Abstract
Carbonic anhydrases (CAs, EC 4.2.1.1) began to be investigated in detail in pathogenic bacteria, in the search for antibiotics with a novel mechanism of action, since it has been demonstrated that in many bacteria CAs are essential for the life cycle of the organism. The presence of CAs in pathogenic bacteria allows the development of anti-infectives with a new mechanism of action, less explored to date. Here, novel quinazoline derivatives crowned with sulfonamide functionality at position-2 were tested for their ability to inhibit the bacterial γ-CA (PgiCA), identified in the genome of Porphyromonas gingivalis. Six compounds were highly effective, nanomolar inhibitors of the pathogenic enzyme γ-PgCA. Three of them were also highly effective sub-nanomolar inhibitors of the cytosolic human isoform II (hCAII). The best γ-PgCA inhibitor was compound 8c, with a KI of 3.53 nM and selectivity ratio of 24.5 and 24.8 against hCA I and hCA II, respectively. Many of these new compounds showed a high selectivity for bacterial enzyme respect to the mammalian CA isoforms (hCAI and hCAII). These results suggest that sulfonamides with quinazoline scaffold could be considered as suitable candidates for further derivatization to better understand the role of bacterial CAs in pathogenesis.
Introduction
Infectious diseases and development of hospital acquired bacterial resistance is a major health problem, which concerns the medical community all over the worldCitation1. Unfortunately, drug design and development activities in this vital area of locating new lead molecules for combating serious bacterial diseases have declined, and at the same time, infections from resistant bacteria are currently prevalent with inadequate treatment optionsCitation2. It was estimated that in the USA, every year about two million people are infected with antibiotic-resistant strains, moreover at least 23 000 are dying out of such threatCitation3. Another unexpected but induced source for the development of bacterial resistance was recently recognized to be due to mutation of non-pathogenic organisms, which made the situation more badlyCitation4.
Throughout the past few years, efforts to combat multiple resistant microorganisms were mainly focused in Gram-positive bacteria and the pharmaceutical companies have succeeded to develop new drugs against such organismsCitation5. Unfortunately, no similar efforts towards controlling the growing problem of Gram-negative resistance were doneCitation6. Researches were not parallel to discover antibiotics to treat infections caused by these bacteria leading to the emergence of a disturbing perspective in addition to a considerable number of untreatable Gram-negative infectionsCitation7. Consequently, there is an urgent need to develop lead compounds based on novel mechanisms of action and it is time to intensify attention to Gram-negative bacterial resistanceCitation8.
Recently, carbonic anhydrases (CAs, EC 4.2.1.1) began to be investigated in detail in pathogenic bacteria, in the search for antibiotics with a novel mechanism of action, since it has been demonstrated that in many bacteria CAs are essential for the life cycle of the organismCitation9. CA enzymes have been investigated in detail in pathogenic (as well as non-pathogenic) bacteria such as Brucella suis, Mycobacterium tuberculosis, Streptococcus spp., Helicobacter pylori, Salmonella enterica, Sulfurihydrogenibium yellowstonense, Sulfurihydrogenibium azorense, Vibrio cholerae and othersCitation10–13. The presence of CAs in these pathogenic microorganisms allows the development of anti-infectives with a new mechanism of action, less explored to dateCitation11–15.
CAs are a family of enzymes widely distributed in a diversity of organisms and catalyze the reversible hydration of carbon dioxideCitation11. So, they are involved in a variety of simple but essential physiological process within the living cell such as in the transport of CO2 or , in supplying CO2 or
for the biosynthetic reactions (and thus metabolisms), in pH regulation and also in cyanate degradation as in Escherichia coli, as well as in the survival of intracellular pathogens within their host. Five genetically distinct CA families are known to date, α-, β-, γ-, δ- and ζ-CAs, and all of them are metalloenzymes, using Zn(II), Cd(II), or Fe(II) at their active siteCitation8. In bacteria, genes encoding CAs from three classes, α-, β- and γ-CAs were identifiedCitation15. The inhibition of CAs is well-known processes, with most types of inhibitors binding to the metal center. Sulfonamides and their bioisosteres, such as sulfamates and sulfamides are the most investigated types of organic inhibitors, having various biomedical applications as diuretics or as drugs for the treatment or prevention of a variety of disorders such as anti-glaucoma drugs, anti-convulsants, anti-obesity, anti-cancer, anti-pain and anti-infective agentsCitation15–18.
Recently, our group reported a new poorly characterized enzyme belonging to the γ-CA, the Porphyromonas gingivalis. A Gram-negative bacterium isolated from the oral cavity which is the main causative agent of periodontitisCitation19. Recent studies proved that an infection caused by this periodontal pathogen increases the risk for myocardial infarctionCitation19. The genome of P. gingivalis encodes for a β- and a γ-CAs. Recently, our group produced the γ-CA (named PgiCA) and the β-CA (named PgiCAb) from this pathogenic bacterium by the recombinant DNA technology. PgiCA and PgiCAb were shown to possess a significant catalytic activity for the reaction that converts the CO2 to bicarbonate and protons (PgiCA: kcat of 4.1 × 105 s−1 and kcat/Km of 5.4 × 107 M−1 × s−1; PgiCAb: kcat of 2.8 × 105 s−1 and a kcat/Km of 1.5 × 107 M−1 × s−1)Citation20–23.
Sulfonamides and their bioisosteres sulfamates are the most well known and extensively studied CA inhibitorsCitation8–18. Quinazolines are 1,3-benzodiazine derivatives representing an important class among heterocyclic compounds of medical, biological, and industrial interest. They attracted the attention of many researchers since the discovery of Thymitaq, Ralitrexed, Gefitinib and Erlotinib as approved anti-tumor drugs in addition to Prazosin, the second-line drug of choice for the management of elevated blood pressureCitation24–27. Here, we test a new series of sulfonamides containing quinazoline scaffolds, recently developed in our laboratories, for their ability to inhibit the PgiCA enzyme.
Materials and methods
Construct preparation, protein expression and purification
The synthetic P. gingivalis gene encoding PgiCA was designed and produced by Life Technologies (Invitrogen, Milan, Italy), a company specialized in the gene synthesis. The gene contained an NdeI and XhoI restrictions sites at the 5′ and 3′ ends, respectively. The synthetic PgiCA was ligated into the expression vector pET15-b (Novagen, Milan, Italy) by T4 DNA ligase to form the expression vector pET15-b/PgiCA. Competent E. coli BL21 (DE3) cells were transformed with pET15-b/PgiCA, grown at 37 °C and induced with 1 mM IPTG. 0.5 mM ZnSO4, which was added after 30 min incubation for uptake in the expressed protein. At 5 h post-induction, cells were harvested and disrupted by sonication at 4 °C. Following centrifugation, the supernatant was loaded onto HIS-Select HF Nickel Affinity Gel (Sigma-Aldrich, Milan, Italy) and the protein was eluted with 200-mM imidazole. At this stage of purification the enzyme was at least 95% pure.
SDS-PAGE
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described previouslyCitation24, using 12% gels.
Determination of the enzyme kinetic parameters
An Applied Photophysics stopped-flow instrument has been used for assaying the CA catalysed CO2 hydration activityCitation25. Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 20 mM Hepes (pH 7.5, for α-CAs) or 20 mM TRIS (pH 8.3 for the β- and γ-CAs) as buffers, and 20 mM NaClO4 (for maintaining constant the ionic strength), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and inhibition constants. For each inhibitor at least six traces of the initial 5–10% of the reaction have been used for determining the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of inhibitor (10–50 mM) were prepared in distilled-deionized water and dilutions up to 0.01 µM were done thereafter with the assay buffer. Inhibitor and enzyme solutions were preincubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the E-I complex or for the eventual active site mediated hydrolysis of the inhibitor. The inhibition constants were obtained by non-linear least-squares methods using PRISM 3, as reported earlierCitation1, and represent the mean from at least three different determinations. All CA isoforms were recombinant ones obtained in-house as reported earlierCitation26–30.
Results and discussion
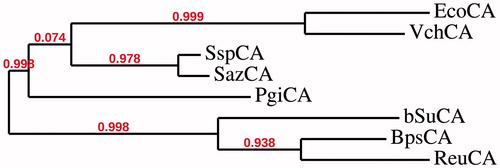
IPTG induction of E. coli BL21 (DE3) cells transformed with the plasmid pET15-b/PgiCA resulted in the production of the recombinant PgiCA. The new γ-CA was isolated and purified to homogeneity from E. coli (DE3) cell extract. Most of the CA activity was recovered in the soluble fraction of cell extract after sonication and centrifugation. Using the affinity column (His-select HF Nickel affinity gel), PgiCA was purified to apparent homogeneity, as indicated by a single protein band after SDS-PAGE (data not shown). The open reading frame of the P. gingivalis gene encodes a 192 amino acid polypeptide chain. showed the amino acid sequence alignment of the γ-CAs from different Gram-negative bacteria: PgiCA, VchCA and bSuCA. The metal ion ligands (see for details) are all conserved. We have constructed a phylogenetic tree including most γ-CAs sequences available from Gram-negative bacteria, in order to better understand the relationships of the P. gingivalis enzyme with those of different other γ-CAs (). Data of show that the P. gingivalis enzyme (PgiCA) is more related to SazCA, SspCA, VchCA and EcoCA (see for details) than bSuCA, BpsCA, ReuCA. These finding suggested that in Gram-negative bacteria γ-CAs apparently seemed to belong at two distinctly phylogenetic groups (Scheme 1).
Figure 1. Amino acid sequence alignment of the γ-CAs from Porphyromonas gingivalis (PgiCA), Vibrio cholerae (VhCA) and Brucella suis (bSuCA). The metal ion ligands are indicated in red. The multialignment was performed with the program Clustal W. The asterisk (*) indicates identity at all aligned positions; the symbol (:) relates to conserved substitutions, while (.) means that semi-conserved substitutions are observed.

Figure 2. Phylogenetic trees of the amino acid sequences of γ-CAs from different Gram-negative bacteria. The tree was constructed using the program PhyML 3.0. Legend: EcoCA, Escherichia coli; VchCA, Vibrio cholerae; SspCA, Sulfurihydrogenibium yellowstonense; SazCA, Sulfurihydrogenibium azorense; PgiCA, Porphyromonas gingivalis; bSuCA, Brucella suis; BpsCA, Burkholderia pseudomallei; ReuCA, Ralstonia eutropha.
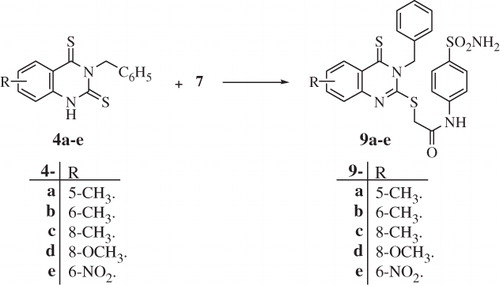
Scheme 1. Synthesis of 3,5,6 and/or 8-substituted-2-thio-4-oxoquinazoline and 2-thio-4-thioxoquinazoline derivatives 3a–h and 4a–e.
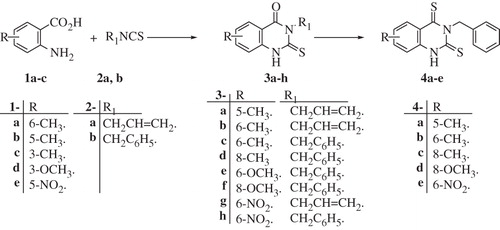
In the present article, novel quinazoline derivatives crowned with sulfonamide functionality at position-2 were tested for their ability to inhibit the bacterial γ-CA (PgiCA), identified in the genome of P. gingivalis. The obtained results were compared with those acquired for the human CAs (hCA I and hCA II). The targeted quinazoline carrying sulfonamide CAIs 8a–h and 9a–e incorporate the classical benzenesulfonamide group (which binds to the Zn(II) ion from the enzyme active site through coordination with the deprotonated sulfonamide moiety)Citation11. The mercaptoacyl group was inserted as a flexible spacer linking the 4-oxo and/or thioxo-2-quinazolinyl functionality to the 4-aminobenzenesulfonamide. The quinazoline scaffold was equipped with allyl and/or benzyl moiety at the 3-position in addition to methyl, methoxy and/or nitro at positions 6,7 and/or 8. In this way a suitable degree of chemical diversity has been achieved in the congeneric series of fully characterized compounds 8a–h and 9a–e investigated here as γ PgiCA inhibitors (Schemes 2 and 3).
Scheme 2. Synthesis of substituted sulfa derivative 7 and 2,3,5,6 and/or 8-substituted-4-oxoquinazoline derivatives 8a–h.
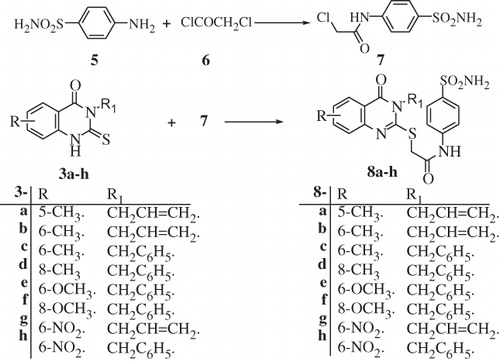
Inhibition data of the above mentioned compounds and acetazolamide AAZ (as a standard drug), against the Gram-negative enzyme PgiCA, as well as the cytosolic CA isozymes hCA I and II (h = human) and the corresponding ratios, are shown in . The following structure–activity relationship (SAR) can be observed from data of :
(i) All sulfonamides 8a–h and 9a–e investigated here significantly inhibited PgiCA, with inhibition constants in the range of 3.5–377.5 nM. Four compounds were mostly equipotent or less active compared to the standard drug acetazolamide AAZ (5-acetamido-1,3,4-thiadiazole-2-sulfonamide). AAZ with a KI of 324 nM is one of the best sulfonamide inhibitors detected until now against γ PgiCA in the unique sulfonamide inhibition study reported so far against this enzymeCitation20.
(ii) Few compounds among the investigated series, namely 8e, 8f and 9a showed KI s in the range of 377.5–344.6 nM against the γ-class bacterial enzyme PgiCA. The least effective PgiCA inhibitor detected here was 9e which inhibited the enzyme with KI over 425 nM. Although the last derivatives represent the less effective CAIs of the series, they still have inhibition potency similar to that showed by AAZ (324 nM).
(iii) Quinazoline-sulfonamide derivatives 8b, 8c, 8g, 9b–9d were very effective PgiCA inhibitors, with KIs in nanomolar range (3.5–39.4 nM). It was clear that compound 8c, the 4-oxoquinazoline with methyl group at position-6 and benzyl group at position-3 was the most active compound with KI 3.5 nM. Interestingly, replacing the 3-benzyl group with allyl (8b KI 18.9 nM), reduces the activity and selectivity towards CA I to some extent but it inverted the selectivity towards CA II. However, keeping the 3-benzyl moiety and moving the methyl group from position-6 (8c KI 3.5 nM) to position-8 (8d KI 248.7 nM) dramatically reduces not only the activity but also the selectivity ratio.
(iv) It is also interesting to notice that the presence of an electron withdrawing group such as NO2 at position-6 leads to less active compounds compared to the presence of benzyl group at position-3 (8h with KI 292.6 nM and 9e with KI > 425.0 nM, respectively). Keeping NO2 group at position-6 and allyl group at position-3 produced highly potent compound 8g with KI of 23.8 nM, compared with the standard drug AAZ with KI 324 nM.
(v) The most active compound was the derivative 8c (3-benzyl-4-oxo-6-methyl-quinazoline) which belongs to the 4-oxoquinazoline series. It was also very selective for the inhibition of the bacterial enzyme over the human ones showing a selectivity ratio for inhibiting PgiCA over hCA I of 24.7, and of PgiCA over hCA II of 24.8 (for acetazolamide the selectivity ratios are of 0.77 and 0.03, respectively, ). Among the 4-thioxoquinazoline series the compounds 9b–9d with KIs values in the range of 30.7–39.4 nM, and selectivity ratio between 20.4 and 56.4 against CA I showed to be also PgiCA-selective inhibitors. Although, they were not very selective inhibitors over the human isoform CA II showing low selectivity ratios in the range of 1.7–3 against this isoform, they still represent a better solution compared to the standard the clinically used sulfonamide AAZ which is a poor and low selective PgiCA inhibitor over hCAII.
Table 1. Inhibition data of sulfonamides 8a–h, 9a–e and acetazolamide AAZ (as standard inhibitor) against human (h) isoforms hCA I, II (cytosolic) and bacterial enzyme PgiCA by a stopped-flow CO2 hydrase assayCitation18.
Conclusions
We located interesting molecules with remarkable low nanomolar inhibitory activity against PgiCA. Most of the newly tested benzenesulfonamides obtained and evaluated in this study showed low nanomolar or sub-nanomolar inhibitory activity. Five compounds showed high selectivity ratio against the Gram-negative bacterial γ-CA over the cytosolic hCA I and II. Some γ-CA selective inhibitors were also detected in the present study. Further studies should be done to explore in detail the exact mechanism of action and to broaden the SAR for this interesting class of quinazoline sulfonamide CAIs.
Declaration of interest
This research was financed by the National Plan of Science, Technology and Innovation (Grant No. 11-MED1874-02), King Saud University, Riyadh (to A. F.). Authors declare no conflict of interest.
References
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs - antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87
- Balsalobre LC, Dropa M, Matté MH. An overview of antimicrobial resistance and its public health significance. Braz J Microbiol 2014;45:1–5
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria inhibitors? J Enzyme Inhib Med Chem 2014 (in press)
- Wright GD. Antibiotic resistance in the environment: a link to the clinic. Curr Opin Microbiol 2010;13:589–94
- Supuran CT. Structure-based drug discovery of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2012;27:759–72
- Supuran CT. Carbonic anhydrases: from biomedical applications of the inhibitors and activators to biotechnologic use for CO2 capture. J Enzyme Inhib Med Chem 2013;28:229–30
- Allouche F, Chabchoub F, Carta F, Supuran CT. Synthesis of aminocyanopyrazoles via a multi-component reaction and anti-carbonic anhydrase inhibitory activity of their sulfamide derivatives against cytosolic and transmembrane isoforms. J Enzyme Inhib Med Chem 2013;28:343–9
- Supuran CT, Carta F, Scozzafava A. Metalloenzyme inhibitors for the treatment of Gram-negative bacterial infections: a patent review (2009–2012). Expert Opin Ther Pat 2013;23:777–88
- Supuran CT. Carbonic anhydrases. Bioorg Med Chem 2013;21:1377–8
- Ekinci D, Fidan I, Durdagi S, Kaban Ş, Supuran CT. Kinetic and in silico analysis of thiazolidin-based inhibitors of α-carbonic anhydrase isoenzymes. J Enzyme Inhib Med Chem 2013;28:370–4
- McKenna R, Supuran CT. Carbonic anhydrase inhibitors drug design. Subcell Biochem 2014;75:291–323
- Nishimori I, Minakuchi T, Morimoto K, et al. Carbonic anhydrase inhibitors: DNA cloning and inhibition studies of the alpha-carbonic anhydrase from Helicobacter pylori, a new target for developing sulfonamide and sulfamate gastric drugs. J Med Chem 2006;49:2117–26
- Alafeefy AM, Abdel-Aziz HA, Vullo D, et al. Inhibition of carbonic anhydrases from the extremophilic bacteria Sulfurihydrogenibium yellostonense (SspCA) and S. azorense (SazCA) with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. Bioorg Med Chem 2014;22:141–7
- Del Prete S, Isik S, Vullo D, et al. DNA cloning, characterization, and inhibition studies of an α-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. J Med Chem 2012;55:10742–8
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704
- Maresca A, Carta F, Vullo D, Supuran CT. Dithiocarbamates strongly inhibit the β-class carbonic anhydrases from Mycobacterium tuberculosis. J Enzyme Inhib Med Chem 2013;28:407–11
- Ekinci D, Karagoz L, Ekinci D, et al. Carbonic anhydrase inhibitors: in vitro inhibition of α isoforms (hCA I, hCA II, bCA III, hCA IV) by flavonoids. J Enzyme Inhib Med Chem 2013;28:283–8
- Maresca A, Vullo D, Scozzafava A, Supuran CT. Inhibition of the alpha- and beta- carbonic anhydrases from the gastric pathogen Helycobacter pylori with anions. J Enzyme Inhib Med Chem 2013;28:388–91
- Pussinen PJ, Alfthan G, Tuomilehto J, et al. High serum antibody levels to Porphyromonas gingivalis predict myocardial infarction. Eur J Cardiovasc Prev Rehabil 2004;11:408–11
- Del Prete S, Vullo D, De Luca V, et al. A highly catalytically active γ-carbonic anhydrase from the pathogenic anaerobe Porphyromonas gingivalis and its inhibition profile with anions and small molecules. Bioorg Med Chem Lett 2013;23:4067–71
- Del Prete S, De Luca V, Vullo D, et al. Biochemical characterization of the gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis, PgiCA. J Enzyme Inhib Med Chem 2014;29:532–7
- Vullo D, Del Prete S, Osman SM, et al. Sulfonamide inhibition studies of the gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis. Bioorg Med Chem Lett 2014;24:240–4
- Del Prete S, Vullo D, Osman SM, et al. Sulfonamide inhibition study of the carbonic anhydrases from the bacterial pathogen Porphyromonas gingivalis: the β-class (PgiCAb) versus the γ-class (PgiCA) enzymes. Bioorg Med Chem 2014;22:4537–43
- Gangjee A, Jain HD, Kurup S. Recent advances in classical and non-classical antifolates as antitumor and antiopportunistic infection agents: part I. Anticancer Agents Med Chem 2007;7:524–42
- Zain JM, Marchi E. Pralatrexate – from bench to bedside. Drugs Today (Barc) 2010;46:91–9
- Ford HE. Gefitinib for oesophageal cancer: a cog in need of a wheel? Lancet Oncol 2014;pii:S1470–2045
- Writer BW, Meyer EG, Schillerstrom JE. Prazosin for military combat-related PTSD nightmares: a critical review. J Neuropsychiatr Clin Neurosci 2014;26:24–33
- Carta F, Temperini C, Innocenti A, et al. Polyamines inhibit carbonic anhydrases by anchoring to the zinc-coordinated water molecule. J Med Chem 2010;53:5511–22
- Koz O, Ekinci D, Perrone A, et al. Analysis of saponins and phenolic compounds as inhibitors of α-carbonic anhydrase isoenzymes. J Enzyme Inhib Med Chem 2013;28:412–17
- Maresca A, Scozzafava A, Vullo D, Supuran CT. Dihalogenated sulfanilamides and benzolamides are effective inhibitors of the three β-class carbonic anhydrases from Mycobacterium tuberculosis. J Enzyme Inhib Med Chem 2013;28:384–7