Abstract
Isocitrate dehydrogenase (IDH) gene from Staphylococcus aureus ATCC12600 was cloned, sequenced and characterized (HM067707). PknB site was observed in the active site of IDH; thus, it was predicted as IDH may be regulated by phosphorylation. Therefore, in this study, PknB, alkaline phosphatase III (SAOV 2675) and IDH genes (JN695616, JN645811 and HM067707) of S. aureus ATCC12600 were over expressed from clones PV 1, UVPALP-3 and UVIDH 1. On passing the cytosloic fractions through nickel metal chelate column, pure enzymes were obtained. Phosphorylation of pure IDH by PknB resulted in the complete loss of activity and was restored upon dephosphorylation with SAOV 2675 which indicated that phosphorylation and dephosphorylation regulate IDH activity in S. aureus. Further, when S. aureus ATCC12600 was grown in BHI broth, decreased IDH activity and increased biofilm units were observed; therefore, this regulation of IDH alters redox status in this pathogen favouring biofilm formation.
Introduction
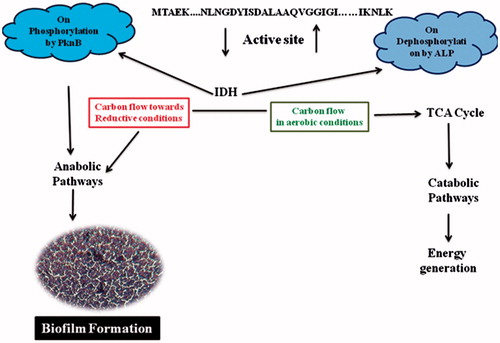
Bacteria have positioned the metabolic requirement of TCA cycle according to their redox status. This cycle is largely regulated by the expression of isocitrate dehydrogenase (IDH)Citation1. In bacteria, the activity of IDH is controlled through phosphorylation and dephosphorylationCitation2–4. In Staphylococcus aureus, the expression of virulence factors, toxins and biofilm formation requires active functioning of TCA cycle and serine/threonine protein kinases (PknB)Citation5–11. Krebs cycle in S. aureus is regulated by IDH whose expression regulates the carbon flow; thus, enabling the organism to adapt and form biofilms in different nichesCitation7,Citation9,Citation12–14. Earlier, we have cloned, sequenced, expressed and characterized IDH gene from S. aureus ATCC12600, the sequence revealed the presence of PknB site in the active site of IDHCitation13.
PknB is also implicated in the cell wall and polysaccharide intercellular adhesin (PIA) synthesis, functioning of various metabolic pathways, resistance to antibacterial agents and autolysis which are all required for biofilm formation. This pathogen survives in the human host mostly as biofilms contributing resistance to several antibiotics making S. aureus as a multidrug resistant strainCitation11,Citation15–20. Further, in this organism, PknB is actively present in the external secretions, and along with phosphatases they interact with vast range of host tissue proteins aiding this pathogen to colonize in any anatomical locales in human hostCitation21,Citation22.
In S. aureus, the dephosphorylation is carried out by phosphatases which are present along with phosphokinases. There are four different alkaline phosphatases which play distinct role in the organism, Pho R and Pho P genes are involved in signal transduction, while SAOV 255 and SAOV 2675 genes are involved in the regulation of metabolic processCitation23. Thus, we predicted that in S. aureus IDH is probably regulated by phosphorylation and dephosphorylation. To establish this hypothesis, PknB, alkaline phosphatase (ALP) III (SAOV2675) and IDH genes from S. aureus ATCC12600 were cloned, expressed and characterized. Furthermore, the effect of phosphorylation and dephsophorylation on IDH activity was demonstrated.
Methods
Bacterial cultures
Staphylococcus aureus ATCC12600 was grown on modified Baird Parker agar. A black shiny colony with clear zone was picked and streaked on modified Baird Parker (BP) agar plate containing 0.001% phenolphthalein bisphosphate tetrasodium salt (Sigma) and 10% NaCl (w/v), as described by Barber and KuperCitation24. After incubation at 37 °C for 2 days, the pink-coloured colonies indicate the expression of phosphatases. Single colony from the above plate was inoculated in brain heart infusion (BHI) and Luria–Bertani (LB) broths. These cultures were grown overnight at 37 °C. After overnight incubation, the grown culture was subjected to isolation of chromosomal DNACitation25.
Biofilm assay
The staphylococcal biofilm assay was carried out as described earlierCitation14.
Alkaline phosphatase enzyme assay
The enzyme reaction mixture contained 0.1 M carbonate–bicarbonate buffer, pH 10, 5 mM p-Nitro phenyl phosphate and mixed thoroughly just before the incubation at 37 °C crude or pure ALP enzyme was added. The absorbance was measured at 405 nm against blank (carbonate–bicarbonate buffer), and the kinetic parameters Vmax and Km were calculated from the Hanes–Woolf plot [[S] versus ([S]/V)]Citation26.
IDH assay
Pure IDH assay was obtained from the cytosolic fraction of UVIDH1 clone, and the enzyme assay was carried out as described earlierCitation13.
Serine/threonine protein kinase (PknB) assay
PknB activity was determined at 30 °C using novel non-radiolabeled protein kinase spectrophotometric assay with synthetic peptide which acts as substrate in a Cyberlab spectrophotometer (Mill bury Town, MA). Serine/threonine protein kinase (PknB) assay mixture contained 0.1 M Tris-HCl at pH 7.5, 0.1 M ATP, 11.8 µM (30 µg/µl) peptide (stpks = NLCNIPCSALLSSDITASVNCAK), and 1 µg/µl enzyme fraction (pure His tag PknB) was mixed and incubated at 30 °C for 10 min. The phosphorylated peptide was purified by passing through Sephadex G-25 column (1 cm× 15 cm), the fractions were eluted with 0.1 M Tris-HCl pH 7.5 and 150 mM NaCl. The enzyme fraction appeared in the void volume, while phosphorylated peptide was obtained in the elution volume. The phosphate covalently bound to the proteins were estimated by adding freshly prepared reagent A: 3.4 mM of ammonium molybdate dissolved in 0.5 mM H2SO4, 10% SDS, 0.6 M l-ascorbic acid mixed in 6:1:1 (v/v/v) ratio and incubated at 30 °C for 15 min, and the absorbance was recorded at 820 nm against blank (0.1 M Tris-HCl, pH 7.5, 150 mM NaCl and reagent A)Citation27. The enzyme activity was measured as amount of phosphorous added per microgram peptide at 30 °C/min/ml. For this, the calibration curve was developed using standard KH2PO4 for the estimation of inorganic phosphate, and free phosphate was determined by adding reagent ACitation28. The phosphorylated peptide was further demonstrated by fractionating the eluted peptide in 12.5% SDS-PAGE. Upon staining the gel with reagent A, the blue-coloured band appeared in the gel indicating the peptide was phosphorylated by the enzyme fraction. Similarly, the auto-phosphorylation property of PknB was also determined for this, the reaction mixture composition was same, except stpks was not added. The enzyme activity was measured as amount of phosphorous added per microgram enzyme at 30 °C/min/ml. Substrate level phosphorylation was performed by taking different substrate concentrations of 10–120 µM of synthetic peptide keeping the ATP concentration constant, and the corresponding velocities were calculated and a graph of [[S] versus [S/V]] was plotted; from the graph obtained, Km and Vmax were determined. Similarly, the Km and Vmax for auto-phosphorylation of PknB was determined by the Hanes–Woolf plot. Protein concentrations in all steps were determined by the Bradford methodCitation29.
PknB gene amplification and sequencing
The PknB gene was amplified using the primers designed from the sequence of S. aureus USA 300 (). The PCR was performed using Eppendorf (Master gradient) thermocycler. The 50 -µl PCR reaction mixture contained 0.5 µg of chromosomal DNA, 100 µM of dNTPs mix, 100 pM of forward and reverse primers, 1× reaction buffer that contained MgCl2 supplied by the manufacturer (Merck Biosciences, Bangalore, India) and 1 U of Hotstart Taq DNA polymerase (Merck Biosciences). PCR was performed with the following conditions: initial denaturation at 94 °C for 10 min, and 40 cycles of denaturation at 94 °C for 60 s of denaturation, 32 °C for 45 s of annealing, 72 °C for 90 s amplification followed by final elongation at 72 °C for 10 min. The amplified PCR products were resolved in 1% agarose gel along with supermix DNA ladder (Merck Bioscience), and resolved DNA bands were recorded in Vilber Lourmat gel documentation system. The purified PknB PCR product was sequenced by dye terminating method at commercial sequencing facility of Xcelris Labs Ltd, Ahmadabad, India. The obtained sequences were analyzed and deposited at NCBI-Gen Bank (http://www.ncbi.nlm.nih.gov/nuccore/JN695616).
Table 1. Primers used for PCR amplification.
ALP gene amplification and sequencing
The ALP gene was amplified from the chromosomal DNA using the primers designed manually from the ALP gene sequence of S. aureus Mu 50 strainCitation30 (). The cocktail mixture contained 100 pM of each primer, 100 μM of dNTPS mix, Tris with 15 mM MgCl2, 1 U of hot start Taq DNA polymerase (Merck Bioscience Pvt. Ltd) and 0.5 µg chromosomal DNA. Thermocycler conditions included an initial denaturation step for 10 min at 94 °C; 35 cycles of each having denaturation at 94 °C for 60 s, annealing at 39 °C for 35 s and amplification at 72 °C for 90 s which was followed by a final extension step at 72 °C for 10 min. Amplicons were purified with NP-PCR purification kit (Taurus Scientific, Cincinnati, OH) and sequenced by chain terminating method at MWG Biotech India Ltd. Thus, obtained ALP gene sequence was deposited at Gen Bank (http://www.ncbi.nlm.nih.gov/nuccore/JN645811).
Multiple sequence alignment
The multiple sequence alignment was performed between S. aureus alp, Mycobacterium tuberculosis (NCBI ID: WP_006242595.1), Bacillus subtilis (NCBI ID: WP_009879253.1) Streptococcus (NCBI ID: YP_002746311) and Homo sapiens (NCBI ID: NP_000469.3) using ClustalX software. Similarly, using the same software, the multiple sequence alignment was performed between S. aureus PknB, M. tuberculosis (NCBI ID: NP_214528.1), B. subtilis (NCBI ID: NP_00748102.1), Streptococcus (NCBI ID: NP_720924.1) and H. sapiens (NCBI ID: AAQ_02456.1)Citation14.
Cloning and expression of PknB and alkaline phosphatase III genes
The amplified PCR products of 2 kb and 1.425 kb corresponding to PknB and alkaline phosphatase III genes were electroeluted from the agarose gel and were made into proper blunt ends using Klenow fragment (New England Bio Labs, Ipswich, MA) following the manufacturer’s protocol. The blunt-ended PCR products were cloned into the SmaI site of plasmid pQE-30 (QIAGEN, Valencia, CA) and were transformed into the E. coli DH5α. Thus, obtained clones were named as PV 1 and UVP-ALP3. These clones were induced with 0.1, 0.2, 0.5, 0.75, 1 and 1.5 mM IPTG and we found at 0.75 mM IPTG concentration PV 1 clone cells survived and expressed PknB similarly, at 1 mM IPTG concentration UVP-ALP3 cells survived, and optimum expression of alkaline phosphatase III was achieved. The recombinant enzymes were purified by passing the cytosolic fractions of PV1 and UVP-ALP3 clones through nickel metal chelate agarose column. The pure recombinant alp was analyzed in 10% SDS-PAGE, and kinetics of purified recombinant proteins were determined as described earlierCitation13,Citation31,Citation32.
In vitro regulation of IDH
Phosphorylation of IDH with PknB was performed in the assay mixture containing 0.1 M Tris-HCl, pH 7.5, 0.1 M ATP, 1 µg/ml pure IDH and 3 µg/ml pure PknB, and incubated at 30 °C for 10 min. The phosphorylated IDH was purified by passing through Sephadex G-25 column (GE Healthcare Bio-Sciences, Pittsburgh, PA) (1 cm× 15 cm), the fractions were eluted with 0.1 M Tris-HCl, pH 7.5 and 150 mM NaCl. The enzyme fractions appeared in the void volume, the bound phosphorous was estimated by adding freshly prepared reagent A and incubated at 30 °C for 15 min, and the absorbance was read at 820 nm against blank (0.1 M Tris-HCl, pH 7.5, 150 mM NaCl and reagent A). The phosphorylated IDH was used to carry out the enzyme assay as described earlier. Similarly, the phosphorylated IDH was incubated with pure alkaline phosphatase III enzyme, and this dephosphorylated IDH was used for enzyme assayCitation13.
Results
Cloning, sequencing and expression of ALP gene
In the present study, all phosphatase-producing colonies of S. aureus ATCC12600 exhibited pink colour, and from this culture the chromosomal DNA was extracted and used to amplify SAOV 2675 gene. The 1.425 kb PCR product was sequenced (JN645811) and the sequence when BLAST searched showed complete homology with (SAOV 2675) alkaline phosphatase III present in all the strains of S. aureus. This gene was cloned in the SmaI site of pQE 30 in “-1” frame and transformed into E. coli DH5α, the resultant clone UVP-ALP3. The expression of alkaline phosphatase III was induced with 1 mM IPTG, and the recombinant alp3 was purified from the cytosolic fraction of UVP-ALP3 by passing through Nickel metal chelate column. The enzyme kinetics of recombinant protein showed Vmax 4.703 ± 0.62 µM/mg/ml/min, Km 7.1 ± 0.2 mM and exhibited single band in SDS-PAGE with molecular weight of 52 kDa (; ). The annotated protein sequence of S. aureus (SAOV 2675) when scanned in PROSITE showed one active site that contains following amino acids 104I, 105T, 106D, 107S, 108A, 109A, 110G, 111G and 112T which are also conserved in human alkaline phosphatase (halp) (PDB ID: 2GLQ). Multiple sequence alignment using Clustal X showed very low-sequence identity with human and other bacterial alkaline phosphatase ().
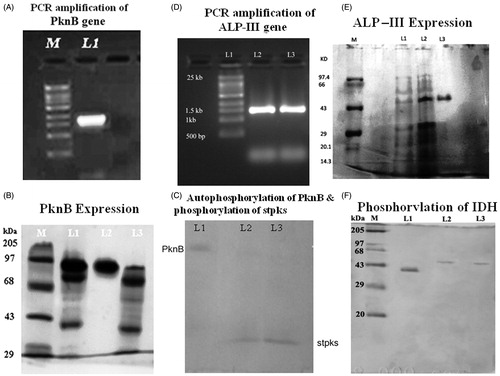
Figure 1. (A) Lane 1 PCR amplification of PknB gene (2.0 kb) from the chromosomal DNA of S. aureus ATCC12600, Lane M supermix DNA ladder obtained from Merck Bioscience Pvt. Ltd. (B) SDS-PAGE (10%) analysis of rPknB expression. Lane 1 cytosolic fraction of IPTG-induced PV1 clone, Lane 2 Nickel metal agarose column purified PknB and Lane 3 cytosolic fraction of uninduced PV1 clone and Lane M molecular size marker obtained from Merck Bioscience Pvt. Ltd. (C) Separation of pure phosphorylated PknB identified by reagent-A. L1: Pure phosphorylated PknB. L2 & L3: phosphorylated stpks. (D) Lanes 2 and 3 PCR amplification of alp gene (1.425 kb) from the chromosomal DNA of S. aureus ATCC12600, L1 supermix DNA ladder obtained from Merck Bioscience Pvt. Ltd. (E) SDS-PAGE (10%) analysis of rALP-III expression. Lane 1 uninduced cell lysate of UVPALP-3 clone, L2 IPTG-induced cell lysate of UVP-ALP3 clone and L3 pure rUVP-ALP3 eluted from nickel metal chelate agarose chromatographic column, and Lane M molecular size marker obtained from Merck Bioscience Pvt. Ltd. (F) Separation of pure phospharylated IDH identified by reagent-A. L1: Pure IDH. Lane M: Marker. L2 & L3: phosphorylated IDH.
Cloning, sequencing and expression of PknB gene
The gene encoding PknB (2.0 kb) was amplified from S. aureus ATCC 12600 chromosomal DNA and sequenced, and the sequence showed complete homology with PknB of several S. aureus strains in the reported databases. The sequence analysis of PknB enzyme consisting of catalytic domain was distributed between 10th and 267th residues which contain 12 specific Hanks motifs, they contained both ATP and substrate-binding regions which are also conserved in eukaryotic protein kinases. Transmembrane domain was followed by three different duplicated forms of PASTA domains: PASTA1 distributed between 377th and 440th residues, PASTA2 distributed between 445th and 508th residues, PASTA3 distributed between 514th and 577th residues in the annotated protein sequence of PknB gene, and between these two domains is a single transmembrane segment. These unique characters are the features exhibited by a several PknB enzymes expressed in different strains of S. aureus ().
The PknB gene was cloned in pQE 30 vector in “-1” frame and the cloned named as PV 1. The insert in the clone was confirmed by sequencing (JN695616); the PknB gene was expressed with 0.75 mM IPTG and purified by passing through nickel metal chelate column chromatography. The molecular weight of the pure rPknB was found to be 73 kDa, which corresponds to the inserted cloned and is equivalent to the monomeric form of PknB protein (). The enzyme kinetics of PknB is shown in . IDH of S. aureus ATCC 12600 was expressed from UVIDH 1 cloneCitation12, and the kinetics of IDH are shown in . The rate of S. aureus biofilm formation was higher in BHI broth compared to LB broth with decreased IDH activity ().
Table 2. Enzyme kinetics of rPknB and rALP III.
Table 3. Enzyme kinetics of S. aureus IDH grown in LB and BHI media.
Regulation of IDH by phosphorylation and dephosphorylation
The deduced amino acid sequence of IDH showed the presence of PknB site in the active site of enzyme where NADP binds and regulates the catalysis process (), IDH was phosphorylated with PknB and the bound phosphate was identified by its ability to react with reagent A, which was identified spectrophotometrically. On fractionating these phosphorylated enzymes in 10% SDS-PAGE and immersing in reagent A, appearance of blue-coloured bands indicated IDH was phosphorylated (). The mobility of phosphorylated IDH was lower than the native pure IDH (). The phosphorylated IDH lost its activity completely, and the activity was restored on treatment with ALP III (). Thus, it is clear that in S. aureus IDH is regulated by phosphorylation and dephosphorylation.
Table 4. IDH regulation by phosphorylation and dephosphorylation.
Discussion
Phosphorylation and dephosphorylation of proteins are the most conserved biochemical process used for intercellular and intracellular communication in both prokaryotes and eukaryotesCitation33. This plays crucial role in the regulation of various metabolic processes in the organism as well as expression of various virulence factors and by passing the host immune system, thus helping the pathogen in colonization in any anatomical locales in human host. This condition is further aided by the presence of secretary PknB and alkaline phosphataseCitation21–23.
In S. aureus cell wall, exopolysaccharide and PIA molecule biosynthesis are primarily regulated by PknB. In anaerobic conditions, increased PknB and PIA expression downregulates TCA cycle and purine biosynthesis in S. aureus, wherein it facilitates rapid biofilm formation. The results of the present study too concur with low-IDH activity and increased biofilm formation in anaerobic conditions (). Such features are commonly observed in all multi-drug resistant strains of S. aureus which also show high resistance to vancomycinCitation8,Citation11,Citation14–17.
In the present study we have cloned, sequenced, expressed and characterized PknB and alkaline phosphatase III (SAOV 2675) genes of S. aureus ATCC12600. The PknB sequence showed well-conserved PASTA domains which sense environmental signals by triggering eukaryote-like N-terminal catalytic domain to autophosphorylate and hence activate different downstream signalling cascades which help to regulate physiology of the organismCitation11,Citation15–17,Citation21,Citation34,Citation37. The results of the current study also explain unambiguously how IDH become completely inactive on phosphorylation with PknB and are restored upon dephosphorylation with alp III (). This shift in phosphorylation and dephosphorylation plays vital role in maintaining the redox status in S. aureus, which in turn has profound effect in the formation of biofilmsCitation11,Citation15–17,Citation35,Citation36,Citation38,Citation39 ().
Conclusion
In S. aureus, high-reductive condition coupled with increased expression of PknB is one of the prime reasons for increased rate of biofilm formation. In this study IDH enzyme expression was assessed in S. aureus ATCC12600 grown in anaerobic and aerobic media, which revealed decreased IDH activity and increased biofilm formation correlated with complete loss of IDH activity on phosphorylation with PknB. Thus, explaining in reductive conditions, biosynthesis phase is higher than energy generation leading to the increased cell wall, PIA and exopolysaccharide synthesis which facilitated increased rate of biofilm formation, which is one of the key pathogenic factors of S. aureus.
Declaration of interest
Authors report no conflict of interest. We sincerely acknowledge Sri Venkateswara Institute of Medical Sciences University for providing funds and facilities under SBAVP scheme (SBAVP/Ph. D/02) to carry out this work, and this paper is a part of Ph.D. work going to be submitted to SVIMS University.
References
- Steen IH, Madern D, Karlstrom M, et al. Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state and phylogenetic affiliation. J Biol Chem 2010;276:43924–31
- Hurley J, Thorsness PE, Ramalingam V, et al. Structure of a bacterial enzyme regulated by phosphorylation isocitrate dehydrogenase. Proc Natl Acad Sci USA 1989;86:8635–9
- Oudot C, Jaquinod M, Cortay JC, et al. The isocitrate dehydrogenase kinase/phosphatase from Escherichia coli is highly sensitive to in vitro oxidative conditions. Role of cysteine 67 and cysteine108 in the formation of a disulfide-bonded homodimer. Eur J Biochem 1999;262:224–9
- Zheng J, Jia Z. Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase. Nature 2010;465:961–5
- Caiazza NC, O'Toole GA. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J Bacteriol 2003;185:3214–17
- Somerville GA, Chaussee MS, Morgan CI, et al. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 2002;70:6373–82
- Somerville GA, Cockayne A, Dürr M, et al. Synthesis and deformylation of Staphylococcus aureus delta-toxin are linked to tricarboxylic acid cycle activity. J Bacteriol 2003;185:6686–94
- Cui L, Ma X, Sato K, et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol 2003;41:5–14
- Zhu Y, Xiong YQ, Sadykov MR, et al. Tricarboxylic acid cycle-dependent attenuation of Staphylococcus aureus in vivo virulence by selective inhibition of amino acid transport. Infect Immun 2009;77:4256–64
- Burnside K, Lembo A, de Los Reyes M, et al. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One 2010;5:e11071
- Tamber S, Schwartzman J, Cheung AL. Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA. Infect Immun 2010;78:3637–46
- Zhu Y, Weiss EC, Otto M, et al. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect Immun 2007;75:4219–26
- Prasad UV, Vasu D, Kumar YN, et al. Cloning, expression and characterization of NADP-dependent isocitrate dehydrogenase from Staphylococcus aureus. Appl Biochem Biotechnol 2013;169:862–9
- Yeswanth S, Nanda Kumar Y, Prasad UV, et al. Cloning and characterization of l-lactate dehydrogenase gene of Staphylococcus aureus. Anaerobe 2013;24:43–8
- Beltramini AM, Mukhopadhyay CD, Pancholi V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun 2009;77:1406–16
- Débarbouille M, Dramsi S, Dussurget O, et al. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol 2009;191:4070–81
- Donat S, Streker K, Schirmeister T, et al. Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J Bacteriol 2009;91:4056–69
- Foster TJ. The Staphylococcus aureus “superbug”. J Clin Invest 2004;114:1693–6
- Ippolito G, Leone S, Lauria FN, et al. Methicillin resistant Staphylococcus aureus: the superbug. Int J Infect Dis 2010;14:S7–11
- Liu Q, Fan J, Niu C, et al. The eukaryotic-type serine/threonine protein kinase STK is required for biofilm formation and virulence in Staphylococcus epidermidis. PLoS One 2011;6:e25380
- Miller M, Donat S, Rakette S, et al. Staphylococcal PknB as the first prokaryotic representative of the proline-directed kinases. PLoS One 2010;5:e9057
- Lowy FD. Staphylococcus aureus infections. N Engl J Med 1998;339:520–32
- Guinane CM, Ben Zakour NL, Tormo-Mas MA, et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol Evol 2010;2:454–66
- Barber M, Kuper SW. Identification of Staphylococcus pyogenes by the phosphatase reaction. J Pathol Bacteriol 1951;63:65–8
- Hari Prasad O, Nanda Kumar Y, Reddy OV, et al. Cloning, expression, purification and characterization of UMP kinase from Staphylococcus aureus. Protein J 2012;31:345–52
- Sarma PVGK, Prasad OH, Prasad UV, et al. Purification and characterization of human intestinal alkaline phosphatase and its role in the colonization of Helicobacter pylori in the duodenum. Curr Trends Biotechnol Pharm 2009;3:389–95
- Clore JN, Stillman J, Sugerman H. Glucose-6-phosphatase flux in vitro is increased in type 2 diabetes. Diabetes 2000;49:969–74
- Fiske CH, Subbarao Y. The colorimetric determination of phosphorous. Biol Chem 1925;66:375–400
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248e54
- Ohta T, Hirakawa H, Morikawa K, et al. Nucleotide substitutions in Staphylococcus aureus strains, Mu50, Mu3, and N315. DNA Res 2004;11:51–6
- Sambrook J, Russell DW. Molecular cloning. A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5
- Fischer EH, Krebs EG. Commentary on the phosphorylase b to a converting enzyme of rabbit skeletal muscle. Biochim Biophys Acta 1989;1000:297–301
- Ruggiero A, Squeglia F, Marasco D, et al. X-ray structural studies of the entire extracellular region of the serine/threonine kinase PrkC from Staphylococcus aureus. Biochem J 2011;435:33–41
- Vuong C, Kidder JB, Jacobson ER, et al. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J Bacteriol 2005;187:2967–73
- Cramton SE, Ulrich M, et al. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus eidermidis. Infect Immun 2001;69:4079–85
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 1988;241:42–52
- Islam N, Kim Y, Ross JM, Marten MR. Proteomic analysis of Staphylococcus aureus biofilm cells grown under physiologically relevant fluid shear stress conditions. Proteome Sci 2014;12:21. doi: 10.1186/1477-5956-12-21
- Prasad UV, Swarupa V, Yeswanth S, et al. Structural and functional analysis of Staphylococcus aureus NADP-dependent IDH and its comparison with bacterial and human ADP dependent IDH. Bioinformation 2014;10:81–6