Abstract
Although it is now common to look at the information carried by the genome as a linear sequence of nucleotides, such representation does not say much about the organization of the genetic information within the cell nucleus. How the genome accommodates the tight packing needed to fit in the cell nucleus and at the same time maintains the accessibility necessary for specific expression is one of the open questions in modern biology. In eukaryotic cells, DNA is wrapped and packaged into chromatin through the binding of histones assembled into nucleosomes. In addition to bundling DNA, the nucleosomes also facilitate communication between distant genomic sites, such as enhancers and promoters found at the ends of protein-mediated loops. In order to understand the physical and chemical basis of such processes, we have begun to investigate chromatin organization and looping. We have developed a mesoscale model of chromatin at a resolution of a single base pair and used Monte Carlo numerical strategies to understand how the presence of nucleosomes on DNA can influence and possibly control chromatin looping. We have validated this model by successfully reproducing experimental measurements of gene expression on nucleosomal arrays (Kulaeva et al., 2012). Our results show a wide variety of chromatin organization depending on the way nucleosomes are positioned on DNA (i.e. on the spacing between successive nucleosomes as illustrated in the pictures below) and also on chemical details at the histone level, such as modifications of the N-terminal tails. This diversity in chromatin organization, which extends beyond the conventional solenoid and zigzag models, comes along with very different physical and mechanical properties and looping propensities. Furthermore, our simulations reveal some surprising properties of chromatin: for example, we found that tightly packed chromatin fragments are highly flexible, a remarkable feature for a material that needs to fit inside a cell nucleus. In conclusion, our work uncovers parts of a rich and dynamic picture of chromatin where the DNA sequence influences the positioning of nucleosomes, which in turn shape the genomic material and have an impact on the accessibility and expression of the genetic message.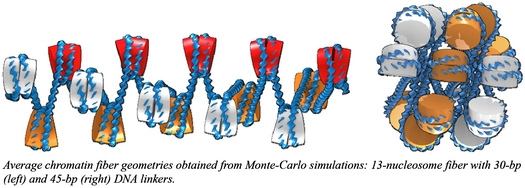
Average chromatin fiber geometries obtained from Monte-Carlo simulations: 13-nucleosome fiber with 30-bp (left) and 45-bp (right) DNA linkers.
This research has been supported by USPHS Research Grant GM 34809.
References
- Kulaeva , O. I. 2012 . Internucleosomal interactions mediated by histone tails allow distant communication in chromatin . Journal of Biological Chemistry , 287 : 20248 – 20257 .