ABSTRACT
Grassland is one of the most important terrestrial ecosystems for carbon (C) and nitrogen (N) cycling. However, while CO2 fixation by phototrophic bacteria is relatively well studied, little is known about microbial CO2 fixation without light by chemoautotrophic bacteria in grassland soils. Therefore, in this study, the isotope 14C-CO2 was used to investigate the CO2-fixing process in grassland soils. Soil samples were collected from both fenced and adjacent continuous grazing grassland sites in Inner Mongolia and then incubated for 120 days under dark conditions. Meanwhile, the cbbL genes (red- and green-like) were analyzed to isolate chemoautotrophic bacteria, which are responsible for CO2 fixation. After incubation, 14C was fixed into soil organic carbon (14C-SOC) and microbial biomass carbon (14C-MBC) were found in both the fenced and grazing soils, and the fixation rate of 14C-SOC in the fenced soils (48.55‰) was significantly higher than in the grazing soils (22.11‰). The fixation rate of 14C-MBC in the fenced soils (14.05‰) was higher than in the grazing soils (7.08‰), but the difference was not significant. The red-like cbbL genes could be detected in all the soil samples, but the green-like cbbL genes could not be amplified. A greater number of identified operational taxonomic units were observed in the fenced soils compared with the grazing soils. The chemoautotrophic bacteria were mainly affiliated with Alphaproteobacteria and Actinobacteria. However, Chloroflexi was detected in only the fenced soils. The results suggested that CO2 fixation by chemoautotrophic bacteria might be significant in carbon cycling in grassland.
Introduction
Grassland ecosystems constitute about 40% of the global land area and play a significant role in the global terrestrial C cycle (Wang & Fang Citation2009). Due to the relatively high C sequestration rates (mainly associated with below-ground C pools) and extensive area, grasslands are recognized for their great potential as net sink for atmospheric CO2 and climate change regulator (Follett & Reed Citation2010). Marginal changes in soil C sequestration rates in grasslands can have significant impacts on atmospheric CO2 concentrations (Follett & Reed Citation2010). Globally, soils contain 1500 Pg C, which is twice the amount of atmospheric C pool (Schlesinger & Andrews Citation2000), with grasslands containing ∼12% of the overall terrestrial soil C pool (Schlesinger Citation1977). However, grassland management can alter the quantity and quality of litter inputs and, subsequently, impact the amounts and stability of C stored in the soil (Dubeux et al. Citation2006). Conversely, adoption of improved grassland management practices such as proper grazing and soil nutrient management can also have positive impacts on soil C sequestration (Dubeux et al. Citation2006).
CO2 is a greenhouse gas which is with the potential threat of global climate change. The concentration of atmospheric CO2 dramatically increase from about 280 to more than 380 parts per million (ppm) over the last 250 years because of the consumption of fossil fuels. And it will rise up to 570 ppm in the atmosphere by the year 2100 predicted by the International Panel on Climate Change (Song Citation2006). Nowadays, it is an urgent need to reduce atmospheric CO2 concentration and slow down the effects of global warming. Now many techniques (physical fixation, chemical fixation and biofixation of CO2) are developed to alleviate atmospheric CO2, of which the biofixation of CO2 by microorganisms was considered an economical, effective and sanitary method. Microbial biofixation of CO2 employs the capacity of autotrophic microorganisms, including photoautotrophs and chemoautotrophs, for CO2 fixation (Yousuf et al. Citation2012), which are known to contribute significantly to CO2 assimilation in aquatic environment. Photoautotrophs are capable of synthesizing their own food from inorganic substances by using light as an energy source. And the chemoautotrophs can support carbon fixation in the absence of light by using the energy from oxidizing H2, H2S, ,
, NO2 and Fe2+ (Santoro et al. Citation2013). Photoautotrophs (e.g. microalgae and cyanobacteria) can grow much faster than terrestrial plants and live in harsh conditions due to their structure. Microalgal cells absorb CO2 to support their growth by converting solar energy into biomass, which can be converted to secondary products with high commercial value (Beer et al. Citation2009). In addition, microalgae are predominant carbon-fixing agents in water with efficiency being 10 times greater than that of terrestrial plants (Usui & Ikenouchi Citation1997). Like algae, cyanobacteria are also efficient at the carbon fixation owing to their simpler structure (Oliver et al. Citation2014). They are considered to play a key role among photosynthetic organisms, accounting for 20–30% of Earth’s primary photosynthetic activity (Pisciotta et al. Citation2010).
In general, CO2 can be fixed by five different pathways (Herter et al. Citation2002; Fan Citation2008), but the Calvin–Benson–Bassham (CBB) cycle is the major and most widely distributed pathway using autotrophic microorganisms to fix CO2 (Yuan, Ge, Chen et al. Citation2012). The key enzyme of the CBB cycle is the ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO) (Ellis Citation1979). It is a bifunctional enzyme controlling the reduction of CO2 and the oxygenolysis of ribulose-1,5-bisphophate. RubisCO exists in four forms (I, II, III and IV), which differ in structure. And the most distributed type of RubisCO is the form I, which occurs in plants as well as in autotrophic (photoautotrophic and chemoautotrophic) bacteria (Selesi et al. Citation2005). The form I RubisCO is composed of eight large and eight small subunits (L8S8) (Tabita Citation1999). Phylogenetic studies based on these cbbL sequences have shown that form I RubisCO had to be subdivided into two major groups: green-like and red-like (Watson & Tabita Citation1997). The green-like RubisCO is divided into two types, IA and IB, and found cbbL sequences from plants, green algae, and Alpha-, Beta- and Gammaproteobacteria as well as from Cyanobacteria. The red-like RubisCO is also divided into two types, IC and ID, which is found in many non-green algea and Alpha-, and Betaproteobacteria (Videmšek et al. Citation2009).
Microbial assimilation of CO2 is a ubiquitous process in soils. Global estimates of microbial CO2 fixation in soils range between 0.6 and 4.9 Pg C year−1 (Yuan, Ge, Chen et al. Citation2012). Nowak et al. (Citation2015) have estimated that in the wetland soils up to 27% of soil organic matter (SOC) in the 0–10 cm layer was derived from autotrophic (photo- and chemoautotrophic) microbial fixation of CO2. The highest CO2 fixation by autotrophic microorganisms is found in the 0–1 cm layer and 14C labeled SOC concentration in the paddy soils is higher in both 0–1 and 1–5 cm layers than those in the upland soils after incubated with continuous 14CO2 for 110 days (Wu et al. Citation2014). In addition, chemoautotrophic acetogenes can assimilate CO2 with exogenous H2 in rice field soil incubated at 50°C (Liu & Conrad Citation2011). High abundance of autotrophic CO2 fixation bacteria is shown in arid soil in northwest China and in grassland soils near natural springs with high CO2 concentration (Videmšek et al. Citation2009). However, only limited knowledge is available on the CO2 fixation by chemoautotrophic bacteria in grassland soils under the dark condition. The aims of this study were (i) to test the ability of CO2 fixation by grassland soils; (ii) to compare the CO2 fixation rate between the fenced and grazing grassland soils and (iii) to determine the composition of these cbbL types (green- and red-like) in soil bacteria isolated from both the fenced and grazing grassland soils in a semi-arid region in Inner Mongolia, China.
Materials and methods
Study sites and soil sampling
The study area is located in Baarin Right Banner of Chifeng (43°12′55″–44°27′52″ N, 118°12′09″–120°01′42″ E) in the southeastern part of Inner Mongolia. This area has a temperate, semi-arid continental monsoon climate with dry and windy winters and springs, and hot, humid summers followed by short and cool autumns. The mean annual precipitation is around 360 mm, with 60–70% of the rainfall from June to September. The annual mean temperature is around 4.9°C, with a minimum monthly mean temperature of −13°C in January and a maximum of 22.2°C in July. The annual frost-free period is approximately 125 days. From north to south it stretches 139 km, while from east to west it stretches 154 km. Elevations decrease from a high of more than 1000 m in the northwest to less than 400 m in the southeast.
All field sampling was carried out in early August 2014. A continuously grazed grassland site (GG) and an adjacent fenced grassland site (FG) with grazing exclusion of more than 20 years were selected. Five random quadrats were established at each site for soil research. One soil sample was taken from five points in each quadrat (four corners and the center of the quadrat) at depths of 0–5 cm and mixed into one sample. After carefully removing the surface organic materials and fine roots, each mixed sample was divided into two parts. One part was air-dried for analysis of soil physicochemical properties and the other was sifted through a 2 mm sieve for microbial assays and stored at −20°C. Three aboveground plant sampling quadrats (1.0 × 1.0 m) were set up in both the grazed and fenced grassland sites. After botanical composition, plant height and canopy cover were measured, the remaining biomass in each quadrat was clipped and weighed after being dried at 80°C for 48 h. The characteristics of the plant community in this study site are summarized in .
Table 1. Description of plant characteristics of the study sites.
Incubation experiments with labelled CO2
Two sets of 25 g fresh soil of each sample was added to glass bottles (5.5 cm diameter and 11 cm height) and placed into containers (30 cm diameter and 17 cm depth). In one set as a control, the soils were fumigated under a vacuum with CHCl3 for 24 h. In another set, the soil samples were just added to a bottle without any treatment. In each container, one bottle was filled with sterile distilled water to maintain the moisture level in the container, one bottle contained equal NaH14CO3 (specific activity 310.8 MBq mmol−1, 2.31 MBq) and the lid was closed after HCl (1 M) was added to the NaH14CO3. All the containers were then incubated for 120 days at 20°C in a plant growth chamber (LH-100RD; NKS, Tokyo, Japan) under dark conditions. After incubation, each soil sample was then mixed thoroughly and divided into two portions. One portion was oven-dried at 70°C to a constant weight to determine the fixation rate of 14C-SOC fixed from 14C-CO2 in the air, and the other was stored at 4°C to determine 14C-MBC.
Analytical methods
Soil pH was measured using a pH meter (F-21; Horiba, Japan) at a soil-to-water ratio of 1:5 after shaking for 1 h. Electrical conductivity (EC) was measured after the pH measurement using an EC meter (CM-14P; TOA Electronics Ltd., Japan). Soil moisture was measured after being oven-dried at 105°C for 24 h. Particle size distribution was determined using the pipette method (Gee & Bauder Citation1986). Part of each sample was air-dried and finely ground to pass through a 0.1 mm sieve and analyzed for total N (TN) using the dry combustion method with an NC-Analyzer (Sumigraph Nc-80; Sumika Chemical Analysis Service Co., Tokyo, Japan). Soil organic carbon (Corg) was determined using the Walkley and Black method (Walkley Citation1947). The available phosphorus (Avail. P) was determined using the Bray II method (Kuo Citation1996), where soil samples were extracted with an extraction solution (1 M NH4F and 0.5 M HCl) and color-developing reagent was added. The Avail. P was then determined using the absorbance measurement with a spectrophotometer at a wavelength of 710 nm (UV-142-02; Shimadzu, Kyoto, Japan). Exchangeable cations (Ca2+, Mg2+, K+ and Na+) were extracted three times with 1 M ammonium acetate at pH 7.0, and the concentration was measured using an atomic absorption spectrophotometer (AA-6800; Shimadzu, Kyoto, Japan). Microbial biomass C (Cmic) and N (Nmic) was estimated using the fumigation extraction method (Tanaka et al. Citation1998).
The 14C-SOC was measured according to Ge et al. (Citation2013). In brief, about 0.50 g of soil (dry soil) was added to 10 mL 0.2 M potassium dichromate and 20 mL of a mixture of concentrated H2SO4 and H3PO4 (5:1, v:v). 14C-CO2 was trapped in 25 mL NaOH (0.5 M) after digesting at 165°C for 8 min with O2 continuously replenished. And then the mixture containing 1 mL NaOH and 5 mL Ultima Gold XR (PerkinElmer, 940 Winter Street Waltham, MA 02451, USA) was measured in an automated liquid scintillation counter (LS-6500; Beckman) for 10 min. The 14C-SOC fixation rate (‰) fixed by per kilogram soil was calculated as follows:where F1 represents the factor to convert the counting volume (1 mL from 25 mL); Rs, Ro and Rp are 14C-radioactivity (Bq) for a trapped solution, 14C in the natural soil of the study sites and 14C-CO2 generated from NaH14CO3 in the container, respectively; and W, the weight (kg) of soil.
The 14C-MBC was measured according to Ge et al. (Citation2013). Moist soil samples (8 g) were fumigated for 24 h in the dark, followed by extraction with 32 mL K2SO4 (0.5 M). The 14C-radioactivity in the extractant (1 mL) of fumigated soil sample, together with that (1 mL) extracted from equivalent unfumigated portions, was measured as above. The 14C-MBC rate (‰) per kilogram soil was calculated as follows:where F2 represents the factor to convert the counting volume (from 1 mL to the volume of 32 mL plus soil water volume in mL); Rf and Ruf are 14C trapped in fumigated and unfumigated soil extractant, respectively; Rp, radioactivity of 14C-CO2 generated from NaH14CO3 in the container; and W, the weight (kg) of soil.
Microbial DNA extraction
Total DNA was extracted from 0.5 g of grassland soil using an ISOIL for Beads Beating kit (Nippon Gene Co., Ltd., Japan) according to the manufacturer’s instructions. Bacterial cells in the soil sample were lysed for 45 s at 4500 rpm of the fast prep bead beating instrument (Tomy Micro Smash MS-100; Tomy Seiko Co., Ltd., Japan). The extracted pellet was dissolved in 100 μl TE and the DNA concentration determined using a spectrophotometer (Nanodrop ND-1000; Thermo Fisher Scientific Inc., USA). DNA was stored at −20°C until further use.
Design of cbbL primers
New primer sets for amplification of cbbL (green- and red-like) sequences were designed manually. All cbbL nucleotide sequences, which were available from the National Center for Biotechnology Information (NCBI) sequence database, were used to establish a cbbL database by using CLC Sequence Viewer 7. The sequences were first translated into amino acids using Genetyx ver. 10, and the deduced amino acid sequences were then aligned using CLC Sequence Viewer 7. Amino acid alignments were performed manually and nucleotide sequences were aligned accordingly. Based on these data, we designed two primer sets specific for the selected cbbL sequences of the red-like and green-like groups. The primers cbbLRA and cbbLRB, used to amplify the red-like cbbL gene, were designed from multiple sequence alignment data for the cbbL genes of Ralstonia eutropha H16, Ralstonia eutropha megaplasmid pHG1, Sinorhizobium meliloti WSM419, Rhodobacter sphaeroides HR and Rhodopseudomonas palustris CGA009. The primers cbbLGA and cbbLGB, used to amplify the green-like cbbL genes, were designed from multiple sequence alignment data for the cbbL genes of Nitrobacter vulgaris T3, Acidithiobacillus ferrooxidans ATCC 23270, Hydrogenophaga pseudoflava DSM1083, Thiobacillus denitrificans ATCC 25259 and Nitrospira sp. strain TCH716. The primers designed and used for this study are listed in .
Table 2. Primers used for amplification of cbbL gene.
PCR amplification, cloning and sequencing
The previously designed primers of the cbbL (green- and red-like) genes were used for amplification. In brief, each individual PCR mixture contained approximately 150 ng soil DNA, 2 μl 10×Ex Taq buffer, 1 U Ex Taq polymerase (TaKaRa, Japan), 200 μM deoxynucleoside triphosphates, 2.5 μM of each cbbL primer and made up to 20 μl with sterilized H2O. The thermos-cycle conditions were as follows: 4 min of initial denaturation at 95°C, followed by 40 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 50°C for the red-like and 52°C for the green-like cbbL primers and 1 min of elongation at 72°C. The reaction was completed by a final extension for 10 min at 72°C. PCR products were checked in 0.8% (wt/vol) agarose gels (PeqLab Biotechnology GmbH, Erlangen, Germany) using horizontal gel electrophoresis at 75 V for 45 min. DNA was observed using UV excitation after staining with ethidium bromide.
PCR products were purified using a Wizard SV Gel and PCR Clean-Up System kit (Promega, USA) and ligated into p3 T Vector (MoBiTec, Germany), and the resulting ligation products were used to transform Escherichia coli DH12S (Invitrogen, Japan) competent cells. The inserted products were amplified with primers (M13-47 and RV-M) and were sequenced directly from white colonies grown in LB supplemented with ampicillin and X-gal. The sequences were compared with known cbbL gene sequences from the GenBank (NCBI) database using BLAST. Sequence data have been submitted to the DDBJ database under accession numbers LC195753 to LC195828.
Statistical analysis
Data were processed using Excel 2013 for the means and the standard errors. The Student’s t-test was used to compare the difference in soil physicochemical characteristics between the fenced and grazing sites. Multiple comparisons of significant differences were made using one-way analysis of variance followed by a Tukey test (P < .05). All analyses were performed using SPSS 19.0 for Windows XP.
Results
Soil properties changes
According to the results, grazing significantly decreased soil moisture and EC compared with the fenced grassland soil (P < .01) (). The soil was slightly acidic (6.84–6.97), and the differences in soil pH between the fenced and grazing sites were not significant (P > .05). There was a significant difference in soil Corg and TN between the fenced and grazing sites (P < .01). The results revealed that the mean Corg and TN significantly increased from 18.81 g kg−1 in the grazing site to 26.39 g kg−1 in the fenced site and from 2.00 g kg−1 in the grazing site to 3.10 g kg−1 in the fenced site, respectively (P < .01). However, there was no difference in the C-to-N ratio between the fenced and grazing sites (P > .05). The soil silt and clay content decreased from 16.68% and 26.45% in the fenced site to 14.43% and 23.52% in the grazing site, while the soil sand content increased from 56.87% in the fenced site to 62.05% in the grazing site. There were no significant differences between the mean value of exchangeable Mg2+ and Na+ for the fenced and adjacent grazing grassland (P > .05), but the mean value of exchangeable Ca2+ and K+ decreased significantly from 19.81 cmolc kg−1 in the fenced site to 11.03 cmolc kg−1 in the grazing site and 1.04 cmolc kg−1 in the fenced site to 0.77 cmolc kg−1 in the grazing site, respectively (P < .01). The results showed that the mean value of Avail.P decreased from 39.21 mg kg−1 in the fenced site to 32.34 mg kg−1 in the grazing site. And the mean value of Cmic and Nmic significantly decreased from 1218.12 and 159.37 mg kg−1 in the fenced site to 704.99 and 96.63 mg kg−1 in the grazing site, respectively (P < .01).
Table 3. Soil physicochemical characteristics of study sites in 0–5 cm layer.
14C-CO2 fixation rate in the fenced and grazing soils
After 120 days of incubation under dark conditions, 14C in the soils was detected in all the samples (). The highest 14C-SOC fixation rate appeared in the fenced grassland soils (48.55‰), which was significantly higher than in the grazing grassland soils (22.11‰) (P < .05). After fumigation, the fixation rates in the soils decreased to 7.68‰ and 4.18‰ in the fenced and grazing grassland soils, respectively, and the difference was not significant. However, these rates were all significantly lower than in the unfumigated soils.
Figure 1. The fixation rate of 14C-SOC and 14C-MBC in soils after incubation in darkness for 120 d.
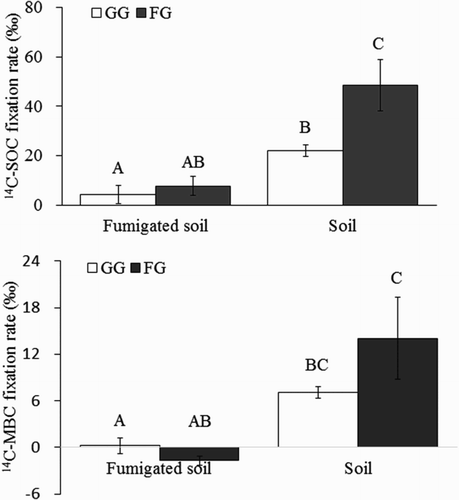
As with the 14C-SOC fixation ratio, the highest 14C-MBC fixation rate also appeared in the fenced grassland soils (14.05‰), which was higher than in the grazing grassland soils (7.08‰). The 14C-MBC fixation ratios in the fumigated grazing and fenced grassland soils were 0.24‰ and −1.66‰, respectively. The 14C-MBC fixation rates in the unfumigated soils were significantly higher than in the fumigated soils.
Phylogenetic analysis of the cbbL gene clones from soils
The red-like types of RubisCO cbbL genes were detected in both the grazing and fenced grassland soil samples, but the green-like cbbL genes were not detected with the used primers. 30 and 46 red-like cbbL operational taxonomic units (OTUs) were retrieved from the grazing and fenced soil samples, respectively (). The unique phylotypes were distributed into four phyla – Proteobacteria (46.1%), Actinobacteria (38.1%), Chloroflexi (7.9%) and Cyanobacteria (7.9%) – with two phyla dominating (). Proteobacteria was dominated by the subgroup Alphaproteobacteria (88.5%), followed by Gammaproteobacteria (8.6%) and Betaproteobacteria (2.9%).
Figure 2. Neighbor-joining phylogenetic tree of red-like cbbL genes in the grazing and fenced grassland soils.
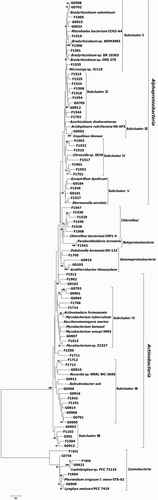
These sequences in the Alphaproteobacteria were tentatively grouped into five subclusters. Subcluster I was grouped with Bradyrhizobium valentinum (accession number WP_057851213), Rhizobiales bacterium CCH2-A4 (WP_068078378), Bradyrhizobium sp. WSM3983 (WP_027529094), Bradyrhizobium sp. BR 10303 (WP_066503379), Bradyrhizobium sp. ORS 375 (WP_009031241) and Microvirga sp. JC119 (WP_046868905). Close resemblance (100% similarity) of one phylotype was observed with Bradyrhizobium sp. WSM3983. Subcluster III was grouped with Inquilinus limosus (WP_034847255), Azorhizobium doebereinerae (WP_051356539) and Acidisphaera rubrifaciens HS-AP3 (GAN77260). Subcluster IV was grouped with Citreicella sp. SE45 (WP_008884071). Clones in this cluster were all from the fenced grassland soil samples. Subcluster V was grouped with Azospirillum lipoferum (WP_014188404) and Skermanella aerolata (WP_044434920). Both Betaproteobacteria and Gammaproteobacteria were grouped into one subcluster, which was affliated with Betaproteobacteria Paraburkholderia terrestris (SAL14411) with 99% resemblance and Gammaproteobacteria Acidiferrobacter thiooxydans (WP_065968746) and Dokdonella koreensis DS-123 (ANB18496), respectively. The clones of Proteobacteria in the fenced grassland soil samples accounted for 68.6% of the total, whereas only 31.4% corresponded to the grazing grassland soil samples.
The sequences in the Actinobacteria were tentatively grouped into three subclusters. Subcluster VI was grouped with Actinomadura formosensis (WP_067800405), Mycobacterium tuberculosis (CNE30029), Saccharomonospora marina (WP_009153612), Mycobacterium kansasii (WP_063467152), Mycobacterium xenopi 3993 (EUA52020) and Mycobacterium sp. E2327 (WP_068106460). Subcluster VII was grouped with Nocardia sp. NRRL WC-3656 (WP_030514398) and Solirubrobacter soli (WP_028064187). The clones from the fenced grassland soil samples accounted for 44.8% of the total, whereas 55.2% corresponded to the grazing grassland soil samples.
There was one subcluster in Chloroflexi, which was grouped with Chloroflexi bacterium CSP1-4 (KRT63449). The clones were all from the fenced grassland soil samples. There was one subcluster in Cyanobacteria, which was grouped with Leptolyngbya sp. PCC 73110 (BAE80672), Phormidium irriguum f. minor ETS-02 (CBL80832) and Lyngbya aestuarii PCC 7419 (BAE80673).
Discussion
Soil physicochemical properties changes
We found significantly higher soil moisture, Corg, EC, TN, Ca2+, K+, Cmic and Nmic in the fenced grassland soils compared with the grazing grassland soils (). Fencing significantly increased soil moisture in the present study (), which was consistent with previous studies (Deng et al. Citation2014). In this study, fences enhanced aboveground biomass and coverage () that could decrease soil evaporation. Moreover, due to continuous grazing and trampling by cattle, the ground surface at the grazing site became bare and could easily become hotter than the covered ground, which caused a decrease in soil moisture and an increase in soil erosion risk (Wang et al. Citation2015). There was no difference in soil pH between the fenced and grazing areas (P > .05) and was constant at about 7.0 (). Similar results were reported about grazing intensity that did not affect soil pH in a Mediterranean rangeland (Akhzari et al. Citation2015) and in the Stipa grandis and Stipa bungeana steppe in northern China (Xie & Wittig Citation2004). The mean value of EC in the fenced grassland soils was remarkably higher than in the grazing grassland in terms of its effect on soil texture (). Soil clay content decreased after long-term grazing (). Soil leaching decreased while soil clay content increased, which can reduce natural soil drainage and conserve water. These lead to the accumulation of salts and minerals in the surface soil, which cause EC to increase.
Our results showed that soil Corg and TN significantly increased in the 0–5 cm layer after long-term fencing (). A similar increase in soil Corg and TN following fencing was reported in other arid and semi-arid rangelands (Steffens et al. Citation2008). In grassland, soil Corg is determined by carbon input from plant productivity, litter decomposition, root turnover and animal excreta, and carbon output through soil respiration, soil erosion and leaching (Cui et al. Citation2005; Wen et al. Citation2013; Zuo et al. Citation2015). Aboveground litter accumulates on the soil surface after fencing and vegetation grows better and develops a better root system compared with grazing plots and is conducive to SOC formation and accumulation (Su et al. Citation2004). However, there were no significant differences in soil C/N ratios between the grazing site and fenced site, indicating the rate of change for C and N after grazing exclusion is the same.
Particle size distribution showed more silt and clay and less sand in the soils of the fenced site compared with the soils of the grazing site (), but the difference was not significant. Grazing and trampling leads to a decrease in ground cover, enlarging patches of bare ground (Ludwig & Tongway Citation1995) and leaving the land surface directly exposed to strong wind erosion, which causes loss of fine soil particles and degradation of soil structure (Gomes et al. Citation2003).
Grazing exclusion increased the concentration of Avail. P, but the difference was not significant (). This result may be due to the runoff from soil erosion (Vadas et al. Citation2015), especially in the grazing site where the soils lacked protection from vegetation because of grazing. In addition, livestock grazing can cause energy and nutrient loss from the ecosystem (Miao et al. Citation2015). The decrease in nutrient feedback from the litter might be related to the decrease in Avail. P in the grazing site. The mean concentrations of Ca2+ and K+ were significantly higher in the fenced site than in the grazing site, but the difference in mean concentrations of Mg2+ and Na+ was not significant. All four of the major cations are subject to loss by leaching (Phillips & Burton Citation2005), but among the exchangeable cations, Ca2+ is usually dominant, often amounting to 60–85% of the total in non-acid soil (Domagała-Świątkiewicz & Sady Citation2011), and when Ca2+ is the dominant cation in the soil, it may be the highest amount leached (Whitehead Citation2000), which was consistent with this study. In addition, potassium, magnesium and calcium appear to compete with each other in the uptake by plants (Evangelou et al. Citation1994), and because sodium is not essential to plant biochemistry, plants exclude it when taking up water and other cations, which can explain the constant Na+ concentration in the fenced and grazing sites.
In the present study, Cmic and Nmic were found to significantly decrease in the grazing site compared with the fenced site, which indicated that continuous grazing was deleterious to microbial growth. This finding was similar to those of many other studies (Northup et al. Citation1999; Wang et al. Citation2008). Soil organic carbon is the major source of energy for soil microorganism growth. In the fenced site, higher organic matter input from plant litter and root exudates may have enhanced the rate of Cmic production in the soil (Bird et al. Citation2002; Ge et al. Citation2011).
The differences in 14C-CO2 fixation rates between the fenced and grazing sites
Labelled 14C was detected in the SOC in both the fenced and grazing grassland soils after 120 days’ incubation under dark conditions (). This result was in agreement with previous studies in which CO2 fixation was found in two artificial soils after incubation in the dark for 14 days (Šantrůčková et al. Citation2005), in agricultural soil in northeast Georgia (Shimmel Citation1987) and in synthetic soil after incubation up to 91 days in the dark (Miltner et al. Citation2005). However, this finding was inconsistent with the study conducted by Ge et al. (Citation2013), who used 14C to incubate subtropical upland and paddy soils for 110 days, but no 14C was fixed in the soils incubated in continuous darkness.
This result indicates that fixed 14C was mainly derived from chemoautotrophic processes and chemoautotrophic microorganisms that sequester atmospheric CO2 in the grassland soils. Our results also showed that 14C-SOC and 14C-MBC fixation rates were significantly higher in the fenced soils than in the grazing soils (). In addition, the 14C-SOC and 14C-MBC fixation rates in the fumigated soil samples significantly decreased compared with those in the unfumigated soil samples in both the fenced and grazing sites. A previous study reported that the red-like cbbL genes were found only in small clay and silt fractions and not in coarse particle fractions (Selesi et al. Citation2007). Moreover, it was found that there is a close link between bacterial cell numbers and smaller silt and clay fractions (Van Gestel et al. Citation1996; Kandeler et al. Citation2000). In this study, clay and silt content in the fenced site was around 12.0% higher than in the grazing site (). In addition, both the amount of the red-like cbbL genes and RubisCO activity had a significant positive relationship with SOC content (Yuan, Ge, Wu et al. Citation2012; Yuan et al. Citation2013). In our study, the mean amount of Corg in the fenced soils increased by 28.7% compared with the grazing soils (). These findings can explain the high fixation rate in the fenced site.
The effect of grazing on the red-like cbbL genes
Grassland is widespread, accounting for 40% of the land area in China and one-third of the world terrestrial area, and is one of the most important ecosystems for C and N cycling (Kang Citation2012). Until now, most previous studies on the distribution and quantification of cbbL gene-containing bacteria have focused on aquatic systems (Yuan, Ge, Wu et al. Citation2012) or terrestrial agricultural bulk soil (Selesi et al. Citation2005, Citation2007), with few studies on grassland soil. The most common pathway of CO2 fixation used by most photoautotrophic and chemoautotrophic bacteria is via the Calvin cycle with the key enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), the large subunit of which is encoded by the cbbL gene. In this study, no green-like bacteria was found in the fenced and grazing sites, which was consistent with previous studies (Miltner et al. Citation2005; Videmšek et al. Citation2009; Yousuf et al. Citation2012). Miltner et al. (Citation2005) concluded that the growth of obligate lithotrophs carrying the green-like cbbL genes was suppressed by the input of readily available carbon sources, and the absence of the green-like cbbL genes could be attributed to primer specificity bias in the rhizospheric soil of groundnut (Yousuf et al. Citation2012).
In contrast to the green-like cbbL genes, the red-like cbbL genes were detected in both the fenced and grazing grassland sites. Our results indicate that the composition of microbial communities carrying red-like cbbL genes was affected by continuous grazing (). Compared with the 30 OTUs detected in the grazing site, there were 46 OTUs detected in the fenced site. In particular, in the cluster Alphaproteobacteria, 22 OTUs were identified from the fenced site compared with 9 OTUs identified from the grazing site. The cluster Alphaproteobacteria contained five subclusters, mainly Bradyrhizobium valentinum, Microvirga sp. and Azospirillum lipoferum bacteria, which are known to promote plant growth and fix nitrogen and CO2 (Videmšek et al. Citation2009). In addition, a high diversity of red-like cbbL sequences was found in grassland soils close to natural carbon dioxide springs, but there was no difference in the number of OTUs between low and high CO2 concentration (Videmšek et al. Citation2009). The amount of the cbbL genes was found to increase by applying straw and chemical fertilizers (Yuan, Ge, Wu et al. Citation2012), and Selesi et al. (Citation2007) found a positive relationship between the amount of the red-like cbbL genes and SOC content. The study by Tang et al. (Citation2015) revealed the amount of cbbL and 16S rRNA genes were lowest in the soils under Cleistogenes chinensis where SOC and pH were lowest. The number of OTUs showed no difference between the fenced and grazing sites in the cluster Actinobacteria, mainly with Mycobacterium sp. and Solirubrobacter soli (). Also, phototrophic Cyanobacteria was detected in both the fenced and grazing soils. However, phototrophic Chloroflexi was found in only the fenced soils. Previous studies showed that the microbial autotrophic community is significantly affected by edaphic factors (Selesi et al. Citation2005; Tolli & King Citation2005; Nigro & King Citation2007; Videmšek et al. Citation2009; Yuan, Ge, Wu et al. Citation2012). The absence of Chloroflexi in the grazing soils indicated continuous grazing shaping the fixation bacterial communities in the grassland soils.
Disclosure statement
No potential conflict of interest was reported by the authors.
Notes on contributors
Jun Yang obtained his master’s degree from the School of Forestry, Northeast Forestry University of China in 2008. He is now studying in the United Graduate School of Agriculture Science of Ehime University for his doctoral degree under the supervision of Professor Yumei Kang. His research is centered on the restoration of the degraded grassland soil in Inner Mongolia, China
Dr Yumei Kang is a professor in the Faculty of Agriculture and Marine Sciences of Kochi University in Japan. She has coauthored over 50 publications including Soil Science and Plant Nutrition, Water Air & Soil Pollution, Acta Agriculturae Scandinavica Section B – Soil & Plant Science, Global Change Biology and so on. Her current research include: (1) soil and water pollution (recovery and utilization of polluted soil and water) and (2) research on the grassland system degradation mechanism and development of grassland recovery methods.
Additional information
Funding
References
- Akhzari D, Pessarakli M, Eftekhari Ahandani S. 2015. Effects of grazing intensity on soil and vegetation properties in a Mediterranean rangeland. Commun Soil Sci Plant Anal. 46:2798–2806. doi: 10.1080/00103624.2015.1089272
- Beer LL, Boyd ES, Peters JW, Posewitz MC. 2009. Engineering algae for biohydrogen and biofuel production. Curr Opin Biotechnol. 20:264–271. doi: 10.1016/j.copbio.2009.06.002
- Bird SB, Herrick JE, Wander MM, Wright SF. 2002. Spatial heterogeneity of aggregate stability and soil carbon in semi-arid rangeland. Environ Pollut. 116:445–455. doi: 10.1016/S0269-7491(01)00222-6
- Cui XY, Wang YF, Niu HS, Wu J, Wang SP, Schnug E, Rogasik J, Fleckenstein J, Tang YH. 2005. Effect of long-term grazing on soil organic carbon content in semiarid steppes in inner Mongolia. Ecol Res. 20:519–527. doi: 10.1007/s11284-005-0063-8
- Deng L, Zhang ZN, Shangguan ZP. 2014. Long-term fencing effects on plant diversity and soil properties in China. Soil Tillage Res. 137:7–15. doi: 10.1016/j.still.2013.11.002
- Domagała-Świątkiewicz I, Sady W. 2011. Effect of nitrogen fertilization on P, K, Mg, Ca and S content in soil and edible parts of white cabbage. J Elem. 16:177–193.
- Dubeux JCB, Jr., Sollenberger LE, Comerford NB, Scholberg JM, Ruggieri AC, Vendramini JMB, Interrante SM, Portier KM. 2006. Management intensity affects density fractions of soil organic matter from grazed bahiagrass swards. Soil Biol Biochem. 38:2705–2711. doi: 10.1016/j.soilbio.2006.04.021
- Ellis RJ. 1979. The most abundant protein in the world. Trends Biochem Sci. 4:241–244. doi: 10.1016/0968-0004(79)90212-3
- Evangelou VP, Wang J, Phillips RE. 1994. New developments and perspectives on soil potassium quantity/intensity relationships. Adv Agron. 52:173–227. doi: 10.1016/S0065-2113(08)60624-0
- Fan ZL. 2008. The fifth carbon fixation pathway. Chinese J Nature. 30:93. (in Chinese)
- Follett RF, Reed DA. 2010. Soil carbon sequestration in grazing lands: societal benefits and policy implications. Rangeland Ecol Manage. 63:4–15. doi: 10.2111/08-225.1
- Ge TD, Nie SA, Wu JS, Shen JL, Xiao HA, Tong CL, Huang DF, Hong Y, Iwasaki K. 2011. Chemical properties, microbial biomass, and activity differ between soils of organic and conventional horticultural systems under greenhouse and open field management: a case study. J Soils Sediments. 11:25–36. doi: 10.1007/s11368-010-0293-4
- Ge TD, Wu XH, Chen XJ, Yuan HZ, Zou ZY, Li BZ, Zhou P, Liu SL, Tong CL, Brookes P, Wu JS. 2013. Microbial phototrophic fixation of atmospheric CO2 in China subtropical upland and paddy soils. Geochim Cosmochim Acta. 113:70–78. doi: 10.1016/j.gca.2013.03.020
- Gee GW, Bauder JW. 1986. Particle-size analysis. In: Klute A, editior. Methods of soil analysis. Part 1 – physical and mineralogical methods. 2nd ed. SSSA Book Series No. 5. Madison (WI): SSSA and ASA, p. 383–411.
- Gomes L, Arrúe JL, López MV, Sterk G, Richard D, Gracia R, Sabre M, Gaudichet A, Frangi JP. 2003. Wind erosion in a semiarid agricultural area of Spain: the WELSONS project. CATENA. 52:235–256. doi: 10.1016/S0341-8162(03)00016-X
- Herter S, Busch A, Fuchs G. 2002. L-malyl-coenzyme a lyase/β-methylmalyl-coenzyme a lyase from Chloroflexus aurantiacus, a bifunctional enzyme involved in autotrophic CO2 fixation. J Bacteriol. 184:5999–6006. doi: 10.1128/JB.184.21.5999-6006.2002
- Kandeler E, Tscherko D, Bruce KD, Stemmer M, Hobbs PJ, Bardgett RD, Amelung W. 2000. Structure and function of the soil microbial community in microhabitats of a heavy metal polluted soil. Biol Fertil Soils. 32:390–400. doi: 10.1007/s003740000268
- Kang YM. 2012. Influence of grassland degradation on soil and vegetation characteristics in Inner Mongolia, China. Pedologist. 56:332–342.
- Kuo S. 1996. Phosphorus. In: Sparks DL, editor. Methods of soil analysis. Part 3. Chemical methods. SSSA Book Series No. 5. Madison (WI): SSSA and ASA, p. 869–919.
- Liu FH, Conrad R. 2011. Chemolithotrophic acetogenic H2/CO2 utilization in Italian rice field soil. ISME J. 5:1526–1539. doi: 10.1038/ismej.2011.17
- Ludwig JA, Tongway DJ. 1995. Desertification in Australia: an eye to grass roots and landscapes. Environ Monit Assess. 37:231–237. doi: 10.1007/BF00546891
- Miao RH, Jiang DM, Musa A, Zhou QL, Guo MX, Wang YC. 2015. Effectiveness of shrub planting and grazing exclusion on degraded sandy grassland restoration in Horqin sandy land in Inner Mongolia. Ecol Eng. 74:164–173. doi: 10.1016/j.ecoleng.2014.10.004
- Miltner A, Kopinke F, Kindler R, Selesi D, Hartmann A, Kästner M. 2005. Non-phototrophic CO2 fixation by soil microorganisms. Plant Soil. 269:193–203. doi: 10.1007/s11104-004-0483-1
- Nigro LM, King GM. 2007. Disparate distributions of chemolithotrophs containing form IA or IC large subunit genes for ribulose-1,5-bisphosphate carboxylase/oxygenase in intertidal marine and littoral lake sediments. FEMS Microbiol Ecol. 60:113–125. doi: 10.1111/j.1574-6941.2007.00272.x
- Northup BK, Brown JR, Holt JA. 1999. Grazing impacts on the spatial distribution of soil microbial biomass around tussock grasses in a tropical grassland. Appl Soil Ecol. 13:259–270. doi: 10.1016/S0929-1393(99)00039-6
- Nowak ME, Beulig F, von Fischer J, Muhr J, Küsel K, Trumbore SE. 2015. Autotrophic fixation of geogenic CO2 by microorganisms contributes to soil organic matter formation and alters isotope signatures in a wetland mofette. Biogeosciences. 12:7169–7183. doi: 10.5194/bg-12-7169-2015
- Oliver JWK, Machado IMP, Yoneda H, Atsumi S. 2014. Combinatorial optimization of cyanobacterial 2,3-butanediol production. Metab Eng. 22:76–82. doi: 10.1016/j.ymben.2014.01.001
- Phillips L, Burton E. 2005. Nutrient leaching in undisturbed cores of an acidic sandy Podosol following simultaneous potassium chloride and di-ammonium phosphate application. Nutri Cycl Agroecosyst. 73:1–14. doi: 10.1007/s10705-005-6080-8
- Pisciotta JM, Zou YJ, Baskakov IV, Yang C-H. 2010. Light-dependent electrogenic activity of cyanobacteria. PLoS One. 5:e10821. doi: 10.1371/journal.pone.0010821
- Santoro AL, Bastviken D, Gudasz C, Tranvik L, Enrich-Prast A, Thrush S. 2013. Dark carbon fixation: an important process in lake sediments. PLoS ONE. 8:e65813. doi: 10.1371/journal.pone.0065813
- Šantrůčková H, Bird MI, Elhottová D, Novák J, Picek T, Šimek M, Tykva R. 2005. Heterotrophic fixation of CO2 in soil. Microb Ecol. 49:218–225. doi: 10.1007/s00248-004-0164-x
- Schlesinger WH. 1977. Carbon balance in terrestrial detritus. Annu Rev Ecol Syst. 8:51–81. doi: 10.1146/annurev.es.08.110177.000411
- Schlesinger WH, Andrews JA. 2000. Soil respiration and the global carbon cycle. Biogeochemistry. 48:7–20. doi: 10.1023/A:1006247623877
- Selesi D, Pattis I, Schmid M, Kandeler E, Hartmann A. 2007. Quantification of bacterial RubisCO genes in soils by cbbL targeted real-time PCR. J Microbiol Methods. 69:497–503. doi: 10.1016/j.mimet.2007.03.002
- Selesi D, Schmid M, Hartmann A. 2005. Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl Environ Microbiol. 71:175–184. doi: 10.1128/AEM.71.1.175-184.2005
- Shimmel SM. 1987. Dark fixation of carbon dioxide in an agricultural soil. Soil Sci. 144:20–23. doi: 10.1097/00010694-198707000-00004
- Song CS. 2006. Global challenges and strategies for control, conversion and utilization of CO2 for sustainable development involving energy, catalysis, adsorption and chemical processing. Catal Today. 115:2–32. doi: 10.1016/j.cattod.2006.02.029
- Steffens M, Kölbl A, Totsche KU, Kögel-Knabner I. 2008. Grazing effects on soil chemical and physical properties in a semiarid steppe of Inner Mongolia (P.R. China). Geoderma. 143:63–72. doi: 10.1016/j.geoderma.2007.09.004
- Su YZ, Zhao HL, Zhang TH, Zhao XY. 2004. Soil properties following cultivation and non-grazing of a semi-arid sandy grassland in northern China. Soil Tillage Res. 75:27–36. doi: 10.1016/S0167-1987(03)00157-0
- Tabita FR. 1999. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res. 60:1–28. doi: 10.1023/A:1006211417981
- Tanaka S, Funakawa S, Kaewkhongkha T, Yonebayashi K. 1998. Labile pools of organic matter and microbial biomass in the surface soils under shifting cultivation in northern Thailand. Soil Sci Plant Nutr. 44:527–537. doi: 10.1080/00380768.1998.10414476
- Tang ZX, Fan FL, Wan YF, Wei W, Lai LM. 2015. Abundance and diversity of RuBisCO genes responsible for CO2 fixation in arid soils of Northwest China. Pedosphere. 25:150–159. doi: 10.1016/S1002-0160(14)60085-0
- Tolli J, King GM. 2005. Diversity and structure of bacterial chemolithotrophic communities in pine forest and agroecosystem soils. Appl Environ Microbiol. 71:8411–8418. doi: 10.1128/AEM.71.12.8411-8418.2005
- Usui N, Ikenouchi M. 1997. The biological CO2 fixation and utilization project by RITE (1). Highly-effective photobioreactor system. Energy Convers Manage. 38:S487–S492. doi: 10.1016/S0196-8904(96)00315-9
- Vadas PA, Busch DL, Powell JM, Brink GE. 2015. Monitoring runoff from cattle-grazed pastures for a phosphorus loss quantification tool. Agric Ecosyst Environ. 199:124–131. doi: 10.1016/j.agee.2014.08.026
- Van Gestel M, Merckx R, Vlassak K. 1996. Spatial distribution of microbial biomass in microaggregates of a silty-loam soil and the relation with the resistance of microorganisms to soil drying. Soil Biol Biochem. 28:503–510. doi: 10.1016/0038-0717(95)00192-1
- Videmšek U, Hagn A, Suhadolc M, Radl V, Knicker H, Schloter M, Vodnik D. 2009. Abundance and diversity of CO2-fixing bacteria in grassland soils close to natural carbon dioxide springs. Microb Ecol. 58:1–9. doi: 10.1007/s00248-008-9442-3
- Walkley A. 1947. A critical examination of a rapid method for determining organic carbon in soils-effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci. 63:251–264. doi: 10.1097/00010694-194704000-00001
- Wang CT, Long RJ, Wang QL, Jing ZC, Shi JJ, Du YG, Cao GM. 2008. Changes in soil organic carbon and microbial biomass carbon at different degradation successional stages of alpine meadows in the headwater region of three rivers in China. Chin J Appl Environ Biol. 14:225–230. (in Chinese).
- Wang D, Liu Y, Wu GL, Ding LM, Yang Z, Hao HM. 2015. Effect of rest-grazing management on soil water and carbon storage in an arid grassland (China). J Hydrol. 527:754–760. doi: 10.1016/j.jhydrol.2015.05.036
- Wang W, Fang JY. 2009. Soil respiration and human effects on global grasslands. Glob Planet Change. 67:20–28. doi: 10.1016/j.gloplacha.2008.12.011
- Watson GMF, Tabita FR. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Micorobiol Lett. 146:13–22. doi: 10.1111/j.1574-6968.1997.tb10165.x
- Wen HY, Niu DC, Fu H, Kang J. 2013. Experimental investigation on soil carbon, nitrogen, and their components under grazing and livestock exclusion in steppe and desert steppe grasslands, Northwestern China. Environ Earth Sci. 70:3131–3141. doi: 10.1007/s12665-013-2376-1
- Whitehead DC. 2000. Nutrient elements in grassland: soil-plant-animal relationships. Wallingford (UK): CABI, p. 185.
- Wu XH, Ge TD, Yuan HZ, Li BZ, Zhu HH, Zhou P, Sui FG, O’Donnell AG, Wu JS. 2014. Changes in bacterial CO2 fixation with depth in agricultural soils. Appl Microbiol Biotechnol. 98:2309–2319. doi: 10.1007/s00253-013-5179-0
- Xie YZ, Wittig R. 2004. The impact of grazing intensity on soil characteristics of Stipa grandis and Stipa bungeana steppe in northern China (autonomous region of Ningxia). Acta Oecol. 25:197–204. doi: 10.1016/j.actao.2004.01.004
- Yousuf B, Keshri J, Mishra A, Jha B. 2012. Application of targeted metagenomics to explore abundance and diversity of CO2-fixing bacterial community using cbbL gene from the rhizosphere of Arachis hypogaea. Gene. 506:18–24. doi: 10.1016/j.gene.2012.06.083
- Yuan HZ, Ge TD, Chen CY, O’Donnell AG, Wu JS. 2012. Significant role for microbial autotrophy in the sequestration of soil carbon. Appl Environ Microbiol. 78:2328–2336. doi: 10.1128/AEM.06881-11
- Yuan HZ, Ge TD, Wu XH, Liu SL, Tong CL, Qin HL, Wu MN, Wei WX, Wu JS. 2012. Long-term field fertilization alters the diversity of autotrophic bacteria based on the ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO) large-subunit genes in paddy soil. Appl Microbiol Biotechnol. 95:1061–1071. doi: 10.1007/s00253-011-3760-y
- Yuan HZ, Ge TD, Zou SY, Wu XH, Liu SL, Zhou P, Chen XJ, Brookes P, Wu JS. 2013. Effect of land use on the abundance and diversity of autotrophic bacteria as measured by ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO) large subunit gene abundance in soils. Biol Fertil Soils. 49:609–616. doi: 10.1007/s00374-012-0750-x
- Zuo XA, Zhang J, Zhou X, Zhao XY, Wang SK, Lian J, Lv P, Knops J. 2015. Changes in carbon and nitrogen storage along a restoration gradient in a semiarid sandy grassland. Acta Oecol. 69:1–8. doi: 10.1016/j.actao.2015.08.004
