Abstract
Candida albicans is one of the most common fungal pathogen in humans due to its high frequency as an opportunistic and pathogenic fungus causing superficial as well as invasive infections in immunocompromised patients. An understanding of gene function in C. albicans is necessary to study the molecular basis of its pathogenesis, virulence and drug resistance. Several manipulation techniques have been used for investigation of gene function in C. albicans, including gene disruption, controlled gene expression, protein tagging, gene reintegration, and overexpression. In this review, the main cassettes containing selectable markers used for gene manipulation in C. albicans are summarized; the advantages and limitations of these cassettes are discussed concerning the influences on the target gene expression and the virulence of the mutant strains.
Introduction
Candida albicans is one of the most common opportunistic pathogen in humans that causes a number of clinical diseases ranging from superficial infections to lifethreatening systemic disease in immunocompromised patients,Citation1 such as those infected with HIV, undergoing cancer chemotherapy and organ transplantation, as well as premature infants.Citation2,Citation3 This pathogenic fungus is becoming the leading cause of nosocomial infections.Citation4 In Kunming, southwest China, the oral yeast colonization rate in AIDS patients (49.5%) was higher than that of healthy people (20.7%).Citation5C. albicans constituted the most frequent species, accounting for 82.2% of yeast isolates and is the most common lethal species (48.8%) in patients with cancer,Citation5 endotracheal intubation or hypoproteinemia.Citation6 Azole antifungal agents, especially fluconazole (FLC), are used to treat candidiasis. However, the overuse and long-term treatment with FLC have resulted in the emergence of resistance in C. albicans.Citation7-Citation10 It is remaining largely mysterious toward the drug resistance mechanism and the pathways of virulence and pathogenesis in C. albicans.Citation11-Citation14
In recent years, the Candida Genome Database is established to offer the genome sequences of C. albicans for researchers to study freely.Citation15 Several gene manipulation techniques are available to allow powerful genetic approaches to study function of the genes in C. albicans, including gene disruption,Citation16-Citation19 controlled gene expression,Citation16,Citation20,Citation21 protein tagging, gene reintegration and overexpression.Citation16,Citation18,Citation22,Citation23 The principles of these gene manipulation techniques are all based on homologous recombination between the complementary sequences and the genomic sequences in C. albicans. But their applications and the use of selectable markers in these gene manipulation approaches are different. In this review, the main cassettes used for gene manipulation in C. albicans are summarized (; ); the advantages and limitations are discussed.
Figure 1. Cassettes used for gene manipulation in C. albicans. The figure shows the necessary elements in these cassettes, the light blue color of the elements represents the 5′ and 3′ DNA flanking your target gene (YTG) or the upstream and downstream sequences of YTG. (1) URA blaster cassette.Citation24 (2) PCR amplifiable marker cassettes from non-C. albicans Candida species.Citation45C.d.HIS1, Candida dubliniensis HIS1; C.m.LEU2, Candida maltosa LEU2; C.d.ARG4, Candida dubliniensis ARG4. (3) URA flipper cassette.Citation38 prom, promoter region given for gene; T, termination sequence of given gene. (4) Cre-loxP system.Citation46 (5) MPAR flipper cassette.Citation47 (6) SAT1 flipper cassette.Citation51C.a.SAT1, C. albicans SAT1. (7) “Tet-Off” system.Citation20 (8) “Tet-On”system.Citation54 (9) Promoter exchange cassette.Citation18,Citation60 (10) Cassettes used for gene reintegration.Citation34C.a.SAT1, C. albicans SAT1; C.a.LEU2, C. albicans LEU2; C.d.ARG4, C. dubliniensis. (11) ACT1P-GFP-ACT1T cassette.Citation76 (12) The firefly luciferase selectable marker cassette.Citation77 (13) PCR-mediated gene-tagging cassette.Citation16 (14) Epitope tagging cassette.Citation23
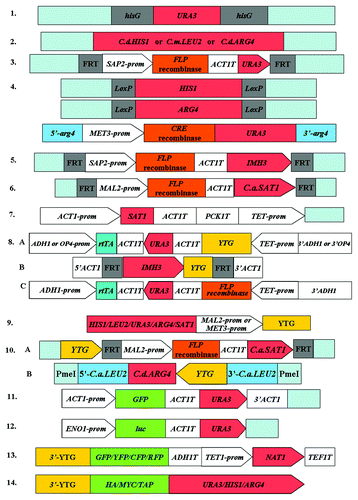
Table 1. Cassettes used for gene manipulation in C. albicans
Cassettes Used in Gene Disruption
The method of targeted gene deletion has enabled the identification of several unique aspects of C. albicans genes that play roles in pathogenesis, virulence, and resistance. Genes that are not essential for survival in C. albicans can be disrupted or replaced by transformation with exogenous DNA. These DNA cassettes used for gene disruption contain the selectable marker, auxotrophic or drug-resistant marker, flanked by sequences homologous to your target gene (YTG).
Gene disruption with nutritional marker for selection
URA blaster cassette
In C. albicans, the URA3 gene encodes the orotidine 5′-monophosphate (OMP) decarboxylase enzyme, catalyzing the conversion of OMP to uridine 5′-monophosphate in the de novo pyrimidine biosynthesis pathway.Citation24 The strain CAI4 (Ura−) () with two alleles’ deletion of URA3 is used as the parental strain. The URA blaster cassette carries two direct repeats of the hisG sequences from Salmonella Typhimurium flanking the URA3 gene (Fig. 1.1). After the strain CAI4 being transformed with the URA blaster cassette, the Ura+ transformants are selected on uracil-deficient medium. The two direct repeats of the hisG sequences can spontaneously recombine, and then loop out the URA3 gene and leave behind a short hisG sequence in the genome. Cells that have lost URA3 by homologous recombination can be selected on medium containing 5-fluoroorotic acid (5-FOA), because 5-FOA is toxic to Ura+ cells. After another round of transformation and homologous recombination, the cells that are null for the target gene can be generated ().Citation24,Citation25
Table 2. The commonly used auxotrophic strains in gene manipulation in C. albicans
Figure 2.URA blaster strategy used for gene disruption in C. albicans.Citation39 YTG, your target gene.
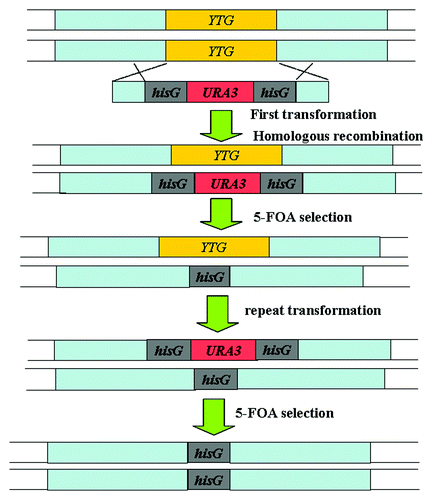
The URA blaster method is a classical and highly effective approach to disrupt the target gene in C. albicans. Several parental strains lacking additional nutritional markers such as RM1000 (Ura3− His1−) and BWP17 (Ura3− His1− Arg4−) () are constructed by this method.Citation25,Citation26 However, the URA blaster method has several shortcomings. First, a portion of the 3′ end of the IRO1 involving in iron utilization was removed during the construction of the parental strain CAI4 (Ura−), which activated the alternative mechanisms of iron uptake in CAI4. The growth of CAI4 is better than that of the wild-type SC5314 strain in an iron-restricted environment.Citation27-Citation29 Second, one copy of hisG sequences left in the chromosome decreases the integration frequency of the disruption cassette at the target locus because of the competition between the endogenous hisG sequences and the successive hisG cassettes in the second round of transformation.Citation30,Citation31 Third, the flanking direct repeats of this cassette integrated at the HWP1 locus can negatively influence the expression of the URA3 in C. albicans and affect the virulence phenotype of the hwp1 null mutants in C. albicans.Citation32 Finally, exposure to 5-FOA is potentially mutagenic and could introduce chromosomal rearrangements.Citation32,Citation33 Particularly, the copy number and the changed chromosomal location of the URA3 gene reduce capability of hyphal morphogenesis, adherence and virulence in C. albicans.Citation24,Citation29,Citation33 To ameliorate this problem, it is recommended to place the remaining URA3 copy at a highly expressed locus, including ENO1, RPS10, ARG4, or at the URA3 itself native locus.Citation29,Citation34-Citation37 Otherwise, researchers also improve the efficiency of the URA3 cassette and have applied the PCR amplifiable URA3 cassette (URA3-dpl200) and the UAU1 cassette (ura3Δ3′-ARG4-ura3Δ5′) to disrupt the target gene in C. albicans.Citation19,Citation26,Citation41,Citation42 The UAU1 cassette on the Tn7 transposon has been used successfully to disrupt several genes in C. albicans.Citation44
PCR amplifiable marker cassettes from non-C. albicans Candida species
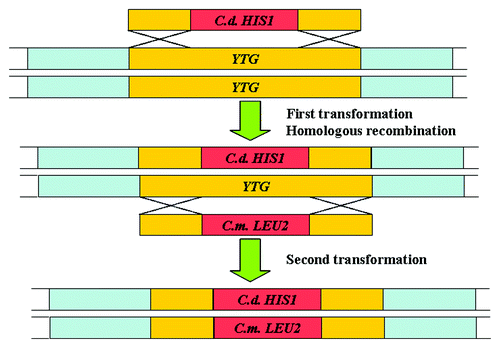
These cassettes contain Candida dubliniensis HIS1, Candida maltosa LEU2, or C. dubliniensis ARG4 as selectable markers flanked by two short repeat sequences homologous to the target gene in C. albicans (Fig. 1.2). The auxotrophic parental strains, including SN87 (Leu2− His1−), SN95 (His1− Arg4−), or SN152 (Leu2− His1− Arg4−) (), are transformed with these cassettes (). These cassettes have been successfully used for large-scale gene deletion studies in C. albicans. The parental strains used for these cassettes display no karyotypic changes and have one copy of URA3 expressed at the native locus, which don’t affect the virulence in C. albicans. So far, there is no evidence showing that the ectopic expression of any of the above nutritional markers would affect the virulence in C. albicans.Citation41,Citation45 However, these cassettes cannot be used to disrupt the genes in the wild-type strains.
Figure 3. Strategy of the fusion PCR and heterologous markers used for gene disruption in C. albicans.Citation45 YTG, your target gene. C. d, Candida dubliniensis; C. m., Candida maltosa.
URA flipper cassette
The URA flipper cassette contains two direct repeats of the minimal FLP recombination target (FRT) sequence flanking the URA3 marker. The site-specific recombinase FLP gene is regulated by secreted aspartyl proteinase (SAP2) promoter (Fig. 1.3). After the insertion of URA3 flipper into the target locus in the parental strain CAI4 (Ura−) (), the transformants are grown in the yeast carbon base and bovine serum albumin (YCB-BSA; pH 4.0) medium () to induce the expression of FLP recombinase which promotes the homologous recombination between the FRT sites to loop out the URA3 gene, making the URA3 marker recyclable for another round of transformation.Citation38
Table 3. The commonly used regulatable promoters in gene manipulation in C. albicans
Compared with the excision of the URA3 gene by spontaneous recombination between the hisG repeats, the FLP-mediated recombination between the FRT sites to loop out the URA3 marker is more efficient.Citation39 The URA flipper method does not need to use 5-FOA to select the ura3− clones, which would avoid mutagenic potentiality. However, this cassette leaves behind a single FRT sequence in the genome after the excision of URA3 marker. The multiple FRT sequences left in the genome might recombine induced by the FLP recombinase, resulting in unexpected deletions or chromosome rearrangements.Citation30,Citation40 The generation of ura3− clones would exclude the possibility of interchromosomal recombination.Citation38,Citation41 Otherwise, the use of the BSA activates the SAP2 promoter to induce the expression of FLP recombinase in the URA flipper cassette, whereas the BSA also represses the expression of other genes of SAP family except for SAP2 gene.Citation38,Citation41
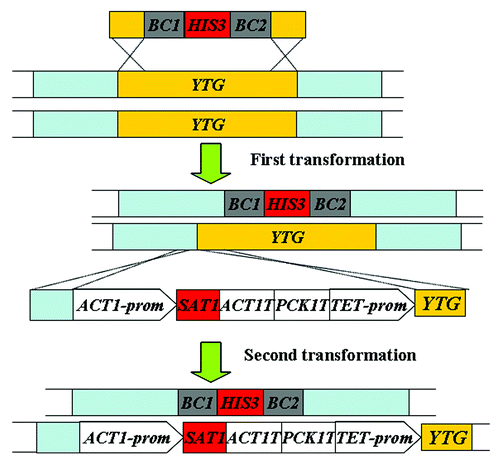
Cre-loxP system
This system is based on the site-specific recombination between two loxP elements which are catalyzed by the Cre recombinase. This system contains three components, including the disruption cassettes (using of HIS1 and ARG4 as auxotrophic markers flanked by 34 bp loxP elements), the resolving cassette (containing a synthetic, codon-optimized cre gene regulated by MET3 promoter and the reintegration URA3 marker gene) (Fig. 1.4). After two separate transformations, the two alleles of the target gene in parent strain BWP17 (Ura3− His1− Arg4−) () are disrupted by HIS1 and ARG4 markers; then these transformants are selected on medium lacking of histidine during the first transformation or histidine/arginine during the second transformation, but containing methionine and cysteine to repress the expression of the MET3-cre fusion ().Citation46 In the third transformation, the segment of the Cre recombinase and the URA3 marker flanked by ARG4 sequences is integrated into the ARG4 gene that has disrupted one copy of the target gene in C. albicans. Then, the transformants that are auxotrophic for HIS1, ARG4, and URA3 were incubated in medium lacking of methionine and cysteine, which can induce the synthetic MET3-cre fusion to produce Cre recombinase and subsequently catalyze the recombination between loxP sites to loop out the HIS1 and the Cre recombinase-URA3 fragment ().Citation46
Figure 4. Strategy of the Cre-loxP system used for gene disruption in C. albicans.Citation46 YTG, your target gene; prom, promoter region given for gene.
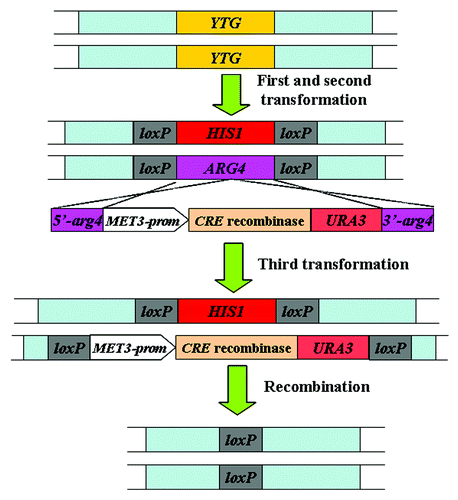
The Cre-loxP system doesn’t need to use mutagen 5′-FOA to select the transformants. The synthetic MET3-cre fusion of this system to loop out the LoxP-marker-LoxP cassettes in C. albicans is efficient and avoids using positive screen method to select for the desired mutants. The knockout strains generated by the Cre-loxP system are auxotrophic, unlike those by the positive selection markers.Citation41,Citation46 Moreover, the future uses of Cre-loxP system in C. albicans are not limited to gene disruptions; it can be used in the tagging of multiple ORF in situ. However, the Cre-mediated recombination of this system might occur in the absence of explicit MET3-cre fusion induction, which will reduce the stability of cre-containing strains. This method leaves a short loxP sequence in the genome after excision of markers. The multiple loxP sequences left in the genome might also recombine induced by the Cre recombinase, which results in unexpected deletions or chromosome rearrangements.Citation30,Citation40
Gene disruption with drug-resistant marker for selection
Mycophenolic acid-resistant (MPAR) flipper cassette
Using the drug-resistant markers, genes of C. albicans can be disrupted in any strains even in clinical isolates. It would be more benefit by using of the drug-resistant marker to study the virulence mechanism of the target genes in C. albicans than using the nutritional markers which might affect the pathways status for the genes of interest.
The inosine monophosphate dehydrogenase (Imh3) directs de novo synthesis of GMP in C. albicans. The mycophenolic acid (MPA) can inhibit the activity of Imh3. The transformants overexpressing IMH3 due to the mutated IMH3 gene are more resistant to MPA. The MPAR flipper cassette contains the mutated IMH3 gene and two FRT direct repeat sequences flanking the genetically engineered FLP gene regulated by SAP2 promoter (Fig. 1.5). After the first transformation, the flanking sequences directly integrate the cassette into the target locus. The transformants are grown in the YCB-BSA medium to activate the SAP2 promoter () which promotes the expression of the FLP gene. Then the FLP recombinase mediates site-specific recombination between FRT sites to loop out the MPAR flipper cassette and makes IMH3 marker recyclable for another round of transformation.Citation47-Citation49
This MPAR flipper method has been successfully used to study the causal relationships between specific genes and drug resistance in clinical isolates.Citation44,Citation47-Citation51 However, this cassette leaves behind a short FRT sequence in the genome after the excision of the MPAR flipper cassette.Citation47,Citation50 Furthermore, It is time-consuming to screen the MPAR flipper cassette integrated at the target site of the MPAR transformants.Citation41,Citation47
SAT1 flipper cassette
This cassette consists of a nourseothricin resistance marker C. albicans SAT1 gene and the FLP-mediated recyclable marker system regulated by the MAL2 promoter (Fig. 1.6). After the first transformation, the flanking sequences direct integration of the cassette into the target locus. The resistant transformants are picked on medium containing nourseothricin. Then, the correct transformants are grown in medium containing either maltose or glucose to active the MAL2 promoter () because the MAL2 promoter contained in the SAT1 flipper cassette is leaky, resulting in the expression of FLP recombinase to induce the homologous recombination between FRT sites and the excision of the FLP recombinase modulate system including the SAT1 marker. The medium containing a low concentration of nourseothricin can be used to select the nourseothricin-sensitive (NouS) mutants. After another round of transformation and selection, the desired homozygous mutants can be generated ().Citation41,Citation51-Citation53
Figure 5. The SAT1 flipper strategy used for gene disruption in C. albicans.Citation51 YTG, your target gene; prom, promoter region given for gene; T, termination sequence of given gene.
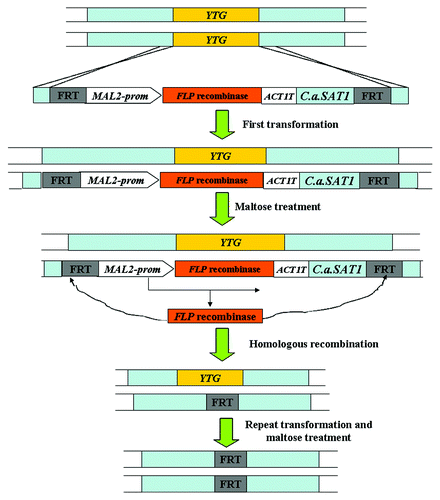
Compared with the MPAR flipper cassette, the integration of the SAT1 cassette into the correct locus occurs with high specificity.Citation52,Citation54 Otherwise, this SAT1 flipper cassette can also be used to reintegrate the intact copy of the target gene into the genome of the null mutant to complement the phenotypes of the mutant.Citation51 However, this cassette leaves a short FRT sequence in the genome after the excision of the marker.
Cassettes Used in Controlled Gene Expression
Usually the inability to obtain homozygous mutants is considered that the target gene might be essential in C. albicans. To test this, one copy of the target gene can be constructed under a promoter and the other allele is disrupted or replaced. The essentiality of the gene is established by measuring survival of C. albicans after a shift to repressive conditions.
“Tet-Off” system
The “Tet-Off” system combines gene replacement and conditional expression (GRACE) to assess gene essentiality. This system contains a chimeric transactivator protein and a tetracycline responsive promoter system. In the first step, the precise gene replacement of one allele of the target gene in the parental strain CaSS1Citation20 (Ura3− His3−) () is made by a cassette containing the HIS3 selectable marker, these transformants were selected on YNB medium to obtain the HIS3 prototrophs which can be used to additional transformation. In the second step, the controllable expression of the remaining allele is made by a Tet regulatable promoter system using a codon optimized SAT1 selectable maker which is driven by the ACT1 promoter (). The desired transformants can be obtained by selecting on medium containing nourseothricin. The transactivator protein binding to the Tet promoter results in constitutive expression of the target gene regulated by the Tet promoter system in the absence of tetracycline. In contrast, in the presence of tetracycline, the association between the transactivator protein and the Tet responsive promoter is disrupted, which leads to repressing of the expression of the target gene ().Citation20
Figure 6. Strategy of the “Tet-Off” system used to repress the expression of the target gene in the presence of tetracycline.Citation20 YTG, your target gene; prom, promoter region given for gene; T, termination sequence of given gene; BC1, up tag; BC2, down tag.
In the GRACE strains, the transactivator and the URA3 marker gene are integrated into the LEU2 locus. Therefore, GRACE strains can be forced to lose the transactivator and the URA3 marker gene on 5-FOA-containing medium. Thus strains that are unable to survive in the medium containing 5-FOA may be identified as those carrying essential gene.Citation20 Furthermore, the strains obtained by this method have been used for high throughput drug screening. However, this method is less suitable for inducing the expression of specific target genes under conditions where they are normally not expressed, because this system is not always feasible to efficiently remove the tetracycline from cells grown in the presence of the drug environment.Citation20,Citation54
“Tet-On” system
The “Tet-On” system contains different elements and can be applied to induce or repress genes in C. albicans. It permits to induce expression of genes in conditions where they are normally not expressed in the wild-type genome. In such case, the Tet-On system contains reverse tetracycline-controlled transactivator (rtTA) regulated by ADH1 or OP4 promoter, the URA3 or SAT1 gene, the target gene regulated by the rtTA-dependent promoter, and the flanking ADH1 sequences (Fig. 1.8A). The cassette would integrate into one copy of the ADH1 in a single transformation step. In the presence of the doxycyline, the binding of the rtTA to the Tet responsive promoter leads to the expression of the target gene.
Tet-inducible gene expression system is also useful to induce the deletion of the essential genes and to create conditional lethal C. albicans mutants. In such case, the “Tet-On” system contains rtTA regulated by ADH1 promoter, the URA3 gene, and the FLP recombinase gene regulated by the rtTA-dependent promoter (Fig. 1.8C). Before using this inducible gene deletion system, the parental strain CAI4 (Ura3−) () is transformed with the URA flipper cassette (Fig. 1.3) to delete one allele of target gene. Then one copy of target gene is reintegrated into the ACT1 gene locus by a cassette that contains the target gene and the MPAR marker flanked by FRT sites (Fig. 1.8B).Citation55 This is followed by the disruption of the remaining allele of target gene by the URA flipper cassette. Subsequently, the “Tet-On” cassette is transformed to integrate into ADH1 locus ().Citation54,Citation55 Providing doxycycline for the cells, the binding of the rtTA to the Tet responsive promoter results in the expression of the recombinase FLP gene, which induces the homologous recombination between the FRT sites to excise the target gene along with the MPAR marker (). The mutants losing the target gene can be identified by the gain of MPA susceptibility. On the contrary, cells remain MPA resistance in the absence of doxycycline, demonstrating that the FLP recombinase is not expressed and that no excision occurs.
Figure 7. Strategy of the “Tet-On” system used to induce the deletion of essential genes.Citation54 YTG, your target gene; prom, promoter region given for gene; T, termination sequence of given gene.
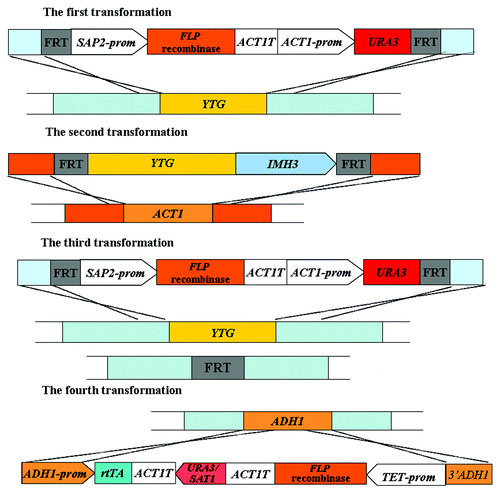
This method does not depend on nutrient changes in the medium but simply supplies an inducible or repressible substance which does not affect metabolism. In contrast to the “Tet-Off” system, the “Tet-On” system also allows an inducer to express the genes of interest in conditions. However, the “Tet-On” system requires higher concentration of doxycycline to induce gene expression.Citation54 Due to the doxycycline is cytotoxic,Citation56 its concentration should be controlled. And dimethyl sulfoxide can be used to enhance doxycycline-dependent protein expression in Tet-On cells.Citation57 Furthermore, the chelation between the doxycycline and iron interferes with iron homeostasis and thus reduces resistance to FLC in C. albicans.Citation58 Otherwise, this system requires more transformation steps to disrupt two copies of the target gene than the “Tet-Off” system does.Citation54
Promoter exchange cassette
The regulatable MAL2 or MET3 promoter following one marker gene (Fig. 1.9) has been successfully used to regulate the expression of target gene in C. albicans heterozygous strains. Marker genes include ARG4, CaHIS1, CdHIS1, CmLEU2, SAT1, and URA3. The strains BWP17 (Ura3− His1− Arg4−), SN148 (Ura− His− Leu− Arg−), and SN152 (His− Leu− Arg−) () are usually used as parental strains. The promoter exchange cassette carries its 5′ end flanking sequences homologous to the non-coding upstream sequences of the target gene and its 3′ end flanking sequences homologous to the starting sequences of the target ORF. After the transformation, the cassette integrates into the genome of heterozygous strain, where the MAL2 or MET3 promoter is right before the target gene. The methionine and cysteine would shut off the MET3 promoter and repress the target gene (). Since the MAL2 promoter is shut down by glucose and activated by maltose, glucose-containing medium is used to repress the target gene (). These promoter exchange modules provide rapid way to study the gene function in C. albicans. Furthermore, this method does not recycle marker gene and is very convenient.Citation18,Citation59,Citation60 In addition, the PG1G1-GFP cassette can be induced by N-acetylglucosamine (GlcNAc) () to express the GFP.Citation61
Cassettes Used for Gene Reintegration and Overexpression
In the process of constructing gene deletion mutants in C. albicans, the ectopic expression of selectable marker can mislead phenotypes of the null mutants.Citation35,Citation62,Citation63 It is necessary to reintegrate one copy of the intact target gene into the null mutant to restore the wild-type phenotypes. Several integrating vectors such as CIp10, CIp20, and CIp30 have been used efficiently for generating the complemented strains.Citation46 These plasmids carrying the HIS1, URA3, and ARG4 markers are usually used for reintegrating the markers or target genes into the RP10 locus of the auxotrophic strains.Citation34 Otherwise, the SAT1 flipper cassette containing the complete ORF as well as upstream and downstream flanking sequences of the target gene can also be applied to reintegrate one allele of target gene into the native locus of the homozygous mutants (Fig. 1.10A).Citation51 Furthermore, the technique of the PCR-directed recombination in S. cerevisiae can be used to construct the integrating cassette which reintegrates one copy of the intact target gene into the LEU2 locus of the null mutants in C. albicans (). The homologous recombination of the PCR fragments and the linearized pRS316 vector in S. cerevisiae can be constructed as the integrating cassette containing (5′ to 3′) a PmeI restriction site, the upstream sequences of LEU2 ORF in C. albicans, the ARG4 gene of C. dubliniensis (used as a selectable marker), the intact target gene ORF plus its promoter sequences, the downstream sequences of the LEU2 ORF in C. albicans, and another PmeI site (Fig. 1.10B).Citation64-Citation66 Meanwhile, the specific gene overexpression strains created by using of the highly active promoter to replace the endogenous promoter of the target gene is important as well for studying the gene function in C. albicans. The highly active promoters such as NAD-linked glyceraldehyde-3-phosphate dehydrogenase (TDH3) promoter, PEP carboxykinase (PCK1) promoter, and hexosaminidase (HEX1) promoter are often fused with the ORF of target gene to promote the expression of the target gene in C. albicans.Citation65,Citation67,Citation68
Cassettes Used in Protein Tagging
The molecular techniques of epitope tags are usually applied to protein purification, localization and detection.Citation23,Citation69-Citation72 The cassettes used for tagging proteins with yellow, cyan, and green fluorescent proteins (YFP, CFP, and GFP) at C-terminus has been applied to analyze the expression and localization of target protein within living cells in C. albicans.Citation18,Citation73 Tagging at the N-terminus usually replace the native promoter with a regulatable or constitutive promoter cassette.Citation74 The small epitopes poly-His, V5, GST, HA, MYC, and FLAG are applied to research the assemble process of target protein complexes in C. albicans ().Citation18,Citation23,Citation75
Table 4. The commonly used tagging in C. albicans
ACT1P-GFP-ACT1T cassette
This cassette contains GFP gene regulated by ACT1 promoter and URA3 used for selectable marker (Fig. 1.11). The flanking sequences of the cassette insert into the target locus through homologous recombination. Then the GFP gene of the integrative transformants exhibits a homogeneous, constitutive fluorescent phenotype, which can be applied to study the gene expression and evaluate the gene activation during pathogen–host cell interactions. The major advantage of this cassette is that gene induction can be monitored in a single cell of living organisms.Citation76
The firefly luciferase selectable marker cassette
This cassette contains the firefly luciferase gene regulated by ENO1 promoter and the URA3 gene as a selectable marker gene (Fig. 1.12). The cassette inserts into the 5′ end of one allele of the target gene in C. albicans through homologous recombination. Then the expression of the firefly luciferase increases the light emission of the transformed colonies. The major feature of this cassette is that it can be used as a sensitive reporter to analyze gene function both in laboratory and clinical isolates of C. albicans. Furthermore, the expression of luciferase might be used as a selection strategy to introduce other genes into C. albicans cells to investigate the effect of these genes in the cells. Otherwise, the strains that constructed by this cassette can be used in animal infection models to monitor the location of the infection by the virtue of their bioluminescence.Citation77,Citation78
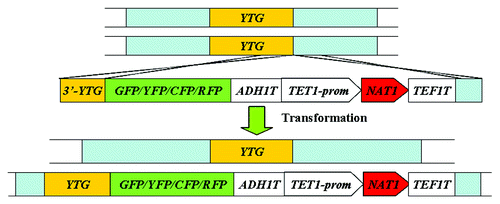
PCR-mediated gene-tagging cassette
The PCR-mediated gene-tagging cassette is based on combing the RFP, CFP, YFP, and GFP variants with the nourseothricin resistance marker CaNAT1 regulated by the heterologous TEF1 promoter and terminus (Fig. 1.13). The parental strain CAI4 (Ura−) or NGY152 (Ura−) () is transformed with this cassette which inserts into the 3′ end of the target gene directly. Then the strains grow in selective medium to select for nourseothricin-resistant transformants ().Citation16 The major feature of this cassette is to be applied to study protein functional in C. albicans. Furthermore, this cassette requires the using of lower concentrations of nourseothricin. However, similar to the use of the SAT1 marker, the expression of the CaNAT1 marker takes time.Citation16
Figure 8. Strategy of protein tagging at the C-terminus of the target gene used in C. albicans.Citation16 YTG, your target gene; prom, promoter region given for gene; T, termination sequence of given gene.
Epitope tagging cassette
The construct for in vivo protein tagging in C. albicans combines the selectable marker genes such as URA3, HIS1, and ARG4 with the protein tagging of TAP, HA, and MYC (Fig. 1.14) by PCR-mediated homologous recombination.Citation23 The insertion of epitope tags enables detection of target gene by immunodetection. The use of the tagged proteins is to perform chromatin immunoprecipitation coupled with microarray analysis (ChIP-ChIP) or tandem affinity purification (TAP). The MYC and HA epitope tags can be used for co-immunoprecipitation (co-IP) or immunoprecipitation (IP) experiments to validate formation of protein complex, while the TAP tag is often applied to obtain high purified protein samples for mass spectrometry analysis of protein complexes. In such case, the strain BWP17 (Ura3− His1− Arg4−) or SN76 (Ura− His1− Arg−) () can be used as parental strain. After the transformation, the tag is integrated into 3′ terminus of one allele of the target gene. This PCR tagging cassettes allow the rapid biochemical analysis of tagged proteins and have been successfully used to analyze the genome-wide location in C. albicans.Citation23,Citation79
Conclusion
Nowadays, several cassettes have been used to manipulate the C. albicans genome to study the function of the genes. The URA blaster cassette is the first used method. However, the expression of selectable auxotrophic marker URA3 affects the virulence, adhesion, and morphogenesis in C. albicans. The ura3 null mutant strains such as CAI4, RM1000, and BWP17 are deficient in virulence.Citation24,Citation29,Citation32,Citation33,Citation63,Citation80,Citation81 The ectopic expression of URA3 results in 30% of published virulence-deficient mutants having misattributed to the deletion of genes.Citation29 To overcome these problems, researchers proposed other solutions which are the reason that generated many other gene manipulation cassettes, such as the PCR amplifiable markers from non-C. albicans Candida species. This method has reintegrated the URA3 gene into its native locus, so the constructed strains by this method have full virulence such as SN95 (His− Arg−), SN87 (His− Leu−), SN100 (His−), and SN152 (His− Leu− Arg−) ().Citation45 Among the use of URA3 as a selectable marker for gene deletion, the UAU1 cassette is the prominent method, because it is useful to create C. albicans null mutants with a single transformation step without leaving behind any chromosomal rearrangements. Nevertheless, the use of ARG4, LEU2, and HIS1 as selectable markers for deleting the gene of interest in C. albicans is the best way among avoid using URA3 as nutritional selectable marker, because this method needs two transformation steps to create C. albicans null mutants and doesn’t need additional steps to excise the markers. So far, there is no evidence showing that the ectopic expression of the remaining nutritional markers LEU2, HIS1, or ARG4 affects the virulence of C. albicans. However, the auxotrophic markers in laboratory strains can still be restrictive because of the target genes are in pathways affected by nutritional status. Therefore, the nutritional markers don’t apply to analysis clinical C. albicans isolates which are not auxotrophic directly. In such case, drug-resistant markers, such as MPAR flipper cassette and SAT1 flipper cassette, can be applied to clinical isolates without creating any auxotrophic mutations. Moreover, if the target gene has three alleles in the parental strains such as CAI4 (Ura−) and SGY-243 (Ura−) which carry three copies of chromosome 1 (Chr 1),Citation82,Citation83 researchers should apply recyclable markers, including URA blaster and drug-resistant markers, and reuse three cycles to create knockout strains, because disrupting triploid with two different auxotrophic markers will not succeed in acquiring homozygous mutants.Citation84 Furthermore, the “Tet-On” system and “Tet-Off” system can be both used to conditionally control the essential genes expression in C. albicans. Otherwise, the protein tagging cassettes are used to locate the target protein or analyze the target protein complex. Therefore, researchers should pay attention to select the cassette fitting for their purpose of research.
On the other hand, researchers should keep an eye on the chromosomal aneuploidy in C. albicans mutants. It is reported that approximately 35% of over 100 published microarray data sets of C. albicans mutants have chromosomal aneuploidy.Citation85 Aneuploidy can influence the virulence of C. albicans.Citation82 For example, the commonly used laboratory strains CAI4 has three alleles of Chr 1, as well as BWP17 and RM1000 which shows a heterozygous deletion in the right arm of Chr 5.Citation86 However, the strains SN76, SN95, SN152, and SN148 are initially free of aneuploidies.Citation87 There are many factors leading to aneuploidies of strains. The strains grown in l-sorbose can often lead to lose a copy of Chr 5 while grown in d-arabinose may result in Chr 6 trisomy.Citation88,Citation89 In addition, the strains exposed to 5-FOA can lead to lose one copy of Chr 1.Citation82 The genome of C. albicans has remarkable tolerance for a wide variety of different chromosomal aneuploidies and changes in chromosomal copy numbers often arise as a response to stress.Citation85,Citation87,Citation90-Citation93 Therefore, C. albicans mutants should be tested for aneuploidy before being used in further studies.
In general, the perfect cassette of gene manipulation should be that the expression of selectable markers do not affect the virulence of strains, the constructed parent strains have no karyotypic changes, as well as the use of reagents do not generate the potential mutant.
To modify and improve the available methods is the first way to conceive the perfect method. Gene expression at an ectopic site is lower relative to that at the native locus, which could lead to virulence defect and growth defects.Citation94-Citation96 However, Gerami-Nejad et al.Citation97 have identified a neutral intergenic region NEUT5L, which facilitates the integration and expression of ectopic genes. They constructed a series of integrated shuttle vectors containing 550 bp sequences homologous to NEUT5L and three selectable markers (NAT1, the recyclable URA3-dpl200 or URA3). Ectopic genes integrated at NEUT5L by these vectors do not influence growth rates and allow native-locus expression levels. Thus the NEUT5L is an ideal genomic locus for the integration of exogenous DNA in C. albicans. Otherwise, Lai et al.Citation98 have modified the vector pTET25 into the pTET25M, so that the URA3 gene flanked by dpl200 can be used repetitively. The pTET25M vectors not only allow ectopic expression of target proteins in a “Tet-On” system with either a C-terminal 6× His epitope or N-terminal or a C-terminal GFP tag, but also possess a Ura-blaster cassette to allow reintroducing a URA3 marker.
The second way is to optimize and apply the gene manipulation methods used in other fungal species. For example, RNA interference techniques are used to disrupt the target genes in other fungal species such as Aspergillus fumigatus, Cryptococcus neoformans, and Aspergillus nidulans. Nowadays, this method is also proving to be successful to disrupt the gene of interest in C. albicans.Citation99 Moreover, Vieira et al.Citation100 have constructed three C. albicans integrative vectors such as CIp10, CIp20, and CIp30 which are ligated to 2 µ S. cerevisiae sequences, so that they are able to in vivo maintain and clone by gap repair within S. cerevisiae. These vectors are especially useful for the integration of genes into C. albicans genome that cannot be reproduced in Escherichia coli because of their toxic effects. Furthermore, Su-Kim et al.Citation101 have adopted double-joint PCR with NAT-split markers to construct the gene disruption cassettes in C. neoformans, which generates higher targeted-integration frequency. The NAT-split marker is referred as the two portions of DNAs of the NAT dominated selectable marker containing 200bp overlapping truncations in between, named the 5′-NAT-split marker and the 3′-NAT-split marker. During the double-joint PCR, the 5′-flanking region of target gene joints with the 5′-NAT-split marker and the 3′-flanking region of target gene joints with the 3′-NAT-split marker. Then, the two double-joint PCR fragments are combined and are introduced into the strains. To obtain the desired deletion mutants in C. neoformans, researchers only have to screen a small number of transformants.
More recently, the transcription activator-like effector nucleases (TALENs) which contain a modular DNA-binding domain and a FokI endonuclease monomer, are fusion proteins and work in pairs. When two TALENs bind to their target DNA sequences, the FokI monomers will dimerize and introduce a double-strand DNA breaking within the specific binding site. Then the disrupted DNA can either be repaired by non-homologous endjoining (NHEJ) or homologous recombination, which leads to deletion/insertion mutations, specific site mutations or specific sequence additions. This technology have been successfully used for gene manipulation in zebrafish, Drosophila, Arabidopsis, mice, human cell lines, and so on.Citation102-Citation105 Otherwise, in the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system, the targeting CRISPR RNA (crRNA) fused with the trans-activating crRNA can generate double stranded DNA breaks (DSDBs) in the target site. Then the DSDBs can be repaired either through NHEJ or through homologous recombination, which would result in random deletions or insertions. This system has also been used for genome editing in human cell lines, mouse, zebrafish, and Caenorhabditis elegans, and so on.Citation106-Citation109
In summary, there are many other novel gene manipulation strategies that should be considered for future investigations in C. albicans. We should make full use of our resources to improve the method used in gene manipulation in C. albicans.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
L. Yan is supported by Natural Science Foundation of China (31000079). Y. Jiang is supported by China National 973 Program (2013CB531602) and Natural Science Foundation of China (81330083). Y. Cao is supported by Natural Science Foundation of China (81173100). The authors would like to apologize to all researchers whose important works were unable to be cited because of space limitations.
References
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol 2007; 45:321 - 46; http://dx.doi.org/10.1080/13693780701218689; PMID: 17510856
- Schelenz S, Abdallah S, Gray G, Stubbings H, Gow I, Baker P, Hunter PR. Epidemiology of oral yeast colonization and infection in patients with hematological malignancies, head neck and solid tumors. J Oral Pathol Med 2011; 40:83 - 9; http://dx.doi.org/10.1111/j.1600-0714.2010.00937.x; PMID: 20923440
- Epstein JB, Hancock PJ, Nantel S. Oral candidiasis in hematopoietic cell transplantation patients: an outcome-based analysis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2003; 96:154 - 63; http://dx.doi.org/10.1016/S1079-2104(03)00296-8; PMID: 12931087
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 2007; 20:133 - 63; http://dx.doi.org/10.1128/CMR.00029-06; PMID: 17223626
- Li YY, Chen WY, Li X, Li HB, Li HQ, Wang L, He L, Yang XP, Wang XC, Huang YL, et al. Asymptomatic oral yeast carriage and antifungal susceptibility profile of HIV-infected patients in Kunming, Yunnan Province of China. BMC Infect Dis 2013; 13:46; http://dx.doi.org/10.1186/1471-2334-13-46; PMID: 23356471
- Zhang XB, Yu SJ, Yu JX, Gong YL, Feng W, Sun FJ. Retrospective analysis of epidemiology and prognostic factors for candidemia at a hospital in China, 2000-2009. Jpn J Infect Dis 2012; 65:510 - 5; http://dx.doi.org/10.7883/yoken.65.510; PMID: 23183203
- De Rosa FG, Garazzino S, Pasero D, Di Perri G, Ranieri VM. Invasive candidiasis and candidemia: new guidelines. Minerva Anestesiol 2009; 75:453 - 8; PMID: 19078900
- Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol 2008; 6:187 - 98; http://dx.doi.org/10.1038/nrmicro1835; PMID: 18246082
- Lai CC, Tan CK, Huang YT, Shao PL, Hsueh PR. Current challenges in the management of invasive fungal infections. J Infect Chemother 2008; 14:77 - 85; http://dx.doi.org/10.1007/s10156-007-0595-7; PMID: 18622668
- Cannon RD, Lamping E, Holmes AR, Niimi K, Tanabe K, Niimi M, Monk BC. Candida albicans drug resistance another way to cope with stress. Microbiology 2007; 153:3211 - 7; http://dx.doi.org/10.1099/mic.0.2007/010405-0; PMID: 17906120
- Barnett JA. A history of research on yeasts 12: medical yeasts part 1, Candida albicans. Yeast 2008; 25:385 - 417; http://dx.doi.org/10.1002/yea.1595; PMID: 18509848
- Noble SM, Johnson AD. Genetics of Candida albicans, a diploid human fungal pathogen. Annu Rev Genet 2007; 41:193 - 211; http://dx.doi.org/10.1146/annurev.genet.41.042007.170146; PMID: 17614788
- Magee PT, Gale C, Berman J, Davis D. Molecular genetic and genomic approaches to the study of medically important fungi. Infect Immun 2003; 71:2299 - 309; http://dx.doi.org/10.1128/IAI.71.5.2299-2309.2003; PMID: 12704098
- De Backer MD, Magee PT, Pla J. Recent developments in molecular genetics of Candida albicans. Annu Rev Microbiol 2000; 54:463 - 98; http://dx.doi.org/10.1146/annurev.micro.54.1.463; PMID: 11018135
- d’Enfert C, Goyard S, Rodriguez-Arnaveilhe S, Frangeul L, Jones L, Tekaia F, Bader O, Albrecht A, Castillo L, Dominguez A, et al. CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res 2005; 33:D353 - 7; http://dx.doi.org/10.1093/nar/gki124; PMID: 15608215
- Milne SW, Cheetham J, Lloyd D, Aves S, Bates S. Cassettes for PCR-mediated gene tagging in Candida albicans utilizing nourseothricin resistance. Yeast 2011; 28:833 - 41; http://dx.doi.org/10.1002/yea.1910; PMID: 22072586
- Walther A, Wendland J. PCR-based gene targeting in Candida albicans. Nat Protoc 2008; 3:1414 - 21; http://dx.doi.org/10.1038/nprot.2008.137; PMID: 18772868
- Schaub Y, Dünkler A, Walther A, Wendland J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J Basic Microbiol 2006; 46:416 - 29; http://dx.doi.org/10.1002/jobm.200510133; PMID: 17009297
- Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 2000; 16:65 - 70; http://dx.doi.org/10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M; PMID: 10620776
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 2003; 50:167 - 81; http://dx.doi.org/10.1046/j.1365-2958.2003.03697.x; PMID: 14507372
- Fonzi WA. Role of pH response in Candida albicans virulence. Mycoses 2002; 45:Suppl 1 16 - 21; http://dx.doi.org/10.1111/j.1439-0507.2002.tb04540.x; PMID: 12073557
- Hernday AD, Noble SM, Mitrovich QM, Johnson AD. Genetics and molecular biology in Candida albicans. Methods Enzymol 2010; 470:737 - 58; http://dx.doi.org/10.1016/S0076-6879(10)70031-8; PMID: 20946834
- Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 2008; 9:578; http://dx.doi.org/10.1186/1471-2164-9-578; PMID: 19055720
- Lay J, Henry LK, Clifford J, Koltin Y, Bulawa CE, Becker JM. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect Immun 1998; 66:5301 - 6; PMID: 9784536
- Negredo A, Monteoliva L, Gil C, Pla J, Nombela C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 1997; 143:297 - 302; http://dx.doi.org/10.1099/00221287-143-2-297; PMID: 9043106
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 1999; 181:1868 - 74; PMID: 10074081
- García MG, O’Connor JE, García LL, Martínez SI, Herrero E, del Castillo Agudo L. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast 2001; 18:301 - 11; http://dx.doi.org/10.1002/1097-0061(20010315)18:4<301::AID-YEA672>3.0.CO;2-H; PMID: 11223939
- Ramanan N, Wang Y. A high-affinity iron permease essential for Candida albicans virulence. Science 2000; 288:1062 - 4; http://dx.doi.org/10.1126/science.288.5468.1062; PMID: 10807578
- Brand A, MacCallum DM, Brown AJ, Gow NA, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell 2004; 3:900 - 9; http://dx.doi.org/10.1128/EC.3.4.900-909.2004; PMID: 15302823
- Akada R, Kitagawa T, Kaneko S, Toyonaga D, Ito S, Kakihara Y, Hoshida H, Morimura S, Kondo A, Kida K. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast 2006; 23:399 - 405; http://dx.doi.org/10.1002/yea.1365; PMID: 16598691
- Davidson JF, Schiestl RH. Mis-targeting of multiple gene disruption constructs containing hisG. Curr Genet 2000; 38:188 - 90; http://dx.doi.org/10.1007/s002940000154; PMID: 11126777
- Sharkey LL, Liao WL, Ghosh AK, Fonzi WA. Flanking direct repeats of hisG alter URA3 marker expression at the HWP1 locus of Candida albicans. Microbiology 2005; 151:1061 - 71; http://dx.doi.org/10.1099/mic.0.27487-0; PMID: 15817775
- Cheng S, Nguyen MH, Zhang Z, Jia H, Handfield M, Clancy CJ. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect Immun 2003; 71:6101 - 3; http://dx.doi.org/10.1128/IAI.71.10.6101-6103.2003; PMID: 14500538
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 2000; 16:325 - 7; http://dx.doi.org/10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#; PMID: 10669870
- Sundstrom P, Cutler JE, Staab JF. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect Immun 2002; 70:3281 - 3; http://dx.doi.org/10.1128/IAI.70.6.3281-3283.2002; PMID: 12011025
- Davis D, Edwards JE Jr., Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun 2000; 68:5953 - 9; http://dx.doi.org/10.1128/IAI.68.10.5953-5959.2000; PMID: 10992507
- Ramón AM, Fonzi WA. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot Cell 2003; 2:718 - 28; http://dx.doi.org/10.1128/EC.2.4.718-728.2003; PMID: 12912891
- Morschhäuser J, Michel S, Staib P. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol 1999; 32:547 - 56; http://dx.doi.org/10.1046/j.1365-2958.1999.01393.x; PMID: 10320577
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics 1993; 134:717 - 28; PMID: 8349105
- Delneri D, Tomlin GC, Wixon JL, Hutter A, Sefton M, Louis EJ, Oliver SG. Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene 2000; 252:127 - 35; http://dx.doi.org/10.1016/S0378-1119(00)00217-1; PMID: 10903444
- Samaranayake DP, Hanes SD. Milestones in Candida albicans gene manipulation. Fungal Genet Biol 2011; 48:858 - 65; http://dx.doi.org/10.1016/j.fgb.2011.04.003; PMID: 21511047
- Ganguly S, Mitchell AP. Mini-blaster-mediated targeted gene disruption and marker complementation in Candida albicans. Methods Mol Biol 2012; 845:19 - 39; http://dx.doi.org/10.1007/978-1-61779-539-8_2; PMID: 22328365
- Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol 2000; 182:5730 - 6; http://dx.doi.org/10.1128/JB.182.20.5730-5736.2000; PMID: 11004171
- Nobile CJ, Mitchell AP. Large-scale gene disruption using the UAU1 cassette. Methods Mol Biol 2009; 499:175 - 94; http://dx.doi.org/10.1007/978-1-60327-151-6_17; PMID: 19152049
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 2005; 4:298 - 309; http://dx.doi.org/10.1128/EC.4.2.298-309.2005; PMID: 15701792
- Dennison PM, Ramsdale M, Manson CL, Brown AJ. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol 2005; 42:737 - 48; http://dx.doi.org/10.1016/j.fgb.2005.05.006; PMID: 16043373
- Wirsching S, Michel S, Morschhäuser J. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol Microbiol 2000; 36:856 - 65; http://dx.doi.org/10.1046/j.1365-2958.2000.01899.x; PMID: 10844673
- Goshorn AK, Scherer S. Genetic analysis of prototrophic natural variants of Candida albicans. Genetics 1989; 123:667 - 73; PMID: 2575557
- Köhler GA, White TC, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol 1997; 179:2331 - 8; PMID: 9079920
- Wirsching S, Michel S, Köhler G, Morschhäuser J. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J Bacteriol 2000; 182:400 - 4; http://dx.doi.org/10.1128/JB.182.2.400-404.2000; PMID: 10629186
- Reuss O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004; 341:119 - 27; http://dx.doi.org/10.1016/j.gene.2004.06.021; PMID: 15474295
- Shen J, Guo W, Köhler JR. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun 2005; 73:1239 - 42; http://dx.doi.org/10.1128/IAI.73.2.1239-1242.2005; PMID: 15664973
- Sasse C, Morschhäuser J. Gene deletion in Candida albicans wild-type strains using the SAT1-flipping strategy. Methods Mol Biol 2012; 845:3 - 17; http://dx.doi.org/10.1007/978-1-61779-539-8_1; PMID: 22328364
- Park YN, Morschhäuser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot Cell 2005; 4:1328 - 42; http://dx.doi.org/10.1128/EC.4.8.1328-1342.2005; PMID: 16087738
- Michel S, Ushinsky S, Klebl B, Leberer E, Thomas D, Whiteway M, Morschhäuser J. Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol Microbiol 2002; 46:269 - 80; http://dx.doi.org/10.1046/j.1365-2958.2002.03167.x; PMID: 12366849
- Ermak G, Cancasci VJ, Davies KJ. Cytotoxic effect of doxycycline and its implications for tet-on gene expression systems. Anal Biochem 2003; 318:152 - 4; http://dx.doi.org/10.1016/S0003-2697(03)00166-0; PMID: 12782044
- Oppermann M, Fechner H, Eberle J. Dimethyl sulfoxide enhances doxycycline-dependent protein expression in Tet-On cells. Biotechniques 2007; 42:304 - , 306, 308 passim; http://dx.doi.org/10.2144/000112387; PMID: 17390537
- Fiori A, Van Dijck P. Potent synergistic effect of doxycycline with fluconazole against Candida albicans is mediated by interference with iron homeostasis. Antimicrob Agents Chemother 2012; 56:3785 - 96; http://dx.doi.org/10.1128/AAC.06017-11; PMID: 22564841
- Backen AC, Broadbent ID, Fetherston RW, Rosamond JD, Schnell NF, Stark MJ. Evaluation of the CaMAL2 promoter for regulated expression of genes in Candida albicans. Yeast 2000; 16:1121 - 9; http://dx.doi.org/10.1002/1097-0061(20000915)16:12<1121::AID-YEA614>3.0.CO;2-U; PMID: 10953084
- Gola S, Martin R, Walther A, Dünkler A, Wendland J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 2003; 20:1339 - 47; http://dx.doi.org/10.1002/yea.1044; PMID: 14663826
- Gunasekera A, Alvarez FJ, Douglas LM, Wang HX, Rosebrock AP, Konopka JB. Identification of GIG1, a GlcNAc-induced gene in Candida albicans needed for normal sensitivity to the chitin synthase inhibitor nikkomycin Z. Eukaryot Cell 2010; 9:1476 - 83; http://dx.doi.org/10.1128/EC.00178-10; PMID: 20675577
- Bain JM, Stubberfield C, Gow NA. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol Lett 2001; 204:323 - 8; http://dx.doi.org/10.1111/j.1574-6968.2001.tb10905.x; PMID: 11731143
- Staab JF, Sundstrom P. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol 2003; 11:69 - 73; http://dx.doi.org/10.1016/S0966-842X(02)00029-X; PMID: 12598128
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 1997; 25:451 - 2; http://dx.doi.org/10.1093/nar/25.2.451; PMID: 9016579
- Chen C, Noble SM. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog 2012; 8:e1002956; http://dx.doi.org/10.1371/journal.ppat.1002956; PMID: 23133381
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989; 122:19 - 27; PMID: 2659436
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J 1997; 16:1982 - 91; http://dx.doi.org/10.1093/emboj/16.8.1982; PMID: 9155024
- Niimi M, Niimi K, Takano Y, Holmes AR, Fischer FJ, Uehara Y, Cannon RD. Regulated overexpression of CDR1 in Candida albicans confers multidrug resistance. J Antimicrob Chemother 2004; 54:999 - 1006; http://dx.doi.org/10.1093/jac/dkh456; PMID: 15486081
- Sung MK, Ha CW, Huh WK. A vector system for efficient and economical switching of C-terminal epitope tags in Saccharomyces cerevisiae. Yeast 2008; 25:301 - 11; http://dx.doi.org/10.1002/yea.1588; PMID: 18350525
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953 - 61; http://dx.doi.org/10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U; PMID: 9717241
- Krawchuk MD, Wahls WP. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 1999; 15:1419 - 27; http://dx.doi.org/10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q; PMID: 10509024
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998; 14:943 - 51; http://dx.doi.org/10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y; PMID: 9717240
- Gerami-Nejad M, Berman J, Gale CA. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 2001; 18:859 - 64; http://dx.doi.org/10.1002/yea.738; PMID: 11427968
- Gerami-Nejad M, Hausauer D, McClellan M, Berman J, Gale C. Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast 2004; 21:429 - 36; http://dx.doi.org/10.1002/yea.1080; PMID: 15116343
- Gerami-Nejad M, Dulmage K, Berman J. Additional cassettes for epitope and fluorescent fusion proteins in Candida albicans. Yeast 2009; 26:399 - 406; http://dx.doi.org/10.1002/yea.1674; PMID: 19504625
- Morschhäuser J, Michel S, Hacker J. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol Gen Genet 1998; 257:412 - 20; http://dx.doi.org/10.1007/s004380050665; PMID: 9529522
- Doyle TC, Nawotka KA, Purchio AF, Akin AR, Francis KP, Contag PR. Expression of firefly luciferase in Candida albicans and its use in the selection of stable transformants. Microb Pathog 2006; 40:69 - 81; http://dx.doi.org/10.1016/j.micpath.2005.11.002; PMID: 16427765
- Doyle TC, Nawotka KA, Kawahara CB, Francis KP, Contag PR. Visualizing fungal infections in living mice using bioluminescent pathogenic Candida albicans strains transformed with the firefly luciferase gene. Microb Pathog 2006; 40:82 - 90; http://dx.doi.org/10.1016/j.micpath.2005.11.003; PMID: 16426810
- Toyoda T, Masunaga K, Ohtsu Y, Hara K, Hamada N, Kashiwagi T, Iwahashi J. Antibody-scanning and epitope-tagging methods; molecular mapping of proteins using antibodies. Curr Protein Pept Sci 2000; 1:303 - 8; http://dx.doi.org/10.2174/1389203003381360; PMID: 12369911
- Balish E. A URA3 null mutant of Candida albicans (CAI-4) causes oro-oesophageal and gastric candidiasis and is lethal for gnotobiotic, transgenic mice (Tgepsilon26) that are deficient in both natural killer and T cells. J Med Microbiol 2009; 58:290 - 5; http://dx.doi.org/10.1099/jmm.0.004846-0; PMID: 19208876
- Chibana H, Uno J, Cho T, Mikami Y. Mutation in IRO1 tightly linked with URA3 gene reduces virulence of Candida albicans. Microbiol Immunol 2005; 49:937 - 9; http://dx.doi.org/10.1111/j.1348-0421.2005.tb03686.x; PMID: 16237272
- Chen X, Magee BB, Dawson D, Magee PT, Kumamoto CA. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol Microbiol 2004; 51:551 - 65; http://dx.doi.org/10.1046/j.1365-2958.2003.03852.x; PMID: 14756793
- Riggle PJ, Kumamoto CA. Role of a Candida albicans P1-type ATPase in resistance to copper and silver ion toxicity. J Bacteriol 2000; 182:4899 - 905; http://dx.doi.org/10.1128/JB.182.17.4899-4905.2000; PMID: 10940034
- Rogers KM, Pierson CA, Culbertson NT, Mo C, Sturm AM, Eckstein J, Barbuch R, Lees ND, Bard M. Disruption of the Candida albicans CYB5 gene results in increased azole sensitivity. Antimicrob Agents Chemother 2004; 48:3425 - 35; http://dx.doi.org/10.1128/AAC.48.9.3425-3435.2004; PMID: 15328107
- Arbour M, Epp E, Hogues H, Sellam A, Lacroix C, Rauceo J, Mitchell A, Whiteway M, Nantel A. Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Res 2009; 9:1070 - 7; http://dx.doi.org/10.1111/j.1567-1364.2009.00563.x; PMID: 19732157
- Selmecki A, Bergmann S, Berman J. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol Microbiol 2005; 55:1553 - 65; http://dx.doi.org/10.1111/j.1365-2958.2005.04492.x; PMID: 15720560
- Ahmad A, Kabir MA, Kravets A, Andaluz E, Larriba G, Rustchenko E. Chromosome instability and unusual features of some widely used strains of Candida albicans. Yeast 2008; 25:433 - 48; http://dx.doi.org/10.1002/yea.1597; PMID: 18509849
- Rustchenko EP, Howard DH, Sherman F. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J Bacteriol 1994; 176:3231 - 41; PMID: 8195078
- Kabir MA, Ahmad A, Greenberg JR, Wang YK, Rustchenko E. Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc Natl Acad Sci U S A 2005; 102:12147 - 52; http://dx.doi.org/10.1073/pnas.0505625102; PMID: 16099828
- Rustchenko E. Chromosome instability in Candida albicans. FEMS Yeast Res 2007; 7:2 - 11; http://dx.doi.org/10.1111/j.1567-1364.2006.00150.x; PMID: 17311580
- Coste A, Turner V, Ischer F, Morschhäuser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 2006; 172:2139 - 56; http://dx.doi.org/10.1534/genetics.105.054767; PMID: 16452151
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 2006; 313:367 - 70; http://dx.doi.org/10.1126/science.1128242; PMID: 16857942
- Bouchonville K, Forche A, Tang KE, Selmecki A, Berman J. Aneuploid chromosomes are highly unstable during DNA transformation of Candida albicans. Eukaryot Cell 2009; 8:1554 - 66; http://dx.doi.org/10.1128/EC.00209-09; PMID: 19700634
- Oh J, Fung E, Schlecht U, Davis RW, Giaever G, St Onge RP, Deutschbauer A, Nislow C. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog 2010; 6:e1001140; http://dx.doi.org/10.1371/journal.ppat.1001140; PMID: 20949076
- Pirrotta V, Gross DS. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol Cell 2005; 18:395 - 8; http://dx.doi.org/10.1016/j.molcel.2005.04.013; PMID: 15893722
- Tartof KD. Position effect variegation in yeast. Bioessays 1994; 16:713 - 4; http://dx.doi.org/10.1002/bies.950161004; PMID: 7980474
- Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J. Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans. Microbiology 2013; 159:565 - 79; http://dx.doi.org/10.1099/mic.0.064097-0; PMID: 23306673
- Lai WC, Tseng TL, Jian T, Lee TL, Cheng CW, Shieh JC. Construction of Candida albicans Tet-on tagging vectors with a Ura-blaster cassette. Yeast 2011; 28:253 - 63; http://dx.doi.org/10.1002/yea.1833; PMID: 21360736
- Moazeni M, Khoramizadeh MR, Kordbacheh P, Sepehrizadeh Z, Zeraati H, Noorbakhsh F, Teimoori-Toolabi L, Rezaie S. RNA-mediated gene silencing in Candida albicans: inhibition of hyphae formation by use of RNAi technology. Mycopathologia 2012; 174:177 - 85; http://dx.doi.org/10.1007/s11046-012-9539-6; PMID: 22484810
- Vieira N, Pereira F, Casal M, Brown AJ, Paiva S, Johansson B. Plasmids for in vivo construction of integrative Candida albicans vectors in Saccharomyces cerevisiae. Yeast 2010; 27:933 - 9; http://dx.doi.org/10.1002/yea.1800; PMID: 20602447
- Kim MS, Kim SY, Yoon JK, Lee YW, Bahn YS. An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem Biophys Res Commun 2009; 390:983 - 8; http://dx.doi.org/10.1016/j.bbrc.2009.10.089; PMID: 19852932
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009; 326:1509 - 12; http://dx.doi.org/10.1126/science.1178811; PMID: 19933107
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 2010; 48:419 - 36; http://dx.doi.org/10.1146/annurev-phyto-080508-081936; PMID: 19400638
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 2013; 14:49 - 55; http://dx.doi.org/10.1038/nrm3486; PMID: 23169466
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science 2009; 326:1501; http://dx.doi.org/10.1126/science.1178817; PMID: 19933106
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013; 339:823 - 6; http://dx.doi.org/10.1126/science.1232033; PMID: 23287722
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153:910 - 8; http://dx.doi.org/10.1016/j.cell.2013.04.025; PMID: 23643243
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 2013; 8:e68708; http://dx.doi.org/10.1371/journal.pone.0068708; PMID: 23874735
- Waaijers S, Portegijs V, Kerver J, Lemmens BB, Tijsterman M, van den Heuvel S, Boxem M. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 2013; 195:1187 - 91; http://dx.doi.org/10.1534/genetics.113.156299; PMID: 23979586
- Green CB, Zhao X, Yeater KM, Hoyer LL. Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology 2005; 151:1051 - 60; http://dx.doi.org/10.1099/mic.0.27696-0; PMID: 15817774
- Barelle CJ, Manson CL, MacCallum DM, Odds FC, Gow NA, Brown AJ. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 2004; 21:333 - 40; http://dx.doi.org/10.1002/yea.1099; PMID: 15042593

