Abstract
Background. p53 accumulation and TP53 mutations are known prognostic markers for breast cancer. To clarify their interrelationship and the importance of different TP53 mutation types, these markers were investigated in tumours from 630 patients with breast cancer. Materials and methods. Tumour sections were stained for p53 and scored based on staining intensity and percentages of invasive tumour cells with nuclear staining. TP53 mutations were identified by sequencing. Patient cohorts were from the DBCG (Danish Breast Cancer Cooperative Group) protocols DBCG82 and DBCG89. Results. TP53 was mutated in 29% of the patients. The disease-specific survival (DSS) at 15 years of follow-up for patients with missense mutations directly involved in DNA or zinc binding was 21±8%. Patients with the remaining missense mutations within the structural/conserved domains and patients with null mutations had a DSS of 36±6% and 31±17%, respectively. For patients without TP53 mutations and patients with mutations affecting amino acids outside these domains, the 15 year DSS was 51±3% and 71±10%, respectively. p53 accumulation was successfully scored in 567 patients and categorized into three groups. Tumours with no p53 expression had a high frequency of null mutations (37% compared to 10% in the whole cohort), and tumours with high p53 expression contained 82% of the missense mutations inside structural/conserved domains including those directly involved in DNA or zinc binding. Conclusion. The clinical outcome for breast cancer patients is significantly different for different TP53 mutation types, but further functional studies are required to clarify the exact role of these mutation types. Most of the mutations that lead to mutant p53 protein accumulation can be detected by immunohistochemistry but the specificity is low. Samples showing lack of detectable p53 protein should be considered as an indication of a possible null mutation.
Several studies have evaluated the prognostic role of TP53 mutations and p53 accumulation (determined by immunohistochemistry) in breast cancer. Some of the largest studies are performed by Olivier et al. Citation[1] on TP53 mutations in 1 794 patients with primary breast cancer and by Silvestrini et al. Citation[2] on p53 accumulation in 1 400 node-negative breast cancer patients. Although these and other studies have confirmed that TP53 mutation, and to a lesser extent p53 accumulation, are independent prognostic markers, there are still a number of open questions regarding the role of p53 in breast cancer Citation[3], Citation[4]. Recent studies have shown that different TP53 mutations can influence tumour phenotype and clinical outcome differently, but the biological mechanism behind this is only beginning to be elucidated. How this affects TP53 as a clinical useful marker, is still unclear Citation[3]. Similarly, recent knowledge has questioned the mechanisms behind the accumulation of mutant p53 protein Citation[4].
In the present study, the role of TP53 mutations is explored in further details and correlated with p53 accumulation as determined by immunohistochemistry. Two patient cohorts are investigated: a consecutive cohort of 401 patients treated according to the DBCG89 guidelines Citation[5], Citation[6], and a subgroup of 229 high-risk patients from the DBCG82 b&c trials Citation[7], Citation[8].
Materials and methods
Patients and tumour samples
Tumour material was collected from two series. The first cohort consisted of 401 consecutive patients with early breast cancer diagnosed from January 1990 to 1994 and fulfilled the following criteria: having primary unilateral breast carcinoma with no clinical evidence of metastasis; availability of complete clinical, histopathological and biological information; having no other malignancies; having received radical surgical therapy according to the DBCG89 criteria (for details, see Offersen et al. Citation[5], Citation[6]).
The second cohort consisted of 229 patients from the DBCG82 b&c studies diagnosed from 1982 to 1989 and fulfilled the following criteria: having high-risk breast cancer (defined as either positive lymph nodes and/or tumour size larger than 5 cm and/or invasion of tumour to surrounding skin or pectoral fascia); availability of complete clinical, histopathological and biological information (from frozen tissue); having no other malignancies; having received total mastectomy and a partial axillary dissection; having been randomized to CMF±radiotherapy (DBCG82b, pre-menopausal women) or Tamoxifen 1 year±radiotherapy (DBCG82c, post-menopausal women). For details, see Overgaard et al. Citation[7–9] and Nielsen et al. Citation[10].
TP53 evaluation for gene mutations by DNA sequencing
In the DBCG89 cohort, DNA was extracted from the pellet left over from estrogen receptor analysis Citation[11]. The entire coding region and all exon/intron boundaries of TP53 were analyzed by DGGE (denaturing gradient gel electrophoresis) Citation[12]. Mutant heteroduplex and homoduplex bands were excised and reamplified. Sequencing of PCR products was performed either with 33P-end-labeled primers using ThermoPrime Cycle Sequencing Kit (Amersham/GE Healthcare) or with the BigDye DyeTerminator Cycle Sequencing Kit and analyzed on an ABI PRISM™ 310 (Applied Biosystems). Only excised bands were sequenced.
In the DBCG82 cohort, DNA was extracted from frozen tumour samples using chloroform/phenol extraction followed by ethanol precipitation (Nuclear Acid Extractor 340A; Applied Biosystems) according to standard procedures. The entire coding region and all exon/intron boundaries of TP53 were analyzed by direct sequencing using an ABI PRISM™ 3700 DNA Sequencer (Applied Biosystems).
TP53 mutation types and domain structure of p53 are defined as described by Alsner et al. Citation[12].
p53 evaluation by immunohistochemistry
In the DBCG89 cohort, immunohistochemical staining of the p53 antigen was performed on 4 µm sections from the paraffin-embedded tumours. Antigen retrieval was carried out by microwaving the tissue sections in T-EG buffer (Tris 10 mM + EGTA 0.5 mM, pH 9.0) twice for 5 minutes followed by incubation with a monoclonal anti-p53 antibody (p-53 protein DO-7 code M 7001, Dako, Glostrup, Denmark) diluted 1:600 in Antibody Diluent (Dako, Glostrup, Denmark) overnight at 4°C. Primary antibody was detected by EnVision™ + /HRP Mouse code K4001 (Dako, Glostrup, Denmark) and visualized by Dako Liquid DAB+. Counter staining was done with Mayers hematoxylin. Nuclear p53 staining intensity was defined as low or high. Tumours were semi-quantitatively categorized into three groups as follows: <10% (regardless of intensity), >50% (high intensity only), and 10–49% (regardless of intensity) plus >50% (low intensity) of nuclei staining positive.
In the DBCG82 cohort, tissue microarrays (TMAs) Citation[13] were used for immunohistochemical staining of the p53 antigen Citation[14]. Antigen retrieval was carried out by microwaving the TMA sections in T-EG buffer (Tris 10 mM + EGTA 0.5 mM, pH 9.0) twice for 5 minutes followed by incubation with a monoclonal anti-p53 antibody (p-53 protein DO-7 code K4001, Dako, Glostrup, Denmark) diluted 1:600 in Antibody Diluent (Dako, Glostrup, Denmark) overnight at 4°C. Primary antibody was detected by EnVision™ + /HRP Mouse code K4001 (Dako, Glostrup, Denmark) and visualized with Novared (Vektor, SK-4800). Counter staining was done with Mayers hematoxylin. Nuclear p53 staining intensity was scored on a scale from 0 to 3 and percentages invasive tumour cells with nuclear staining were recorded. Tumours were categorized into three groups as follows: 0% (intensity 0), >50% (intensity 3), and 1–49% (regardless of intensity) plus >50% (intensity 1–2) of nuclei staining positive.
Statistical analysis
A χ2 test was used to investigate the correlations among variables and known clinicopathological parameters. The probability of treatment failure were calculated for the endpoint of disease-specific survival Citation[5] according to the Kaplan-Meier method and the differences among the survival curves were calculated with a log-rank test with a test for trend. Follow-up time was calculated using the date of primary operation as initial value.
Results
p53 accumulation and TP53 mutations, correlations with clinicopathological parameters
The analysis for TP53 mutations was performed in 630 tumours and TP53 was mutated in 180 cases (29%). TP53 mutations were categorized into four groups as described previously Citation[12]. Missense mutations affecting amino acids directly involved in DNA or zinc binding accounted for 16% of all mutations. The remaining missense mutations within the structural/conserved domains, null mutations, and mutations affecting amino acids outside structural/conserved domains accounted for 34%, 37%, and 13% of all mutations, respectively. p53 accumulation measured by immunohistochemistry was successfully scored in 567 patients. As described in further details below, p53 accumulation was categorized into three groups: tumours with no expression (9%), high expression (46%) or intermediate expression (45%).
shows the correlations between p53 accumulation, TP53 mutations and classical prognostic markers in breast cancer in the whole cohort and when separated in the DBCG89 cohort and the DBCG82 cohort. The DBCG82 cohort consisted of high-risk patients. Compared to the DBCG89 cohort, there were significant differences in patient age and menopausal status, tumour size, histological malignancy grade, and lymph node status. p53 accumulation correlated significantly with histological malignancy grade, oestrogen receptor status, and lymph node status. TP53 mutations correlated significantly with patient age, histological malignancy grade, and oestrogen receptor status.
Table I. p53 accumulation and TP53 mutation in relation to clinicopathological parameters in 630 patients diagnosed with breast cancer.
TP53 mutation patterns and prognosis
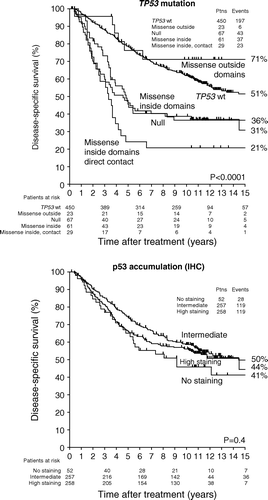
Of the 401 DBCG89 patients presented here, 243 were included in a previous study presented with 5 year follow-up on the heterogeneity in clinical phenotype of different TP53 mutation types Citation[12]. Data on the extended cohort presented with 10 year follow-up are shown in (left). In full agreement with the first report, patients with tumours containing missense mutations directly involved in DNA or zinc binding had the worst outcome with a disease-specific survival (DSS) of 25±10%. Patients with null mutations and patients with the remaining missense mutations within the structural/conserved domains had an intermediate DSS of 49±8% and 41±8%, respectively. Patients without TP53 mutations had a DSS of 67±3%. Finally, a small group of patients with mutations affecting amino acids outside these domains had a DSS of 86±10%. In the study by Offersen et al. Citation[5] comparing a number of biological variables as prognostic markers in the same patient cohort, TP53 mutations were reduced to two groups. As depicted in , “TP53 wt” contains patients without mutations and patients with mutations affecting amino acids outside structural/conserved domains. “TP53 mutation” contains the remaining three groups.
Figure 1. Disease-specific survival as a function of TP53 mutation types (top) and p53 accumulation (bottom) in breast cancer patient cohorts from DBCG89 (left) and DBCG82 (right).
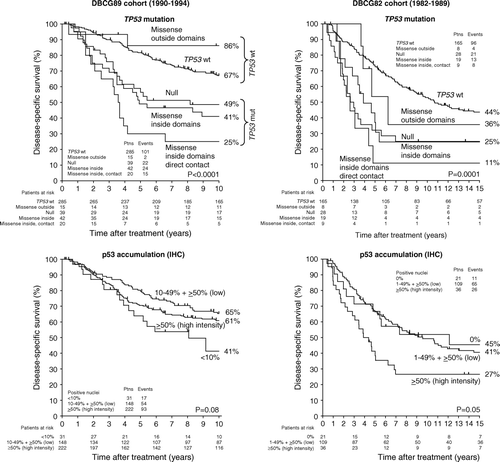
A similar association between TP53 mutation type and outcome was observed in 229 DBCG82 patients presented with 15 year follow-up in (right). Patients with missense mutations directly involved in DNA or zinc binding had again the worst outcome with a DSS of 11±10%. Patients with null mutations and patients with the remaining missense mutations within the structural/conserved domains had an intermediate DSS of 25±8% and 25±10%, respectively. Patients without TP53 mutations had a DSS of 44±4%. Finally, patients with mutations affecting amino acids outside these domains had a DSS of 36±20%.
illustrates the associations between TP53 mutation type and outcome in the combined cohort. For patients without TP53 mutations and patients with mutations affecting amino acids outside these domains, the 15 year DSS was 51±3% and 71±10%, respectively. Patients with missense mutations directly involved in DNA or zinc binding, null mutations, and patients with the remaining missense mutations within the structural/conserved domains had a DSS of 21±8%, 31±17% and 36±6%, respectively.
p53 accumulation and prognosis
The 401 DBCG89 patients were originally semi-quantitatively scored in four groups (<10%, 10–49%, 50–79%, ≥80%) based on the percentage of positive tumour nuclei in single sections. The staining intensity was scored as ‘low’ or ‘high’. Initial analysis showed that the <10% group was associated with the worst outcome, whereas the 10–49% group had the best outcome (data not shown). Based on the associations between TP53 mutations and p53 accumulation (see below), the final analysis presented in (left) includes three groups: <10% (regardless of intensity); >50% (high intensity only); and 10–49% (regardless of intensity) plus >50% (low intensity) of nuclei staining positive. In the DBCG89 cohort, these groups were associated with a 15-year DSS of 41±9%, 61±3%, and 65±4%, respectively.
Of the 229 DBCG82 patients, 166 were analysed for p53 accumulation using tissue microarrays. The percentage of positive tumour nuclei was recorded and the intensity was scored on a scale from 0-3. Only samples with 0% positive nuclei were scored with an intensity of 0. For the analysis in (right), the samples were then categorized into three groups: 0% (intensity 0); >50% (intensity 3 only); and 10–49% (regardless of intensity) plus >50% (intensity 1–2) of nuclei staining positive. In the DBCG82 cohort, these groups were associated with a 15-year DSS of 45±11%, 41±5%, and 27±8%, respectively.
illustrates the associations between p53 accumulation and outcome in the combined cohort. Patients without staining, intermediate staining, and high staining had a 15-year DSS of 41±7%, 50±4% and 44±8%, respectively.
TP53 mutation patterns and p53 accumulation
presents the associations between TP53 mutations and p53 accumulation in the two breast cancer cohorts and in the combined group. A high frequency of null mutations leading to lack of protein expression was observed in the two groups with very low percentage of positive tumour nuclei. Overall, 10% of the patients had a null mutation. In the group of very low percentage positive cells (no staining) that number was increased to 37%. In contrast, the majority of missense mutations inside structural/conserved domains including those directly involved in DNA or zinc binding were found in patients with >50% positive tumour nuclei (high intensity staining). In the combined cohort, 82% (69 of 84 total) of these missense mutations were associated with the >50% (high intensity) group.
Table II. Correlation between p53 accumulation (% positive nuclei) and TP53 mutation types.
Discussion
The present study including a total of 630 patients demonstrate that different TP53 mutation types are associated with different clinical phenotypes. The distribution and frequency of different mutation types were similar in the two cohorts consisting of 401 consecutive DBCG89 patients and 229 DBCG82 patients, respectively. As some of the patient and tumour characteristics were significantly different, the cohorts were analysed both separately and combined. The same associations between TP53 mutation type and outcome were observed in both cohorts. Missense mutations within structural/conserved domains (excluding missense mutations directly involved in DNA or zinc binding) and null mutations were associated with similar but significantly worse outcome compared to patients without mutations. Missense mutations affecting amino acids directly involved in DNA or zinc binding were associated with the worst outcome. Finally, missense mutations not affecting structural/conserved domains were associated with an outcome similar to patients without mutations.
The associations between TP53 mutation types and outcome presented here are in agreement with the largest study published on TP53 mutation types and associations with clinical phenotypes, a multi-centre analysis by Olivier et al. on 1 794 patients screened for mutations within exons 5–8 Citation[1]. Thus, Olivier et al. also found poor outcomes in patients with missense mutations within structural/conserved domains and null mutations. Although only exons 5–8 were analysed in the multi-centre analysis, missense mutations not affecting structural/conserved domains were also associated with an outcome more similar to patients without mutations. Missense mutations directly involved in DNA or zinc binding also seemed to be associated with a very poor outcome.
Even though there seems to be clear associations between clinical outcome and the different TP53 mutation types described here and by Olivier et al. Citation[1], there is still a long way before the impact of TP53 mutations on breast cancer prognosis and outcome is well understood. Some of the open questions include a better understanding of the functional consequences of mutations (loss of transactivation activities, dominant-negative effects, gain-of-function activities), the effect of TP53 haplotypes and loss of alleles, the effect on protein/protein interactions, tissue specificity, and the effect of genetic variation or alteration in other genes in the p53 pathway Citation[3], Citation[4], Citation[15]. In a study on different biological factors and their potential as prognostic factors in 408 consecutive DBCG89 patients, TP53 mutation status (recorded in the 401 patients further described here) was therefore reduced to either ‘wt’ or ‘mutant’ Citation[5].
TP53 mutation status can also be assessed by immunohistochemical (IHC) detection of mutant p53 accumulation, but this association is neither very solid nor fully understood Citation[4]. Besides the biological uncertainty, the stability of the p53 protein is also affected by different fixation methods Citation[16], and the use of antigen retrieval will lower the detection threshold for p53 staining Citation[17]. Thus, no consensus exists regarding how to score p53 accumulation. One of the problems recognized in the early reports comparing TP53 mutation and p53 accumulation was the observation of gene mutations in samples that were negative by IHC Citation[18]. In the present report, the percentage of positive tumour nuclei was recorded together with an estimate of staining intensity. As expected, the majority of missense mutations were identified in the group of patients with a high percentage of intensely stained tumour nuclei, but the specificity was low. Finally, it was also possible to define a small group of patients with undetectable levels of p53, in which a high proportion (37%) had a null mutation. Thus, future studies on p53 accumulation should not only consider intensely stained samples as indicators of TP53 mutations, but also include samples without any indications of p53 protein.
In conclusion, this study demonstrates that clinical outcome for breast cancer patients are associated with different TP53 mutation types. However, further functional studies are required to clarify the exact role of these mutation types. The study also confirms that many of the mutations will lead to mutant p53 protein accumulation detectable by immunohistochemistry, but that samples showing lack of detectable p53 protein should be considered as an indication of a possible null mutation.
Acknowledgements
The authors would like to thank Ms. Inger Marie Thuesen, Mr. Mogens Jøns Johannesen, Ms. Eldri Undlien Due, Ms. Caroline Jevanord Fæster for excellent technical help. Funding has been received from The Danish Cancer Society, The University of Aarhus, The Fund of M. L. Jørgensen and Gunnar Hansen, The Danish Ministry of Health, The Danish Medical Research Council, The Novo Nordisk Foundation, The Norwegian Cancer Society and by EC FP6 funding (Mutant p53 Contract No 502983; Annex II, Article 33 to ALBD).
References
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 2006; 12: 1157–67
- Silvestrini R, Daidone MG, Benini E, Faranda A, Tomasic G, Boracchi P, et al. Validation of p53 accumulation as a predictor of distant metastasis at 10 years of follow-up in 1400 node-negative breast cancers. Clin Cancer Res 1996; 2: 2007–13
- Petitjean A, Achatz MI, Børresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene 2007; 26: 2157–65
- Soussi T. p53 alterations in human cancer: More questions than answers. Oncogene 2007; 26: 2145–56
- Offersen, BV, Alsner, J, Olsen, KE, Riisbro, R, Brünner, N, Sørensen, FB, et al. A comparison among HER2, TP53, PAI-1, angiogenesis, and proliferation activity as prognostic variables in tumours from 408 patients diagnosed with early breast cancer. Acta Oncol 2008;47:618–32.
- Offersen BV, Sørensen FB, Yilmaz M, Knoop A, Overgaard J. Chalkley estimates of angiogenesis in early breast cancer–relevance to prognosis. Acta Oncol 2002; 41: 695–703
- Overgaard M, Jensen MB, Overgaard J, Hansen PS, Rose C, Andersson M, et al. Postoperative radiotherapy in high-risk postmenopausal breast-cancer patients given adjuvant tamoxifen: Danish Breast Cancer Cooperative Group DBCG 82c randomised trial. Lancet 1999; 353: 1641–8
- Overgaard M, Hansen PS, Overgaard J, Rose C, Andersson M, Bach F, et al. Postoperative radiotherapy in high-risk premenopausal women with breast cancer who receive adjuvant chemotherapy. Danish Breast Cancer Cooperative Group 82b Trial. N Engl J Med 1997; 337: 949–55
- Overgaard M, Nielsen HM, Overgaard J. Is the benefit of postmastectomy irradiation limited to patients with four or more positive nodes, as recommended in international consensus reports? A subgroup analysis of the DBCG 82 b&c randomized trials. Radiother Oncol 2007; 82: 247–53
- Nielsen HM, Overgaard M, Grau C, Jensen AR, Overgaard J. Study of failure pattern among high-risk breast cancer patients with or without postmastectomy radiotherapy in addition to adjuvant systemic therapy: Long-term results from the Danish Breast Cancer Cooperative Group DBCG 82 b and c randomized studies. J Clin Oncol 2006; 24: 2268–75
- Hansen LL, Andersen J, Overgaard J, Kruse TA. Molecular genetic analysis of easily accessible breast tumour DNA, purified from tissue left over from hormone receptor measurement. APMIS 1998; 106: 371–7
- Alsner J, Yilmaz M, Guldberg P, Hansen LL, Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin Cancer Res 2000; 6: 3923–31
- Kyndi, M, Sørensen, FB, Knudsen, H, Overgaard, M, Nielsen, HM, Andersen, J, et al. Tissue microarrays compared with whole sections and biochemical analyses: A subgroup analysis of DBCG82 b&c. Acta Oncol 2008;47:591–99.
- Kyndi, M, Sørensen, FB, Knudsen, H, Alsner, J, Overgaard, M, Nielsen, HM, et al. Impact of BCL2 and p53 on post-mastectomy radiotherapy response in high-risk breast cancer. A subgroup analysis of DBCG82 b&c. Acta Oncol 2008;47:608–17.
- Soussi T, Wiman KG. Shaping genetic alterations in human cancer: The p53 mutation paradigm. Cancer Cell 2007; 12: 303–12
- Fisher CJ, Gillett CE, Vojtesek B, Barnes DM, Millis RR. Problems with p53 immunohistochemical staining: The effect of fixation and variation in the methods of evaluation. Br J Cancer 1994; 69: 26–31
- Battifora H. p53 immunohistochemistry: A word of caution. Hum Pathol 1994; 25: 435–7
- Sjögren S, Inganäs M, Norberg T, Lindgren A, Nordgren H, Holmberg L, et al. The p53 gene in breast cancer: Prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst 1996; 88: 173–82