was optimized for the mass range m/z 395–1500; accumulation time 115 ms. Standard deviations for the determined molecular weights of the intact proteins were derived from the calculated molecular weights after charge state deconvolution from multiple samples.
During active-site titration experiments, for more accurate molecular mass information, ESI-MS measurements were performed on a high-resolution Synapt HDMS MS system (Waters, Manchester, UK) without prior separation under the following conditions: spray voltage: 3.8 kV, gas flow, temperature, and ion transmission was optimized for the mass range m/z 400–2500. Molecular masses of the inhibited (SrLip–DCI complex) and noninhibited lipase were derived after charge state deconvolution. Standard deviation for the molecular weight after deconvolution was calculated from 10 sample analyses.
Results and discussion
In our previous study of active site of SrLip, we have shown that the inhibition of this enzyme with high molar excess (600-fold) of DCI is fast, complete, and stable enough for mass spectrometric measurementsCitation34. Low-energy CID peptide sequencing using a MALDI-QIT-RTOF instrument unambiguously identified covalent bond formation between DCI and SrLip active-site serine (Ser-10). However, we have observed that after prolonged time of incubation, inhibited lipase regained its activity. These results prompted us to further investigate reaction between SrLip (highly purified sample) and DCI, i.e. to determine inhibition and reactivation constants, and to determine active SrLip concentration using the active-site titration with DCI.
Inhibited lipase reactivation
In the above-mentioned preliminary experiments (previous paragraph), we observed that the reactivation is slower at higher DCI concentration in the incubation mixture and lower temperature of incubation. Thus, at pH 6.8 and room temperature, reactivation starts after 25 h and 75 h of incubation of SrLip with 30-fold and 60-fold molar excess of DCI, respectively. Study of SrLip inhibited with 30-fold molar excess of DCI at pH 6.8 revealed that reactivation at +4°C starts after 240 h and at room temperature after 25 h.
To determine the reactivation half-time and to see whether this reactivation process could be followed by mass spectrometry (MS), the experiment was set up as described in Materials and methods, Reactivation kinetics. Percentage of active SrLip was calculated as percentage of starting pNPC activity. It was also estimated as the ratio of the peak intensity of unmodified SrLip to the sum of peak intensities of both unmodified and DCI-modified SrLip (%active = Iunmod/(Iunmod + Imod)), with the assumptions that MALDI signal intensities for enzyme and complex are comparable and that correlation between signal intensity and enzyme concentration is linear. Half-times of reactivation were calculated from plots of percentage of active SrLip vs. time of the incubation (). Values obtained from the activity measurements and MALDI as well as ESI mass spectra are in good agreement (11.3 ± 0.2 h, 13.5 ± 0.4 h, and 12.1 ± 0.5 h, respectively), indicating that MS is indeed suitable for following time-course of inhibited enzyme reactivation. It should be pointed out that reactivation half-times could be well compared. However, percentage of active SrLip calculated from mass spectrometric data was consistently higher than the one calculated from regained pNPC activity. A possible explanation could be instability of the inhibitor in sample preparation conditions for MS. MALDI mass spectra taken at different times of incubation are shown in .
Figure 1. Time-course of SrLip–DCI complex reactivation after incubation of 4.55 µM Streptomyces rimosus lipase with the 10-fold molar excess of DCI, followed by activity assay and mass spectrometric (MALDI-MS and ESI-MS) measurements. Curve fitting performed in GraphPad Prism 4.0. DCI, 3,4-dichloroisocoumarin; ESI, electro-spray ionization; MALDI, matrix-assisted laser desorption/ionization; MS, mass spectrometry; SrLip, Streptomyces rimosus lipase; TOF, time-of-flight.
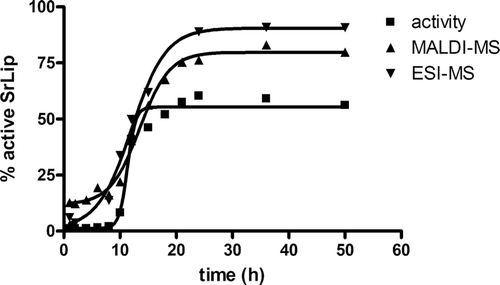
Figure 2. Time-course of SrLip–DCI complex reactivation after incubation of 4.55 µM Streptomyces rimosus lipase with the 10-fold molar excess of DCI, followed by MALDI-TOF-MS. Incubation times are indicated on the right. Only region encompassing doubly protonated ions is shown. DCI, 3,4-dichloroisocoumarin; MALDI, matrix-assisted laser desorption/ionization; MS, mass spectrometry; SrLip, Streptomyces rimosus lipase.
As in our previous workCitation34, doubly protonated molecules of SrLip and SrLip–DCI complex in MALDI mass spectra were more abundant and exhibited significantly higher mass spectrometric resolution (RFWHM) compared with singly protonated molecules. This is due to matrix 2,6-DHAP favouring multiply charged ions. Therefore, the [M+2H]2+ peaks were used for mass determination and calculation of percentage of active SrLip instead of typically used [M+H]+ peaks. However, since mass spectra were calibrated externally, masses of inhibited and noninhibited lipase show slight deviations from theoretical values (noninhibited lipase, measured: 24,170 ± 8 Da, theoretical: 24,166 Da; inhibited lipase, measured: 24,359 ± 10 Da; theoretical: 24,362 Da). For better molecular weight determination, ESI-MS ion trap mass spectra were recorded. Relative ion intensities of analyte ions carrying 23 charges were taken into calculations–m/z 1051.6 for the unmodified and m/z 1060.2 for the modified lipase. After charge state deconvolution, a molecular weight of 24,166 ± 2 Da was determined for the unmodified SrLip (which fits perfectly to the calculated value) and 24,364 ± 3 Da for DCI-modified SrLip (which shows a deviation of 2 Da).
Although DCI was introduced as general mechanism-based irreversible serine-protease inhibitor, the authors immediately noticed that the enzyme–inhibitor complex is unstableCitation5. The authors reported half-times for reactivation of several serine proteases inactivated by DCI (molar excess at least 20-fold) to be from 25 to over 100 h. They reported half-lives for the spontaneous hydrolysis of DCI in HEPES and phosphate buffer, and we have determined half-lives in phosphate buffer in conditions used in this study for incubation method and progress curve method of SrLip inactivation by DCI (), to confirm the stability of the inhibitor during the time needed for inhibition to occur. One should be aware that during spontaneous hydrolysis of DCI, its isocoumarin ring opens. On the other hand, in the process of inhibited enzyme reactivation, hydrolysis of (already opened) isocoumarin in the enzyme active site is mediated by labile acyl moieties ()Citation5. Thus, different chemical processes (consequently in different time-windows) take place.
Table 1. Half-lives and zero-order constants for the spontaneous hydrolysis of DCI in different buffer systems.
The reversibility of enzyme inhibition by DCI can be advantageous if DCI is used as a reversible active-site blocking group for serine proteases. An enzyme (in)activity could be controlled over long period of time by adjusting the inhibitor concentrationCitation5. However, the recent literature has revealed very limited amount of kinetic data based on enzyme inhibition by DCI.
Use of DCI in lipase inhibition – incubation method
The use of DCI as the inhibitor of serine proteases is mostly related to verification of the catalytic residue serine in the active site of an enzymeCitation15,Citation40–43. Data on the inhibition of serine proteases and some other catalytic serine-based hydrolases by DCI are listed in . Among the enzymes listed in , only a few serine proteases reacted very rapidly and strongly with DCI. The covalent binding of DCI to the catalytic serine of an enzyme can be determined reliably by MS methodsCitation34. The 3D structures of the majority of the enzymes and their various complexes, listed in , are deposited in Protein DatabankCitation48. However, the search of Protein Databank revealed a single complex of DCI with the serum serine protease, factor D (at a resolution of 1.8 ÅCitation45) (, pdb code 1dic). The authors of the paper reporting the structure of DCI:D factor complexCitation45 proposed the chemical modification of the inhibitor resulting in the formation of an α-hydroxy acid moiety through the nucleophilic substitution of both chlorine atoms.
Table 2. Inhibition of serine proteases and other enzymes by 3,4-dichloroisocoumarin.
Figure 3. View of the bound inhibitor in the structure of the complex of human factor D with DCI (PDB code 1dic). a) The DCI molecule is covalently bound to the catalytic Ser-195, whereas the catalytic residues His-57 and Asp-102 are out of the pocket with the bound inhibitor. Two hydrogen bond contacts between factor D and DCI are found: a carboxyl oxygen of inhibitor acts as the acceptor to nitrogen of Gly-193 (N–H…O, 2.73 Å) and the carbonyl oxygen is in a weak contact to Gly-216 (3.44 Å). b) The same orientation of the inhibitor depicted against the surface of the protein shaded according to the hydrophobicity of the residues (light to dark with increasing hydrophobicity). DCI, 3,4-dichloroisocoumarin.
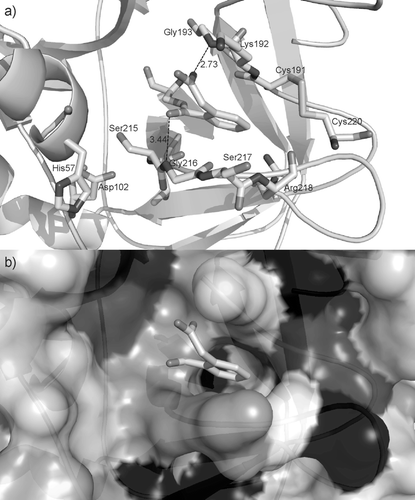
According to the data in the literature on moderate inhibition of factor D by isocoumarinsCitation49, DCI might not be the “ideal model” to study enzyme–inhibitor interactions. Unfortunately, there is no other known 3D structure of the enzyme complex with DCI inhibitor. We decided to extract the relevant structural information on the interactions of DCI inhibitor and the enzyme – human factor DCitation45 and discussed them in view of the reactivation of the enzyme inhibited by DCI observed in our experiments on SrLip inhibition. The relevance of this reasoning lays in the fact that lipases and serine proteases share the topology of active-site triad and general mechanism featuresCitation50.
The inhibitor is covalently bound to the catalytic residue Ser-195 of factor D (numbering according to trypsin, as in ref. Citation45). However, the two catalytic residues His-57 and Asp-102 are not involved in any noncovalent interaction with the inhibitor; the shortest distances between DCI and these catalytic residues are 6.24 and 6.33 Å, respectively. The polar head of DCI is placed at the entrance of a narrow, mostly hydrophilic pocket () and it is stabilized by the two hydrogen bonds: an oxygen atom of the carboxyl group is an acceptor of the peptide NH from Gly-193, whereas the carbonyl oxygen (next to the covalent bond to Ser-195) participates in a weak contact to the peptide NH of Gly-216 (). The hydrophilic pocket, which is the only space available in the vicinity of Ser-195, is not an optimal neighbourhood for the aromatic moiety of the inhibitor. The relative orientation of the phenyl ring is not favourable for C–H…O contacts. Obviously, the location of the inhibitor is not a favourable one. The narrow pocket with the two holes that can serve for the inhibitor exit might be used when the key interaction (the covalent bond to Ser-195) is cleaved.
In our preliminary experiments, we attempted to determine second-order inhibition rate constant (kobs/[I]) by commonly used incubation method. Value obtained from linear part of the inhibition curve – kobs/[I] = 5705 ± 26 M−1 s−1 – shows that SrLip is strongly inhibited by DCI. So far, DCI has shown higher reactivity only with human leukocyte elastase, and comparable reactivity with murine granzyme B, Staphylococcus aureus protease V-8, human proteinase 3, and porcine pancreatic elastase (). Already, Harper et al.Citation5 noted specificity of DCI for serine proteases and particularly human leukocyte elastase, whereas it did not inactivate other tested types of proteases. Later, Powers and KamCitation6 stated high specificity of other substituted isocoumarins for human leukocyte elastase compared to other chymotrypsin-like enzymes. The fact that DCI inhibits trypsin-like enzymes (including several esterases) less effectively and more slowly ()Citation6 indicates similarity of the substrate specificity and active-site architecture between SrLip and elastase (and chymotrypsin), which can be verified when SrLip 3D structure becomes available.
The shape of inhibition curve () confirmed that inhibition of S. rimosus lipase with DCI follows mechanism B depicted in (two-step irreversible inhibition). Namely, according to BiethCitation3, plot of kobs vs. inhibitor concentration can be linear, biphasic, or hyperbolic. Linear plot suggests, depending on the exact equation, one-step reversible or one-step irreversible inhibition. Biphasic plot indicates two-step reversible competitive inhibition. Hyperbolic plot, with the equation ![]() , is a characteristic of two-step irreversible inhibition (). This is valid for inhibitor concentration that is much higher than the enzyme concentration ([I]0 ≥ 10 [E]0). This condition was fulfilled in SrLip–DCI experiments and measured kobs values were fitted to given equation in GraphPad Prism 4.0 program with R2 = 0.99.
, is a characteristic of two-step irreversible inhibition (). This is valid for inhibitor concentration that is much higher than the enzyme concentration ([I]0 ≥ 10 [E]0). This condition was fulfilled in SrLip–DCI experiments and measured kobs values were fitted to given equation in GraphPad Prism 4.0 program with R2 = 0.99.
Figure 4. Inhibition of Streptomyces rimosus lipase by 3,4-dichloroisocoumarin: a) incubation method, b) progress curve method.
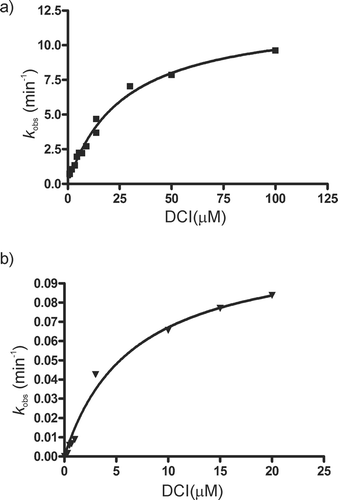
Inhibition kinetics constants were calculated to be k2 = 12.2 ± 0.6 min−1 and Ki* = 26.6 ± 2.8 µM. Dissociation constant of the noncovalent E•I complex, ![]() , is marked with an asterisk because the Km value for the substrate used in these experiments (pNPP) is unknown; SrLip follows interfacial activation kineticsCitation29 and not Michaelis–Menten kinetics. The proper Ki constant and constant calculated in this study are related by:
, is marked with an asterisk because the Km value for the substrate used in these experiments (pNPP) is unknown; SrLip follows interfacial activation kineticsCitation29 and not Michaelis–Menten kinetics. The proper Ki constant and constant calculated in this study are related by: ![]() Citation44.
Citation44.
Lipase inhibition in the presence of substrate
The inhibition of SrLip with DCI was very fast as was the case in most tested enzymes (t1/2 < 1 min, ) and this made the use of incubation method rather impractical. Therefore, we determined inhibition rate also by progress curve method (Mechanism D in ). In this methodCitation38, substrate hydrolysis is monitored in the presence of an irreversible inhibitor, and data analyzed as described in Materials and methods. With this method, inhibition kinetics constants were calculated to be k2 = 0.11 ± 0.01 min−1 and ![]() (). The dissociation constant
(). The dissociation constant ![]() is fourfold lower than the value obtained by incubation method, but inhibition rate constant k2 is over 100-fold lower than the value obtained by incubation method, which results in 27.5-fold decrease in inhibition specificity
is fourfold lower than the value obtained by incubation method, but inhibition rate constant k2 is over 100-fold lower than the value obtained by incubation method, which results in 27.5-fold decrease in inhibition specificity ![]() .
.
A possible explanation for increase in DCI affinity (lower ![]() ) could be in a characteristic lipase property of interfacial activation, i.e. enhanced activity in the presence of water–oil interface. We have previously demonstrated interfacial activation for SrLip and enhanced inhibitory effect of PMSF when it was added during substrate hydrolysisCitation29. Although the affinity of DCI for SrLip active site is high enough to be measured in the absence of substrate (incubation method), one can imagine that in the presence of substrate (progress curve method), active site becomes even more accessible to the inhibitor. The decrease in inhibition rate constant (k2) can probably be explained by kinetic competition of the inhibitor and substrate for the enzyme active site.
) could be in a characteristic lipase property of interfacial activation, i.e. enhanced activity in the presence of water–oil interface. We have previously demonstrated interfacial activation for SrLip and enhanced inhibitory effect of PMSF when it was added during substrate hydrolysisCitation29. Although the affinity of DCI for SrLip active site is high enough to be measured in the absence of substrate (incubation method), one can imagine that in the presence of substrate (progress curve method), active site becomes even more accessible to the inhibitor. The decrease in inhibition rate constant (k2) can probably be explained by kinetic competition of the inhibitor and substrate for the enzyme active site.
Lipase active-site titration
As we have proven that DCI is a covalent inhibitor of SrLip, we attempted enzyme active-site titration of SrLip with this compound. Analogous to reactivation studies, enzyme assay method was compared with MALDI-MS and ESI-MS methods. Using activity assay method under conditions described in Materials and methods, Lipase active-site titration, the active enzyme concentration was determined to be 2.9 µM. This value is in good agreement with protein concentration of 3.0 µM measured with NanoDrop1000 instrument.
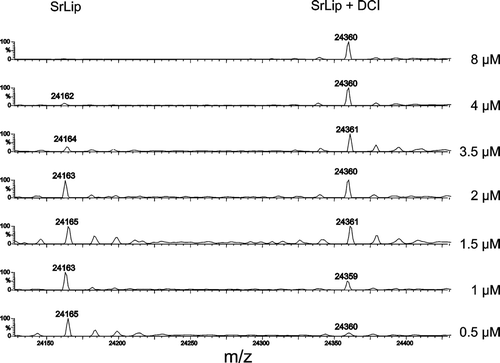
MS was already used, although not extensively, as a tool to determine active-site accessibility in proteinsCitation51 and for active-site titrationCitation52. During active-site titration, a sample of each lipase–inhibitor incubation mixture, after 15 min of incubation, was subjected to ESI- and MALDI-mass spectrometric measurements. The trend of diminishing enzyme activity with increasing inhibitor concentration was clearly detectable; however, in our case, these methods were not accurate enough for precise active enzyme concentration determination. Active enzyme concentrations obtained by ESI-MS and MALDI-MS were 4.2 and 6.1 µM, respectively.
ESI mass spectra taken at different times of incubation are shown in . The analysis of mass spectra was performed as in experiments on SrLip–DCI complex reactivation, and molecular masses determined were 24,168 ± 7 and 24,368 ± 5 Da with MALDI-MS for SrLip and SrLip–DCI complex, respectively, and 24,164 ± 1 and 24,360 ± 1 Da with ESI–high-resolution MS for SrLip and SrLip–DCI complex, respectively (both values show deviation of 2 Da).
Figure 5. Titration of 3 µM SrLip with DCI, followed by ESI-MS after 15 min of incubation of enzyme with increasing concentration of inhibitor. DCI, 3,4-dichloroisocoumarin; ESI, electro-spray ionization; MS, mass spectrometry; SrLip, Streptomyces rimosus lipase.
The use of MS does not require highly purified enzyme in enzyme active-site titration, whereas classical protein concentration determination methods depend on protein purity. However, for the measurement of active enzyme concentration to be accurate and precise, the inhibitor should be stable and firmly bound in the enzyme active site, which is apparently not the case with DCI. It is stable enough for activity measurements, which are performed in “mild” buffer conditions and in short time (so active-site titration was successful with pNPP activity measurements). However, sample preparation conditions and MS measurement are not optimal for DCI when enzyme and inhibitor are in close-to-equimolar concentrations, and inhibitor decomposes in time scale needed to perform the experiment.
Kinetic characterizations of enzymes require knowledge about the concentration of active site in solution. This is especially important in structure–activity studies of genetically engineered enzymes in order to rule out folding or stability problems of the mutants. Probably due to complicated kinetic mechanism caused by interfacial activation, the lipase active-site titrations are seldom performed. However, few methods were developed, mainly using 4-methylumbelliferyl phosphonate- or phosphate-based inhibitorsCitation53,Citation54 or methyl-p-nitrophenyl-n-hexylphosphonateCitation55. To the best of our knowledge, this is the first active-site titration of a lipase by DCI.
Conclusion
In this article, we present the first kinetic study of the lipase (specifically SGNH-hydrolase) inhibition by DCI. Inhibition of SrLip with DCI follows the mechanism of two-step irreversible inhibition, with constants ![]() and k2 = 12.2 ± 0.6 (0.11 ± 0.01) min−1, determined by incubation (or progress curve) method. Determination of the active enzyme concentration using inhibition with DCI and subsequent activity measurements were performed, for the very first time, with a highly purified lipase preparation. These results are in accordance with protein concentration determination using UV absorbance measurement at 280 nm. MS was shown to be a suitable method to follow changes in DCI-modified lipase concentration during the course of inhibited enzyme reactivation or the active-site titration.
and k2 = 12.2 ± 0.6 (0.11 ± 0.01) min−1, determined by incubation (or progress curve) method. Determination of the active enzyme concentration using inhibition with DCI and subsequent activity measurements were performed, for the very first time, with a highly purified lipase preparation. These results are in accordance with protein concentration determination using UV absorbance measurement at 280 nm. MS was shown to be a suitable method to follow changes in DCI-modified lipase concentration during the course of inhibited enzyme reactivation or the active-site titration.
Declaration of interest
The authors thank Prof. J.C. Powers, School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, USA for his information on binding properties of 3,4-dichloroisocoumarin. Financial support by the Croatian Ministry of Science, Education and Sport (grant 098–1191344-2943), the Austrian Federal Ministry of Science and Research (UniInfra IV to G.A.), and a Bilateral Cooperation Grant Croatia-Austria (1/2008 and 1/2010 to I.L.A. and G.A., respectively I.L.A. and M.M.-D.) is gratefully acknowledged.
The authors report no conflicts of interest. The authors are responsible for the paper presented.
References
- Hiratake J. Enzyme inhibitors as chemical tools to study enzyme catalysis: rational design, synthesis, and applications. Chem Rec 2005;5:209–228.
- Cornish-Bowden E. (2004). Fundamentals of enzyme kinetics. London: Portland Press Ltd.
- Bieth JG. Theoretical and practical aspects of proteinase inhibition kinetics. Meth Enzymol 1995;248:59–84.
- Febbraio F, D’Andrea SE, Mandrich L, Merone L, Rossi M, Nucci R et al. Irreversible inhibition of the thermophilic esterase EST2 from Alicyclobacillus acidocaldarius. Extremophiles 2008;12:719–728.
- Harper JW, Hemmi K, Powers JC. Reaction of serine proteases with substituted isocoumarins: discovery of 3,4-dichloroisocoumarin, a new general mechanism based serine protease inhibitor. Biochemistry 1985;24:1831–1841.
- Powers JC, Kam CM. Isocoumarin inhibitors of serine peptidases. Meth Enzymol 1994;244:442–457.
- Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev 2002;102:4639–4750.
- Hameed A, Aslam U, Ying AJ. 3,4-Dichloroisocoumarin serine protease inhibitor induces DNA fragmentation and apoptosis in susceptible target cells. Proc Soc Exp Biol Med 1998;219:132–137.
- Anees M, Steven FS. Inhibition of a tumour protease with 3,4-dichloroisocoumarin, pentamidine-isethionate and guanidino derivatives. J Enzym Inhib 1994;8:213–221.
- Billings PC, Carew JA, Keller-McGandy CE, Goldberg AL, Kennedy AR. A serine protease activity in C3H/10T1/2 cells that is inhibited by anticarcinogenic protease inhibitors. Proc Natl Acad Sci USA 1987;84:4801–4805.
- Finco TS, Beg AA, Baldwin AS Jr. Inducible phosphorylation of I kappa B alpha is not sufficient for its dissociation from NF-kappa B and is inhibited by protease inhibitors. Proc Natl Acad Sci USA 1994;91:11884–11888.
- Megyeri P, Pabst KM, Pabst MJ. Serine protease inhibitors block priming of monocytes for enhanced release of superoxide. Immunology 1995;86:629–635.
- Conseil V, Soête M, Dubremetz JF. Serine protease inhibitors block invasion of host cells by Toxoplasma gondii. Antimicrob Agents Chemother 1999;43:1358–1361.
- Upadhye V, Majumdar A, Gomashe A, Joshi D, Gangane N, Thamke D et al. Inhibition of Mycobacterium tuberculosis secretory serine protease blocks bacterial multiplication both in axenic culture and in human macrophages. Scand J Infect Dis 2009;41:569–576.
- Nikandrov NN, Deshimaru M, Tani A, Chijiwa T, Shibata H, Chang CC et al. Purification, primary structures and evolution of coagulant proteases from Deinagkistrodon actus venom. Toxicon 2005;46:907–917.
- Simons JW, Cox RC, Egmond MR, Verheij HM. Rational design of alpha-keto triglyceride analogues as inhibitors for Staphylococcus hyicus lipase. Biochemistry 1999;38:6346–6351.
- Weibel EK, Hadvary P, Hochuli E, Kupfer E, Lengsfeld H. Lipstatin, an inhibitor of pancreatic lipase, produced by Streptomyces toxytricini. I. Producing organism, fermentation, isolation and biological activity. J Antibiot 1987;40:1081–1085.
- Hochuli E, Kupfer E, Maurer R, Meister W, Mercadal Y, Schmidt K. Lipstatin, an inhibitor of pancreatic lipase, produced by Streptomyces toxytricini. II. Chemistry and structure elucidation. J Antibiot 1987;40:1086–1091.
- Carrière F, Renou C, Ransac S, Lopez V, De Caro J, Ferrato F et al. Inhibition of gastrointestinal lipolysis by Orlistat during digestion of test meals in healthy volunteers. Am J Physiol Gastrointest Liver Physiol 2001;281:G16–G28.
- Ben Ali Y, Chahinian H, Petry S, Muller G, Lebrun R, Verger R et al. Use of an inhibitor to identify members of the hormone-sensitive lipase family. Biochemistry 2006;45:14183–14191.
- Trofa D, Agovino M, Stehr F, Schäfer W, Rykunov D, Fiser A et al. Acetylsalicylic acid (aspirin) reduces damage to reconstituted human tissues infected with Candida species by inhibiting extracellular fungal lipases. Microbes Infect 2009;11:1131–1139.
- Andlauer W, Prunier P, Prim D. Fluorometric method to assess lipase inhibition activity. Chimia 2009;63:695–697.
- McDougall GJ, Kulkarni NN, Stewart D. Berry polyphenols inhibit pancreatic lipase activity in vitro. Food Chem 2009;115:193–199.
- Slanc P, Doljak B, Kreft S, Lunder M, Janes D, Strukelj B. Screening of selected food and medicinal plant extracts for pancreatic lipase inhibition. Phytother Res 2009;23:874–877.
- Kruithof CA, Dijkstra HP, Lutz M, Spek AL, Egmond MR, Gebbink RJMK, van Koten G. Non-tethered organometallic phosphonate inhibitors for lipase inhibition: Positioning of the metal centre in the active site of cutinase. Eur J Inorg Chem 2008;28:4425–4432.
- Cavalier JF, Buono G, Verger R. Covalent inhibition of digestive lipases by chiral phosphonates. Acc Chem Res 2000;33:579–589.
- Malla RK, Bandyopadhyay S, Spilling CD, Dutta S, Dupureur CM. The first total synthesis of (±)-cyclophostin and (±)-cyclipostin P: inhibitors of the serine hydrolases acetyl cholinesterase and hormone sensitive lipase. Org Lett 2011;13:3094–3097.
- Ben Ali Y, Verger R, Carrière F, Petry S, Muller G, Abousalham A. The molecular mechanism of human hormone-sensitive lipase inhibition by substituted 3-phenyl-5-alkoxy-1,3,4-oxadiazol-2-ones. Biochimie 2012;94:137–145.
- Abramić M, Leščić I, Korica T, Vitale Lj, Saenger W, Pigac J. Purification and properties of extracellular lipase from Streptomyces rimosus. Enzyme Microb Technol 1999;25:522–529.
- Leščić I, Vukelić B, Majerić-Elenkov M, Saenger W, Abramić M. Substrate specificity and effects of water-miscible solvents on the activity and stability of extracellular lipase from Streptomyces rimosus. Enzyme Microb Technol 2001;29:548–553.
- Leščić Ašler I, Ivic N, Kovačić F, Schell S, Knorr J, Krauss U et al. Probing enzyme promiscuity of SGNH hydrolases. Chembiochem 2010;11:2158–2167.
- Leščić Ašler I, Zehl M, Kovačić F, Müller R, Abramić M, Allmaier G et al. Mass spectrometric evidence of covalently-bound tetrahydrolipstatin at the catalytic serine of Streptomyces rimosus lipase. Biochim Biophys Acta 2007;1770:163–170.
- Tiss A, Lengsfeld H, Carrière F, Verger R. Inhibition of human pancreatic lipase by tetrahydrolipstatin: further kinetic studies showing its reversibility. J Mol Catal B: Enzym 2009;58:41–47.
- Zehl M, Leščić I, Abramić M, Rizzi A, Kojić-Prodić B, Allmaier G. Characterization of covalently inhibited extracellular lipase from Streptomyces rimosus by matrix-assisted laser desorption/ionization time-of-flight and matrix-assisted laser desorption/ionization quadrupole ion trap reflectron time-of-flight mass spectrometry: localization of the active site serine. J Mass Spectrom 2004;39:1474–1483.
- Vujaklija D, Schröder W, Abramić M, Zou P, Leščić I, Franke P et al. A novel streptomycete lipase: cloning, sequencing and high-level expression of the Streptomyces rimosus GDS(L)-lipase gene. Arch Microbiol 2002;178:124–130.
- Leščić I, Zehl M, Müller R, Vukelic B, Abramić M, Pigac J et al. Structural characterization of extracellular lipase from Streptomyces rimosus: assignment of disulfide bridge pattern by mass spectrometry. Biol Chem 2004;385:1147–1156.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254.
- Tian WX, Tsou CL. Determination of the rate constant of enzyme modification by measuring the substrate reaction in the presence of the modifier. Biochemistry 1982;21:1028–1032.
- Knight CG. Active-site titration of peptidases. Meth Enzymol 1995;248:85–101.
- Tsai TC, Lin W, Yang SH, Cheng WT, Cheng EH, Lee MS et al. Granzyme G is expressed in the two-cell stage mouse embryo and is required for the maternal-zygotic transition. BMC Dev Biol 2010;10:88.
- Bielen A, Cetkovic H, Long PF, Schwab H, Abramić M, Vujaklija D. The SGNH-hydrolase of Streptomyces coelicolor has (aryl)esterase and a true lipase activity. Biochimie 2009;91:390–400.
- Bodvard K, Mohlin J, Knecht W. Recombinant expression, purification, and kinetic and inhibitor characterisation of human site-1-protease. Protein Expr Purif 2007;51:308–319.
- Hou J, Clemmons DR, Smeekens S. Expression and characterization of a serine protease that preferentially cleaves insulin-like growth factor binding protein-5. J Cell Biochem 2005;94:470–484.
- Früh H, Kostoulas G, Michel BA, Baici A. Human myeloblastin (leukocyte proteinase 3): reactions with substrates, inactivators and activators in comparison with leukocyte elastase. Biol Chem 1996;377:579–586.
- Cole LB, Kilpatrick JM, Chu N, Babu YS. Structure of 3,4-dichloroisocoumarin-inhibited factor D. Acta Crystallogr D Biol Crystallogr 1998;54:711–717.
- Morty RE, Authié E, Troeberg L, Lonsdale-Eccles JD, Coetzer TH. Purification and characterisation of a trypsin-like serine oligopeptidase from Trypanosoma congolense. Mol Biochem Parasitol 1999;102:145–155.
- Djaballah H, Harness JA, Savory PJ, Rivett AJ. Use of serine-protease inhibitors as probes for the different proteolytic activities of the rat liver multicatalytic proteinase complex. Eur J Biochem 1992;209:629–634.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235–242.
- Kam CM, Oglesby TJ, Pangburn MK, Volanakis JE, Powers JC. Substituted isocoumarins as inhibitors of complement serine proteases. J Immunol 1992;149:163–168.
- Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci 1998;23:347–352.
- O’Farrell N, Kreiner M, Moore BD, Parker MC. A rapid and direct method for the determination of active site accessibility in proteins based on ESI-MS and active site titrations. Biotechnol Bioeng 2006;95:767–771.
- Bovet C, Zenobi R. Determination of active enzyme concentration using activity-based probes and direct mass spectrometric readout. Anal Biochem 2008;373:380–382.
- Laszlo JA, Jackson M, Blanco RM. Active-site titration analysis of surface influences on immobilized Candida antarctica lipase B activity. J Mol Catal B: Enzym 2011;69:60–65.
- Fujii R, Utsunomiya Y, Hiratake J, Sogabe A, Sakata K. Highly sensitive active-site titration of lipase in microscale culture media using fluorescent organophosphorus ester. Biochim Biophys Acta 2003;1631:197–205.
- Rotticci D, Norin T, Hult K, Martinelle M. An active-site titration method for lipases. Biochim Biophys Acta 2000;1483:132–140.
