Abstract
A series of multifunctional compounds (MFCs) 1a–1e based on 1,8-naphthalimide and [12]aneN3 building blocks were designed and synthesized. They were used as not only fluorescent probes for recognition of Cu2+ ions but also as non-viral gene vectors for DNA and RNA delivery. Furthermore, their complexes with Cu2+ (1-Cu) could also selectively stain lysosome in HeLa cells. In order to achieve high performance multifunctional materials, structure-performance relationship of MFCs 1a–1e was studied. It was found that MFCs 1a–1e exhibited highly selective fluorescence turn-off for Cu2+, without interference by other metal ions in aqueous solution. The fluorescence emission of 1a–1e was quenched by a factor of 10-fold, 47-fold, 6-fold, 64-fold, and 15-fold respectively in the presence of Cu2+ ions. Due to high sensitivity, good water solubility, and low cytotoxicity, MFCs 1a–1d were successfully applied in the recognition of Cu2+ and selectively staining lysosome in HeLa cells. Most importantly, MFCs 1a and 1b had excellent HeLa cell selectivity in RNA delivery, and their performances were far better than lipofectamine 2000 and 25 kDa PEI.
1. Introduction
In recent years, considerable efforts have been paid to develop high performance probes for detecting transition-metal ions or monitoring cellular pH changes (Richter et al., Citation2008; Wu et al., Citation2014; Cotruvo et al., Citation2015; Yue et al., Citation2017; Alam et al., Citation2018; Yang et al., Citation2018). Cu2+ as the third most abundant of the essential nutrients plays an important role in many neurological diseases, including Menke’s disease and prion disease (Karr & Szalai, Citation2007; Zhang et al., Citation2009; Tewari, Citation2010; Eshak et al., Citation2018; Siotto & Squitti, Citation2018). Furthermore, abnormal cellular pH values are also related with some common diseases such as Alzheimer’s diseases and cancer (Izumi et al., Citation2003; Junyan & Kevin, Citation2010). Therefore, sensing of Cu2+ concentration level and monitoring of pH change would give us important information for disease diagnosis. The organelles in eukaryotic cells participate in life activities within various pH ranges. For example, the pH value of lysosome lumen ranges from 4.0 to 6.0, while the cytoplasm is about 7.2. Therefore, many fluorescence probes for the recognition of lysosome or detection of Cu2+ have been reported (Jeppe et al., Citation2013; Khan et al., Citation2015; Behnood et al., Citation2016; Gu et al., Citation2018; Li et al., Citation2018; Shi et al., Citation2018). However, the same compounds used for recognition of both Cu2+ and lysosome are relatively rare. The structure-property relationship of these materials in recognition systems has been seldom investigated (Shuqi et al., Citation2011; Zhu et al., Citation2011; Hao et al., Citation2012). This relationship is usually very important for the designation of high performance fluorescence probes.
It is well known that the development of safe and efficient gene delivery systems is important for gene therapy (Verma & Somia, Citation1997; Dunbar et al., Citation2018; Peng, Citation2018). Viral vectors such as retroviruses and adenoviruses are significantly more efficient for in vitro and in vivo gene delivery (Lundstrom, Citation1995; Marshall, Citation1999; Bloom et al., Citation2018). However, their clinical applications are limited due to high immunogenicity and limited loading capacity (Thomas et al., Citation2003; Luo et al., Citation2015). In contrast, non-viral vectors such as cationic lipids, cationic polymers, and organic functional molecules have received more and more attention due to their easy preparation, low immunogenicity, and good biodegradability (Posocco et al., Citation2013; Yi et al., Citation2014; Ramamoorth & Narvekar, Citation2015; Yang et al., Citation2015; Hu et al., Citation2016; Wang et al., Citation2016; Hardee et al., Citation2017; Wang et al., Citation2017; Luo et al., Citation2018; Tan et al., Citation2018; Li et al., Citation2019; Wang et al., Citation2019). Among these non-viral vectors, organic functional molecules such as organic fluorescent compounds not only show high transfection efficiency but also exhibit good tracking ability (You et al., Citation2014; Gao et al., Citation2015). However, the usage of these organic functional molecules both in non-viral vectors and Cu2+/pH probes has never been reported.
In recent years, we have developed a series of small organic molecules containing rigid fluorescent moieties (naphthalimide, BODIPY, and coumarin) and [12]aneN3 units (Yue et al., Citation2015; Zhang et al., Citation2015; Gao et al., Citation2016; Citation2016; Citation2016; Ding et al., Citation2017). It was found that [12]aneN3 unite exhibited multiple functions such as good water solubility, strong coordination ability with metal ions and excellent DNA binding ability. Therefore, some were used as non-viral vectors to deliver DNA into target cells (Yue et al., Citation2015; Gao et al., Citation2016; Ding et al., Citation2017) and some were applied in fluorescent probes for recognition of metal ions or small bioactive molecules (Zhang et al., Citation2015; Gao et al., Citation2016; Citation2016). However, a functional molecule serving simultaneously as a gene vector as well as the fluorescence probe has not been investigated so far. Furthermore, as the non-viral gene vectors, we mainly focused on DNA delivery, while RNA delivery mediated by these materials has never been investigated.
In this study, we designed and synthesized a series of multifunctional compounds (MFCs) 1a–1e (Scheme 1). They consist of three parts: fluorophore unit naphthalimide, linker benzoyl amine derivative and hydrophilic units triazole and [12]aneN3. The main difference lies in the length of linker or the number of hydrophilic units. 1a, 1b, 1c, and 1d contain different linkers: N,N′- dimethyl-1,2-ethanediamine, ethylenediamine, piperazine and 1,6-hexylenediamine, respectively. Compared to other compounds, 1e linked with 1,6-hexylenediamine only contain one triazole and one [12]aneN3 unit. These structural differences will be beneficial to structure-property relationship investigation. Therefore, their performance on recognition of Cu2+ ions and lysosome as well as gene delivery was systematically investigated. The results show that these multifunctional compounds can not only serve as fluorescent probes for detection of Cu2+ ions and lysosome but also serve as non-viral vectors for DNA and RNA delivery. Most importantly, the structure-property relationship investigation will give further insight to design and synthesize new types of high performance multifunctional compounds.
2. Materials and methods
2.1. Materials and instruments
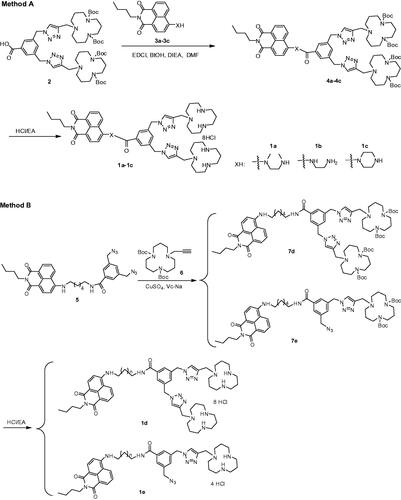
The solvents used for reaction such as tetrahydrofuran (THF) and N,N-dimethylformamide (DMF) were purified by distillation before use. The solvents used for purification such as dichloromethane (DCM), methanol (MeOH), petroleum ether (PE) and ethyl acetate (EA) were directly used without any purification. 1-hydroxybenzotriazole hydrate (HOBt), copper sulfate (CuSO4), diisopropylethylamine (DIEA), 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDCI) and sodium ascorbate (Vc-Na) were purchased from Beijing Ouhe Technology Co. Ltd. (Beijing, China). 3,5-bis((4-((5,9-bis(tert-butoxycarbonyl)- 1,5,9-triazacyclododecan-1-yl)methyl) -1H-1,2,3-triazol-1-yl)methyl) benzoic acid (compound 2), 4-amino-1,8-naphthalimide derivatives (compounds 3a, 3b, and 3c), 3,5- bis(azidomethyl)-N-(6-(2-butyl-1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-6-ylamino)hexyl) benzamide (compound 5) and ditert-butyl 9-(prop-2-ynyl)-1,5,9-triazacyclododecane- 1,5-dicarboxylate (compound 6) were prepared according to the literature (Gao et al., Citation2015; Citation2016; Citation2018).
1H NMR and 13C NMR spectra were collected using a Bruker Avance spectrometer. Mass spectra were obtained on a Waters Quattro Mocro spectrometer and high resolution mass spectra were acquired on a Waters LCT Premier XE spectrometer. The infrared spectra were acquired on a Nicolet 380 spectrometer. Fluorescence spectra were measured on a Hitachi F-4500 Fluorescence Spectrophotometer. Hydrodynamic diameters and Zeta Potentials were collected using a Nano-ZS 3600 ZetaPlus Partical Size and Zeta Potential Analyzer.
2.2. Synthesis of multifunctional compounds
2.2.1. Synthesis of compounds 4a–4c
General procedure: 3,5-bis((4-((5,9-bis(tert-butoxycarbonyl)-1,5,9-triazacyclododecan-1-yl) methyl)-1H-1,2,3-triazol-1-yl)methyl)benzoic acid 2 (1.0 mmol), EDCI (1.1 mmol), DIEA (2.0 mmol) and HOBt (1.1 mmol) were added in anhydrous N,N-dimethylformamide (20 mL) under argon atmosphere and stirred for 0.5 h. 4-Amino-1,8-naphthalimide derivatives 3a/3b/3c (1.0 mmol) was then added slowly to the above reaction mixture, and continued to stir for 12–24 h. After the completion of reaction (monitor by TCL), ice water (50 mL) was added to the mixture. The aqueous phase was extracted with ethyl acetate. The combined organic phase was dried over anhydrous Na2SO4, filtered, and the solvent was evaporated under reduced pressure. The crude material was purified by column chromatography (DCM/MeOH = 10/1) to give products 4a–4c as yellow solid. The detailed analytical data is listed in the Supplementary Information.
2.2.2. Synthesis of compounds 7d and 7e
3,5-bis(Azidomethyl)-N-(6-(2-butyl-1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-6-ylamino)hexyl)benzamide 5 (1.0 mmol) and ditert-butyl 9-(prop-2-ynyl)-1,5,9-triazacyclododecane- 1,5-dicarboxylate 6 (2.0 mmol) were dissolved in the mixture of tetrahydrofuran (10 mL) and water (5 mL). Sodium ascorbate (0.1 mmol) and copper sulfate (0.05 mmol) were added dropwise in 1 mL of water. The mixture was stirred for 8 h at room temperature. After completion of reaction, the solvent was removed by reduced pressure. The solid was washed with CH2Cl2 (3 × 20 mL), the combined organic layer was evaporated under reduced pressure. The crude material was purified by column chromatography (DCM/MeOH = 10/1) to give products 7d and 7e as yellow solid. The detailed analytical data are listed in the Supplementary Information.
2.2.3. Synthesis of multifunctional organic compounds 1a–1e
General procedure: compound 4a/4b/4c/7d/7e (0.1 mmol) was added to a saturated HCl/ethyl acetate solution (5 mL). After string for 1 h, the resulting suspension was filtrated, washed with ether 3 times and dried in vacuum at 60 °C for 24 h to give MFCs 1a–1e as yellow solid.
1a: 95%; 1H NMR (400 MHz, D2O) δ 8.07 - 7.77 (m, 4 H), 7.66 - 7.49 (m, 1 H), 7.31 - 7.17 (m, 1 H), 7.11 - 6.98 (m, 1 H), 6.81 - 6.41 (m, 3 H), 5.30 (s, 2 H), 5.11 (s, 2 H), 4.18 - 4.15 (m, 4 H), 3.73 - 3.42 (m, 5 H), 3.33 - 2.87 (m, 28 H), 2.77 (s, 1 H), 2.36 - 1.91 (m, 14 H), 1.31 (d, J = 6.9 Hz, 1 H), 1.10 (s, 3 H), 0.71 - 0.66 (m, 3 H); 13C NMR (101 MHz, D2O) δ 176.24, 171.88, 171.37, 164.42, 163.66, 154.80, 136.46, 136.23, 135.92, 132.44, 131.03, 129.45, 128.75, 127.95, 126.74, 126.52, 125.01, 123.96, 123.15, 121.04, 114.39, 113.95, 113.04, 112.66, 73.17, 60.25, 58.07, 57.47, 53.26, 53.03, 48.92, 48.24, 47.47, 45.46, 42.35, 42.05, 41.16, 39.98, 37.92, 33.69, 29.49, 20.61, 20.19, 19.81, 17.97, 17.04, 13.27; IR (KBr, cm−1): 3428.01, 2959.04, 2650.00, 1684.94, 1641.57, 1581.93 1451.81, 1384.04, 1356.93, 1234.94, 1053.31, 790.36; HR-MS (ES+) calcd. For C53H77N15O3 (M + H)+: 972.6334, found 972.6427.
1b: 93%; 1H NMR (400 MHz, D2O) δ 7.99 (d, J = 10.7 Hz, 2 H), 7.44 (s, 2 H), 7.35 - 6.95 (m, 4 H), 6.70 - 6.46 (m, 1 H), 5.81 - 5.78 (m, 1 H), 5.40 (s, 4 H), 3.94 (s, 4 H), 3.53 - 2.75 (m, 30 H), 2.06 (s, 4 H), 1.92 (s, 12 H), 1.04 (s, 2 H), 0.94 - 0.82 (m, 2 H), 0.56 (t, J = 7.1 Hz, 3 H); 13C NMR (101 MHz, D2O) δ 176.38, 168.98, 164.30, 163.40, 149.85, 136.44, 136.16, 134.84, 133.60, 131.38, 130.12, 128.04, 127.56, 127.30, 123.44, 119.56, 118.24, 106.46, 103.17, 57.48, 53.32, 48.91, 47.49, 42.82, 42.04, 41.14, 39.74, 38.81, 29.56, 20.54, 20.16, 19.82, 17.97, 16.95, 13.20; IR (KBr, cm−1): 3425.30, 2956.33, 2650.00, 1682.23, 1638.86, 1581.93 1546.69, 1392.17, 1356.93, 1234.07, 1121.08, 1058.73, 776.81; HR-MS (ES+) calcd. For C53H73N15O3 (M + H)+: 944.6021, found 944.6103.
1c: 97%; 1H NMR (400 MHz, D2O) δ 8.25 (s, 2 H), 7.72 - 7.39 (m, 3 H), 7.43 - 7.30 (m, 3 H), 7.04 (s, 1 H), 6.50 (s, 1 H), 5.62 (s, 4 H), 4.40 (s, 4 H), 3.76 (s, 2 H), 3.45 - 3.25 (m, 28 H), 2.88 - 2.76 (m, 4 H), 2.14 - 2.12 (m, 12 H), 1.18 - 1.06 (m, 4 H), 0.67 (s, 3 H); 13C NMR (101 MHz, D2O) δ 170.32, 163.70, 163.25, 154.49, 136.23, 136.04, 135.65, 129.45, 128.20, 126.83, 124.13, 120.87, 114.51, 53.18, 48.97, 47.09, 41.73, 40.85, 29.48, 19.89, 17.66 (s, 8 H), 13.25 (s, 3 H); IR (KBr, cm−1): 3428.01, 2956.33, 2753.01, 1693.07, 1649.70, 1587.35, 1454.52, 1386.75, 1237.65, 1053.31, 787.65; ESI-MS: HR-MS (ES+) calcd. For C53H75N15O3 (M + H)+: 970.6177, found 970.6231.
1d:88%; 1H NMR (400 MHz, D2O) δ 7.77 (s, 2 H), 7.51 (d, J = 6.9 Hz, 1 H), 7.47 (d, J = 7.5 Hz, 1 H), 7.28 (s, 2 H), 7.22 (d, J = 8.1 Hz, 1 H), 7.09 (s, 1 H), 6.85 (t, J = 7.1 Hz, 1 H), 5.76 (d, J = 6.9 Hz, 1 H), 5.26 (s, 4 H), 3.64 (s, 4 H), 3.42 (s, 2 H), 3.16 - 2.99 (m, 18 H), 2.84 (s, 2 H), 2.61 (s, 8 H), 2.02 (s, 4 H), 1.77 (s, 8 H), 1.40 (s, 4 H), 1.19 (s, 6 H), 1.05 - 0.98 (m, 2 H), 0.62 (t, J = 7.1 Hz, 3 H); 13C NMR (101 MHz, D2O) δ 168.23, 164.70, 163.71, 150.41, 138.48, 136.03, 135.34, 134.07, 130.95, 130.56, 128.19, 127.78, 126.97, 126.96, 123.69, 119.90, 118.71, 106.06, 103.34, 53.24, 48.76, 47.31, 43.02, 42.76, 41.77, 40.70, 39.92, 29.76, 28.52, 27.67, 26.22, 26.07, 20.00, 19.94, 19.00, 13.30; IR (KBr, cm−1): 3409.04, 2929.22, 2853.31, 2750.30, 1676.81, 1638.86, 1579.22, 1543.98, 1457.23, 1432.83, 1392.17, 1356.93, 1245.78, 1167.17, 1115.66, 1053.31, 782.23; HR-MS (ES+) calcd. For C55H81N15O3 (M + H)+: 1000.6647, found 1000.6703.
1e:71%;1H NMR (400 MHz, CD3SOCD3) δ 11.23 (s, 1 H), 9.52 (s, 4 H), 8.76 (d, J = 8.3 Hz, 1 H), 8.64 (s, 1 H), 8.51 (s, 1 H), 8.42 (d, J = 6.9 Hz, 1 H), 8.28 - 8.23 (m, 1 H), 7.80 (d, J = 8.7 Hz, 2 H), 7.66 (t, J = 7.6 Hz, 1 H), 7.46 - 7.43 (m, 1 H), 6.76 (d, J = 8.4 Hz, 1 H), 5.74 - 5.67 (m, 2 H), 4.53 (s, 2 H), 4.42 (s, 2 H), 4.20 (s, 6 H), 4.04 - 3.97 (m, 2 H), 3.45 (s, 2 H), 3.26 - 3.16 (m, 8 H), 2.16 - 2.14 (m, 4 H), 1.87 - 1.70 (m, 4 H), 1.64 - 1.26 (m, 12 H), 0.91 (t, J = 7.3 Hz, 3 H); 13C NMR (101 MHz, DMSO) δ 165.74, 164.19, 163.35, 151.13, 137.05, 136.09, 134.71, 131.06, 129.89, 129.23, 127.24, 127.09, 124.59, 122.24, 120.57, 107.82, 104.15, 53.58, 43.19, 30.27, 29.51, 28.22, 26.82, 26.73, 20.27, 14.19; IR (KBr, cm−1): 3400.90, 2923.80, 2853.31, 2099.70, 1679.52, 1638.86, 1579.22, 1546.69, 1462.65, 1397.59, 1384.04, 1354.22, 1272.89, 1237.65, 1115.66, 1056.02, 782.23; HR-MS (ES+) calcd. For C43H58N12O3 (M + H)+: 791.4755, found 791.4841.
2.3. UV or fluorescent spectral measurements
The stock solutions of multifunctional organic compounds (MFCs) 1a–1e (1 mM) and metal ions (10 mM) were prepared in tri-distilled water, and stored at 4 °C for use. The metal salts were AgClO4·H2O, Ca(ClO4)2·4H2O, Cd(ClO4)3·6H2O, Co(ClO4)2·6H2O, Fe(ClO4)2, 3H2O, Hg(ClO4)2·3H2O, KClO4, LiClO4·3H2O, Mg(ClO4)2·6H2O, NaClO4·H2O, Ni(ClO4)2·6H2O, Pb(ClO4)2·3H2O,Zn(ClO4)2·6H2O and Cu(ClO4)2·6H2O. The fluorescence emission and ultraviolet absorption experiments were carried out in Tris-HCl buffer (1 mM, pH 7.2). Test solution was prepared by placing MFCs 1a–1e, Tris-HCl buffer solution and an appropriate volume of each analyte into 3 mL of cuvette. After equilibration for 2 min, an ultraviolet absorption and fluorescence emission spectrum were recorded at 25 °C.
2.4. Cell culture and fluorescence imaging
Cell culture: HeLa cells ((a human cervical carcinoma cell line) were cultured with DMEM containing 100 units/mL penicillin sulfate and streptomycin, medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2 for 24 h.
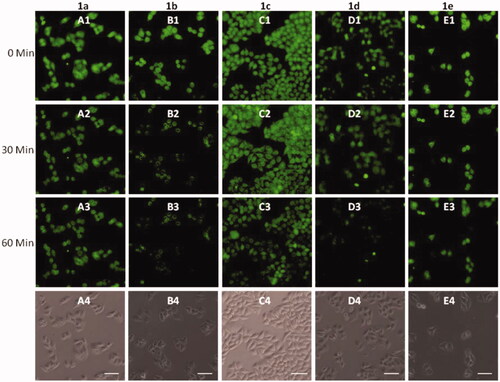
Cell imaging (Cu2+ recognition): cells were seeded in glass bottom cell culture dish (8 × 104 cells), and incubated with 20 μM of MFCs 1a–1e for 0.5 h. After incubation, the cells were washed with PBS 4 times and imaged under a fluorescence microscope. Then 10 equivalent of Cu(ClO4)2 was added to the cells, and fluorescence images were taken one time per 15 minutes under a LEICA DMI8 Inverted Fluorescence Microscope. Fluorescence images were obtained using a 10 × objective lens. Cell images were processed and analyzed using the Image-Pro Plus software. Three repeats were conducted for each sample.
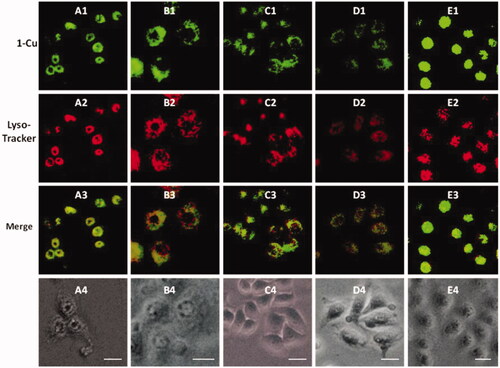
Lysosome imaging: The cells were seeded in 24-well plates at 8 × 104 cells per well and grew for 24 h. After removing the medium, 1a–1e/Cu complexes (20 μM) were added and incubated for 30 minutes in DMEM medium. Then the medium was replaced with fresh medium containing Lyso-Tracker Red (10 μM) and incubated for another 30 minutes. The samples were then imaged by a Inverted Fluorescence Microscope after washing with cell culture medium. Lyso-Tracker Red images were observed in the red channel, 1a–1e/Cu images were observed in the green channel with a 20 × objective lens. Cell images were processed and analyzed using the Image-Pro Plus software. Three repeats were conducted for each sample.
2.5. Dynamic light scattering (DLS) and scanning electron microscope (SEM)
The complexes of MFCs 1a–1e with DNA or RNA were prepared by adding 0.9 μL of pUC18 DNA (200 μg/mL) or 0.7 μL of siRNA (264 μg/mL, 5′-UUCUCCGAACGUGUCACGUTT-3′) to the appropriate volume of the stock solutions of 1a–1e. Then the complex solution was vortexed for 30 s and then diluted up to 0.5 mL by tri-distilled water. The zeta potentials and the hydrodynamic diameters were measured using Nano-ZS 3600 ZetaPlus Partical Size and Zeta Potential Analyzer. The complexes of 1b (20 µM) with RNA or DNA were added dropwise to the silicon slice. The slice was then dried in a vacuum oven at room temperature for 8 h before observation.
2.6. Cell uptake of 1a–1e/RNA (DNA) complexes
The cellular uptake of the complexes of MFCs 1a–1e with Cy5-labeled siRNA or dsDNA was obtained by fluorescence microscopy. HeLa, HepG2 and U2Os cells were cultured with DMEM medium, and MC3T3-E1 cells were cultured with α-MEM medium. The cells were seeded in 24-well plates at 8 × 104 cells per well and grew for 24 h. After washing with DMEM, the cells were incubated with freshly prepared complexes of MFCs 1a–1e with Cy5-RNA (9 μg/mL) or Cy5-DNA (9 μg/mL) and the controls (500 µL). After 4 h of incubation, the cells were washed for 6 times with PBS buffer. Cy5-labeled RNA or DNA was observed in the red channel, and MFCs 1a–1e was observed in the green channel using a Fluorescence Microscope with a 10 × objective lens.
2.7. Cytotoxicity experiment
The cytotoxicity of 1a–1e/RNA or 1a–1e/DNA complexes toward different cell lines (HeLa, U2Os, HepG2, and MC3T3-E1 cells) was tested by MTT assays. The cells were seeded on 96-well plates at densities of 5 × 103 cells in 100 μL α-MEM/DMEM medium. After culture for 24 h, the complexes (1a–1e/RNA or 1a–1e/DNA) were added to cells at various concentrations (10 μM, 15 μM, 20 μM, and 25 μM). After incubation for 4 h, the medium was replaced with 200 μL of fresh medium containing 10% FBS and cells were cultured for another 48 h. Subsequently, 20 µL of MTT (5 mg/mL) solution in PBS was added to each well for additional 4 h incubation. The MTT medium was replaced by 200 µL of DMSO. The absorbance was measured using a microplate spectrophotometer at a wavelength of 490 nm. The cells treated without any complexes were used as controls. The relative viability of the cells was calculated based on the data of five parallel tests by comparing to the controls.
3. Results and discussion
3.1. Synthesis of the MFCs 1a–1e
In this report, five multifunctional compounds were synthesized by two methods (Scheme 1). The compounds linked with short diamine chains (1a, 1b, and 1c, XH with N,N′-Dimethylethylenediamine, ethylenediamine, and piperazine respectively) were easily synthesized according to Method A, and the synthetic route consisted of two steps. Compound 4 was synthesized through acylation of 3 with compound 2 in the presence of (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) (EDC·HCl) and N-hydroxybenzotriazole (HOBt). The title compounds 1a, 1b, and 1c were prepared by de-protection of 4 under acidic condition. The compounds linked with C-6 alky chain (1d with two [12]aneN3 units and 1e with one [12]aneN3 unit) were synthesized according to Method B. Reaction of 5 with propargyl [12]aneN3 6 through click cycloaddition yielded key intermediates 7d and 7e. Further de-protection of Boc under acidic condition resulted in title compounds 1d and 1e. All new compounds were fully characterized by 1H NMR, 13C NMR, IR and MS.
3.2. Spectroscopic properties of 1a–1e
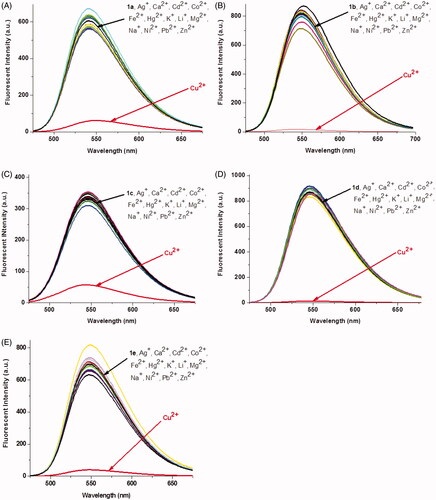
First, the absorption and fluorescence properties of 1a–1e were measured in the water-Tris-HCl buffer (1 mM, pH 7.2). Each compound exhibited a broad absorption peak in the range of 300–500 nm (Figure S1(A)). Compared to other compounds, the maximum absorption wavelength of 1c linked with piperazine shifted toward shorter wavelength, which may be caused by the larger steric hindrance. The fluorescence spectra of 1a–1e showed that there are no obvious changes in fluorescence emission spectra, the maximum emission wavelength of 1a–1e appeared at 542 nm, 550 nm, 546 nm, 546 nm, and 546 nm, respectively (Figure S1(B)). Then, the fluorescence spectra changes of 1a–1e were monitored in the presence of different metal ions (Ag+, Ca2+, Cd2+, Co2+, Fe2+, Hg2+, K+, Li+, Mg2+, Na+, Ni2+, Pb2+, Zn2+, and Cu2+). As shown in , these five materials exhibited high selectivity toward Cu2+ in aqueous solution. The fluorescence intensities of 1a–1e were decreased in different degrees after the addition of Cu2+, about 10-fold, 47-fold, 6-fold, 64-fold, and 15-fold, respectively (Table S1). However, the addition of the other metal ions produced a negligible fluorescence change (). These results suggest that the linker between the naphthalimide unit and the benzoic acid unit has a great influence on the fluorescence recognition of copper ions. The probe linked with a longer alkyl chain exhibited stronger recognition ability to copper ions (1d > 1b). When a bulk group such as N,N′-dimethyl-1,2-ethanediamine or piperazine was used as a linker, the ability to recognize copper ions was decreased dramatically (1b > 1a, 1b > 1c), which may be induced by the steric hindrance effect of substituent group. Besides, the fluorescence changes were also affected by the number of triazole and [12]aneN3. The probes with two triazole and [12]aneN3 units showed stronger Cu2+ recognition ability than these with one unit (1d > 1e). Furthermore, the metal ions competition experiments were also carried out. As shown in Figure S2, the fluorescence emission of compound 1b was nearly quenched after addition of Cu2+ in the presence of other interfering metal ions (except Hg2+), which exhibited its good ability for Cu2+ recognition.
Figure 1. Fluorescence changes of 1a–1e (1 × 10−5M, Tris-HCl buffer, 1 mM) upon the addition of 5 equiv of various metal ions. The excited wavelength of 1a–1e: 465.0 nm, 465.0 nm, 410.0 nm, 465.0 nm, and 465.0 nm, respectively.
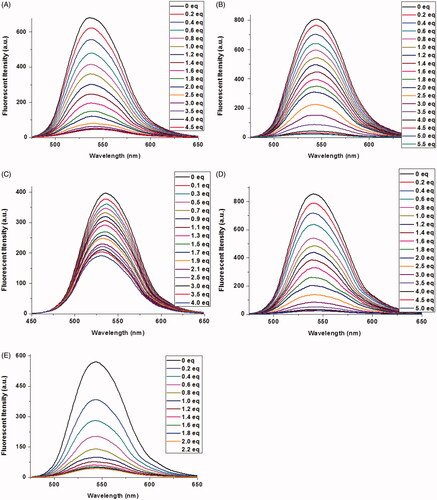
The fluorescence titrations of 1a–1e with Cu2+ were performed in 1 mM Tris-HCl buffer solution. As shown in , the addition of Cu2+ to 1a–1e led to a large quenching effect with the maximum emission at 542 nm, 550 nm, 546 nm, 546 nm, and 546 nm, respectively. A good linear relationship was found between Cu2+ concentration and fluorescent intensities of 1a–1e (1a–1d: 0–2.0 equiv. of Cu2+, 1e: 0–1.0 equiv. of Cu2+). According the formula (LOD = 3σ/k), the limit of detection of 1a–1e was calculated to be 1.21 × 10−8M, 7.48 × 10−9M, 4.40 × 10−8M, 1.23 × 10−9M and 2.36 × 10−9M, respectively (Figure S3). These results indicated that reducing steric hindrance or increasing the length of alkyl chain between naphthalimide unit and benzoic acid unit was beneficial to lower the detection limits of Cu2+. It should be noticed that the limits of detection toward Cu2+by these probes (except 1c) are lower than our previous reports, and far lower than the limit of Cu2+ ions in drinking water (Zhong et al., Citation2013; Zhang et al., Citation2015).
Figure 2. Fluorescence spectra of 1a–1e (10 μM) upon the addition of Cu2+ in aqueous solution (1 mM Tris-HCl, pH 7.2). The excited wavelength of 1a–1e: 465.0 nm, 465.0 nm, 410.0 nm, 465.0 nm, and 465.0 nm, respectively.
To confirm the stoichiometry between 1 and Cu2+, Job’s plot analyses were carried out (Figure S4).
The results indicated that 1a–1d and Cu2+ formed 1:2 stoichiometrical complexes with association constants of Kassoc = 8.2 × 109 M−2, 2.2 × 1010 M−2, 1.1 × 1010 M−2, and 4.4 × 1010 M−2, respectively. Compound 1e and Cu2+ formed a 1:1 stoichiometrical complex with association constant of Kassoc = 3.0 × 1010 M−2. These Job’s plot analyses were in agreement with the fluorescence titration analyses. Furthermore, 1H NMR titration of probe 1b with Cu2+ in DMSO-d6 was undertaken to determine the complexation mode of 1-Cu. As shown in Figure S5, some significant spectral changes are observed in the 1H NMR spectra on addition of Cu2+ ions. For the chromophore naphthalimide region, an amino hydrogen atom (NH) of the naphthalimide undergoes an upfield shift by 8.98–8.69 ppm, while the aromatic proton (H4) shift by 6.81–6.85 ppm. Similarly, in the presence of Cu2+, the aromatic protons of the triazole (H9 and H10) are downfield shifted by 0.05 ppm. These spectral changes suggest that the amino group of naphthalimide and the nitrogen atom of triazole moieties should be involved in the coordination with Cu2+ ions. Based on the titration profile, Job’s plot and structure-variation investigations, we proposed the possible mechanism and binding modes of probes 1a–1f with Cu2+ (Figure S6). Probes 1b and 1d exhibited stronger recognition ability to Cu2+ than 1a and 1c. This phenomenon can be explained by steric effect. Probes 1b (linked with 1,2-ethanediamine) and 1d (linked with 1,6-hexylenediamine) gave smaller steric hindrance than 1a (linked with dimethyl-1,2-ethanediamine) and 1c (linked with piperazine), which made probes 1b and1d have strong coordination affinity with Cu2+. Furthermore, the ability of Cu2+ recognition was also affected by the number of ligands. The probe 1d grafted with two [12]aneN3 moieties exhibited stronger fluorescence quenching effect than 1e modified with one [12]aneN3 moiety. Overall, the amino group linked with naphthalimide played an important role in quenching the fluorescence of the probes through a PET mechanism. The concerted interactions of the triazole-[12]aneN3 unit and the amino group with Cu2+ enhances the fluorescence quenching effect. These results may give us implies to design high performance fluorescence probes to recognition Cu2+.
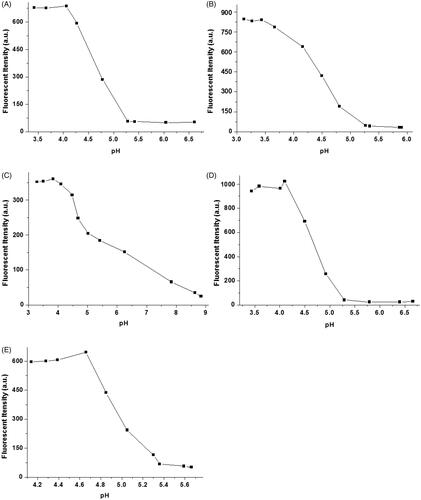
To investigate the effect of pH value on the fluorescence properties of complexes of 1-Cu, we carried out fluorescence emission experiments of complexes of 1-Cu at varying pH. As shown in , pH value has a very strong effect on the complexes of 1-Cu. Under acid pH (pH < 5) the fluorescence emission intensity was significantly enhanced with the increasing acidity of 1-Cu complex solution. When the pH value reached 3.5, no obvious fluorescence change was observed. It is likely that the complete protonation of the nitrogen atoms of probes restricts the binding ability of probes 1a–1e with Cu2+ under strongly acidic conditions. The binding ability of probes with Cu2+ is enhanced under neutral or basic condition, which leads to the fluorescence almost gets quenched and a very negligible change is observed (except 1c) in the presence of Cu2+. Such results make us believe that 1a–1e could become useful Cu2+ ion probes under biological conditions.
3.3. Cell imaging for Cu2+ recognition
To further investigate the biological applications of 1a–1e, the fluorescence microscopy experiment in HeLa cells was carried out. The cells were first treated with probes 1a–1e (20 μM) for 30 min in DMEM medium at 37 °C, and then washed with PBS buffer, significant green fluorescence was observed under the fluorescence microscope (). The fluorescence intensity of 1a–1d was decreased gradually after addition of Cu2+, and no obvious change was observed after 60 min (). However, the fluorescence was not completely quenched, which may be caused by the acidic environment of lysosomes. Strangely, the fluorescence intensity of 1e was almost unaffected by the addition of Cu2+, which was not in accordance with the fluorometric titration spectra measured by fluorescence spectrophotometer (). This phenomenon should be ascribed to the special structure of 1e, which was modified with only one [12]aneN3 unite, while other probes (1a, 1b, 1c, and 1d) were modified with two [12]aneN3 unites. The less number of [12]aneN3 units made 1e have weak coordination ability with Cu2+ as well as poor anti-interference performance especially in a complex intracellular environment, which led to the weak recognition for Cu2+ in the cells.
3.4. Lysosome imaging
It is well known that lysosome is one of important organelles in eukaryotic cells. It plays a key role in the degradation of macromolecules and cell components. The pH value in lysosomes ranges from 4.0 to 6.0, while the cytoplasm is about 7.2. The 1-Cu complexes exhibit strong fluorescence emission under acid conditions, while almost no fluorescence emission under neutral conditions, which make us believe that 1-Cu complexes can be used for lysosome imaging. In order to confirm above idea, lysosome imaging experiment was carried out in HeLa cells. Lyso-Tracker Red, a commercially available lysosome specific staining probe, was used to co-stain HeLa cells with 1-Cu complexes. HeLa cells were firstly incubated with 1-Cu complexes for 30 min, and then the medium was replaced with fresh medium containing Lyso-Tracker Red and incubated for another 30 min. As shown in , the green emission of 1a-Cu, 1b-Cu, 1c-Cu, and 1d-Cu () and the red emission of Lyso-Tracker Red () are nearly overlapping (). The correlation coefficient of overlap between green and red channel has been analyzed by IPP software. The Overlap coefficients of 1a–1e with Lyso-Tracker Red are 0.93, 0.85, 0.88, 0.95 and 0.81, respectively. These results indicate that the 1-Cu complexes can be applied to fluorescence probes lysosome imaging.
3.5. Characterization of 1a–1e/RNA (DNA) complexes
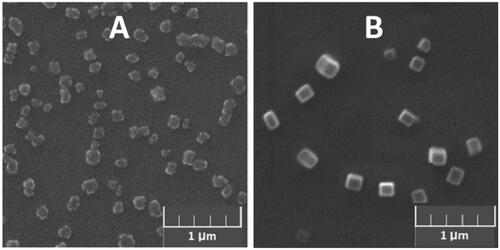
1a–1e/DNA (RNA) complexes were prepared by combining constant amount of DNA or RNA with varying concentrations of MFCs. The changes of particle sizes and zeta-potentials were measured at different concentrations of MFCs 1a–1e ranging from 10 μM to 60 μM via dynamic light scattering (DLS). As shown in Figures S7(A1–E1), these materials can condense DNA/RNA into nanoparticles ranging from 50 nm to 500 nm, and the complexes of 1/RNA particles were slight smaller than 1/DNA particles. SEM was also used to directly visualize the morphology and size of 1b/RNA (DNA) complexes. As shown in b/DNA complexes have homogeneous oblong shape with diameter of about 150 nm, while 1b/RNA complexes have an irregular shape, and it looks something like a cauliflower, the size of which was little smaller than particles formed by 1b/DNA complexes. The zeta potentials of the complexes of 1/RNA (DNA) were also measured via DLS (Figure S6(A2–E2)). The zeta potentials rose along with the increasing of concentration of 1a–1e. Compared to other materials, the complexes of 1e with DNA or RNA showed larger zeta potentials. At the concentration of 20 μM, the surface charges of complexes derived from 1e turned to positive, while other complexes still remained negatively charged. DLS and SEM results suggest that these multifunctional compounds have the potential to be used as nonviral vectors to deliver DNA or RNA.
3.6. Cell uptake of 1a–1e/RNA (DNA) complexes
To examine the possibility of using MFCs 1a–1e as non-viral vectors for gene delivery, cellular uptake experiment was carried out by using Cy5-labeled siRNA and dsDNA, 25 kDa PEI and lipofectamine 2000 were served as positive controls. The Cy5-labeled RNA/DNA emitted red light, while MFCs 1a–1e gave rise to green fluorescence, which enabled us to observe MFCs 1a–1e and RNA/DNA simultaneously under fluorescence microscopy.
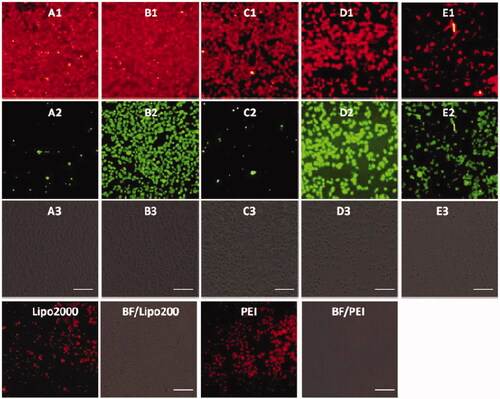
Before a direct comparison on the cellular uptake capacities, the optimal concentration (1d) and w/w ratio (PEI/RNA and lipofectamine2000/RNA) were screened on the HeLa cells. As shown in Figures S7–S9, the MTC 1d exhibited the best RNA delivery ability at the concentration of 20 μM, while the optimized w/w ratios for commercial agents PEI and lipofectamine 2000 were 5/1 and 20/1, respectively. The cellular uptake experiments were then conducted using HeLa cells at the optimal gene delivery condition of each material. As shown in , these materials modified with shorter linker (such as compound 1a and 1b) showed stronger red fluorescence than those with longer linker, and almost all the cells were stained with Cy5-labeled RNA, indicating their excellent RNA delivery ability. The commercial agents 25 kDa PEI and lipofectamine 2000, by contrast, gave much weaker red fluorescence emission than 1a and 1b in HeLa cells. We also found that red fluorescence and green fluorescence did not overlap well. It can be explained from two aspects: on the one hand, the fluorescence intensity of compounds 1a and 1e was much weaker than 1b, 1c, and 1e, which can be observed in . Therefore, much green fluorescence (1a and 1c) was loose when we take pictures at same conditions (1b, 1d, and 1e can show suitable fluorescence) by under fluorescence microscopy. On the other hand, all the images were obtained after 4 h cell uptake, some RNA may be released from the complexes. Therefore, the images from red channel are not overlapped with the green channel very well. The RNA delivery ability of 1a–1e was further evaluated in HepG2, U2Os and MC3T3-E1 cell lines at the concentration of 20 μM. As shown in Figures S10–S12, the intensity of red fluorescence observed in above three cell lines was far weaker than that observed in HeLa cells, indicating that these five compounds, especially for 1a and 1b, have excellent HeLa cell selectivity in RNA delivery. While for lipofectamine 2000 and PEI, they showed almost no cell-selectivity.
Figure 7. Fluorescence microscopy images of HeLa cells transfected with Cy5- labeled RNA (9 μg/mL) by MFCs 1a–1e at the concentration of 20 μM, 20 kD PEI and lipofectamine 2000 as positive control. (A1–E1) red channels, (A2–E2) green channels, (A3–E3) bright field images. The scale bar in the figure is 100 μm.
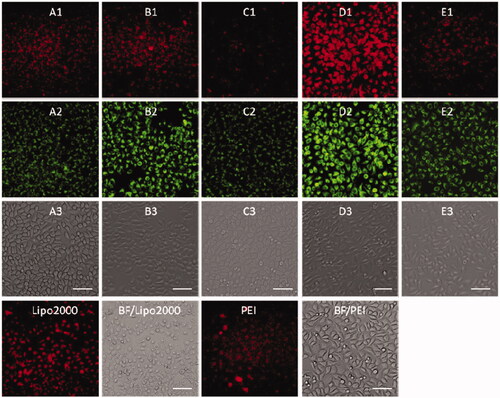
Subsequently, DNA delivery ability of 1a–1e was investigated at the concentration of 20 μM in HeLa and HepG2 cell lines. As shown in and , unlike RNA delivery, longer hydrophobic chain modified MTC 1d gave higher DNA delivery efficiency than shorter chain modified compounds, which was slightly higher than commercial materials lipofectamine 2000 and PEI in HeLa and HepG2 cells. These results indicated that DNA or RNA delivery ability is closely related with the structure of multifunctional compounds. The short chain linked compound 1b has excellent RNA delivery ability, while long chain modified compound 1d exhibits high DNA delivery efficiency, especially in HeLa cells, their performance is superior to lipofectamine 2000 and PEI.
Figure 8. Fluorescence microscopy images of HeLa cells transfected with Cy5-labeled DNA (9 μg/mL) by MFCs 1a–1e at the concentration of 20 μM, 20 kD PEI and lipofectamiine 2000 as positive control. (A1–E1) red channels, (A2–E2) green channels, (A3–E3) bright field images. The scale bar in the figure is 100 μm.
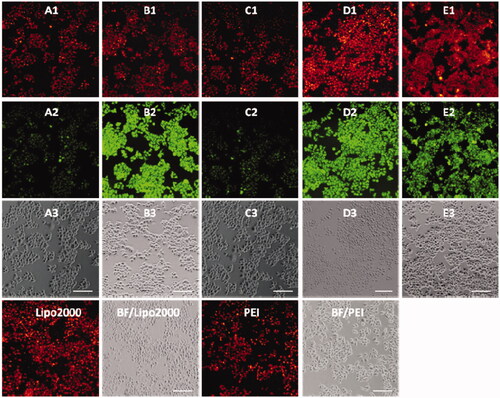
Figure 9. Fluorescence microscope images of HepG2 cells transfected with Cy5-labeled DNA (9 μg/mL) by MFCs 1a–1e at the concentration of 20 μM, 25 kD PEI and lipofectamiine 2000 as positive control. (A1–E1) red channels, (A2–E2) green channels, (A3–E3) bright field images. The scale bar in the figure is 50 μm.
3.7. Cytotoxicity
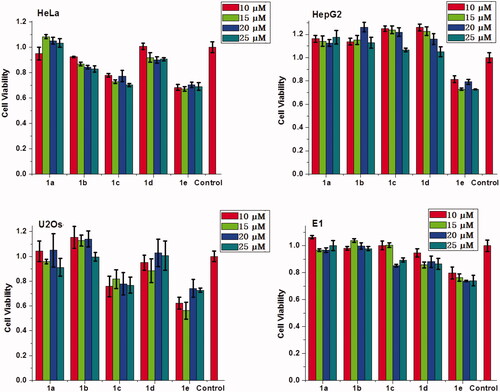
Besides high DNA and RNA delivery efficiency, the biocompatibility of gene vector is also very important in clinical applications. Therefore, MTT assay was performed to evaluate the cytotoxicity of the complexes of 1a–1e/RNA and 1a–1e/DNA in HeLa, HepG2, U2Os, and MC3T3-E1, cells. It should be noted that the concentration of 1a–1e was ranged from 10 μM to 25 μM, which was consistent with cellular uptake experiment. As shown in and S13, these multifunctional compounds (except 1e) have low cytotoxicity, especially for complexes of 1d with RNA or DNA, nearly no cytotoxicity was observed on HepG2 and U2Os cells. These results suggest that these materials can achieve high delivery efficacy and good biocompatibility when used as non-viral gene vectors.
4. Conclusion
A series of [12]aneN3-based naphthalimide MFCs 1a–1e were designed and synthesized. various linkers and different numbers of [12]aneN3 units were employed to facilitate the investigation of the structure-performance relationships. Some of these small organic compounds exhibited excellent performance on detection of Cu2+, lysosomal staining as well as RNA and DNA delivery. It was found that the probes with long hydrophobic chains and low steric hindrance linkers showed higher recognition ability toward Cu2+ (1d > 1b > 1a > 1c). The sensing mechanisms and binding modes of MFCs with Cu2+ were confirmed by fluorescence titration, Job’s plot analyses, and 1H-NMR titration. Besides, their complexes with Cu2+ (1-Cu) could also selectively stain lysosome in HeLa cells due to the acidic environment of lysosome. Cellular uptake experiments revealed that MFCs 1a and 1b have excellent HeLa cell selectivity in RNA delivery, and their performance was far better than lipofectamine 2000 and 25 kDa PEI. While for DNA delivery, long chain modified MFC 1d exhibited higher delivery efficiency than other compounds. These results will give us further insight to design high performance multifunctional compounds for fluorescence probes and non-viral vectors.
Author contributions
All the authors were involved in conducting, drafting or revising the manuscript. Yong-Guang Gao had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study conception and supervision: Yong-Guang Gao, Zhong-Lin Lu, Ai-Rong Qian. Design and preparation of multifunctional compounds: Yong-Guang Gao, Shu-Yuan Huangfu, Analysis of data from cell biology and fluorescent imaging: Suryaji Patil, Quan Tang, Wan Sun.
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alam AU, Qin Y, Nambiar S, et al. (2018). Polymers and organic materials-based pH sensors for healthcare applications. Prog Mater Sci 96:174–216.
- Behnood A, Tittelboom KV, Belie ND. (2016). Methods for measuring pH in concrete: a review. Constr Build Mater 105:176–88.
- Bloom K, Maepa M, Ely A, Arbuthnot P. (2018). Gene therapy for chronic HBV—can we eliminate cccDNA? Genes 9:207.
- Cotruvo JA, Aron AT, Ramos-Torres KM, Chang CJ. (2015). Synthetic fluorescent probes for studying copper in biological systems. Chem Soc Rev 44:4400–14.
- Ding AX, Tan ZL, Shi YD, et al. (2017). Gemini-type tetraphenylethylene amphiphiles containing [12]aneN3 and long hydrocarbon chains as nonviral gene vectors and gene delivery monitors. ACS Appl Mater Interfaces 9:11546–56.
- Dunbar CE, High KA, Joung JK, et al. (2018). Gene therapy comes of age. Science 359:eaan4672.
- Eshak ES, Iso H, Yamagishi K, et al. (2018). Associations between copper and zinc intakes from diet and mortality from cardiovascular disease in a large population-based prospective cohort study. J Nutr Biochem 56:126–32.
- Gao YG, Alam U, Ding AX, et al. (2018). [12]aneN3-based lipid with naphthalimide moiety for enhanced gene transfection efficiency. Bioorg Chem 79:334–40.
- Gao YG, Shi YD, Zhang Y, et al. (2015). A naphthalimide-based [12]aneN3 compound as an effective and real-time fluorescence tracking non-viral gene vector. Chem Commun 51:16695–8.
- Gao YG, Tang Q, Shi YD, et al. (2016). 1,8-Naphthalimide modified [12]aneN3 compounds as selective and sensitive probes for Cu2+ ions and ATP in aqueous solution and living cells. Talanta 152:438–46.
- Gao YG, Uzair A, Tang Q, et al. (2016). Functional lipids based on [12]aneN3 and naphthalimide as efficient non-viral gene vectors. Org Biomol Chem 14:6346–54.
- Gao YG, Zhang Y, Shi YD, et al. (2016). Fluorescent sensors based on [12]aneN3 -modified BODIPY: Discrimination of different biological thiols in aqueous solution and living cells. Bioorg Med Chem 24:1550–59.
- Gu B, Huang L, Xu Z, et al. (2018). A reaction-based, colorimetric and near-infrared fluorescent probe for Cu2+ and its applications. Sensors ActuatB-Chem 273:118–25.
- Hao Z, Jiangli F, Qunli X, et al. (2012). Imaging of lysosomal pH changes with a fluorescent sensor containing a novel lysosome-locating group. Chem Commun 48:11766–8.
- Hardee C, Arévalo-Soliz L, Hornstein B, Zechiedrich L. (2017). Advances in non-viral DNA vectors for gene therapy. Genes 8:65.
- Hu J, Hu K, Cheng Y. (2016). Tailoring the dendrimer core for efficient gene delivery. Acta Biomater 35:1–11.
- Izumi H, Torigoe T, Ishiguchi H, et al. (2003). Cellular pH regulators: potentially promising molecular targets for cancer chemotherapy. Cancer Treat Rev 29:541–9.
- Jeppe M, Irene C, Warren NJ, et al. (2013). Nile Blue-based nanosized pH sensors for simultaneous far-red and near-infrared live bioimaging. J Am Chem Soc 135:14863–70.
- Junyan H, Kevin B. (2010). Fluorescent indicators for intracellular pH. Chem Rev 110:2709–28.
- Karr JW, Szalai VA. (2007). Role of aspartate-1 in Cu(II) binding to the amyloid-β peptide of Alzheimer’s disease. J Am Chem Soc 129:3796–7.
- Khan MA, Ansari U, Ali MN. (2015). Real-time wound management through integrated pH sensors: a review. Sensor Rev 35:183–9.
- Li G, Bai L, Tao F, et al. (2018). A dual chemosensor for Cu2+ and Hg2+ based on a rhodamine-terminated water-soluble polymer in 100% aqueous solution. Analyst 143:5395–403.
- Li W, Yan R, Liu Y, et al. (2019). Co-delivery of Bmi1 small interfering RNA with ursolic acid by folate receptor-targeted cationic liposomes enhances anti-tumor activity of ursolic acid in vitro and in vivo. Drug Deliv 26:794–802.
- Lundstrom K. (1995). Viral vectors in gene therapy. Annu Rev Microbiol 49:807–838.
- Luo TY, He X, Zhang J, et al. (2018). Photoluminescent F-doped carbon dots prepared by ring-opening reaction for gene delivery and cell imaging. RSC Adv 8:6053–6062.
- Luo J, Luo Y, Sun J, et al. (2015). Adeno-associated virus-mediated cancer gene therapy: current status. Cancer Lett 356:347–56.
- Marshall E. (1999). Gene therapy death prompts review of adenovirus vector. Science 286:2244–45.
- Peng L. (2018). Editorial: Current gene therapy on non-viral gene delivery and therapy. Curr Gene Ther 18:2.
- Posocco P, Liu X, Laurini E, et al. (2013). Impact of siRNA overhangs for dendrimer-mediated siRNA delivery and gene silencing. Mol Pharm 10:3262–73.
- Ramamoorth M, Narvekar A. (2015). Non viral vectors in gene therapy- an overview. J Clin Diagn Res 9:GE01–GE06.
- Richter A, Paschew G, Klatt S, et al. (2008). Review on hydrogel-based pH sensors and microsensors. Sensors 8:561–81.
- Shi XM, Mei LP, Zhang N, et al. (2018). A polymer dots-based photoelectrochemical pH sensor: simplicity, high sensitivity, and broad-range pH measurement. Anal Chem 90:8300–3.
- Shuqi W, Zhu L, Jiahuai H, Shoufa H. (2011). Dual colored mesoporous silica nanoparticles with pH activable rhodamine-lactam for ratiometric sensing of lysosomal acidity. Chem Commun 47:11276–8.
- Siotto M, Squitti R. (2018). Copper imbalance in Alzheimer’s disease: Overview of the exchangeable copper component in plasma and the intriguing role albumin plays. Coord Chem Rev 371:86–95.
- Tan E, Lv J, Hu J, et al. (2018). Statistical: versus block fluoropolymers in gene delivery. J Mater Chem B 6:7230–7238.
- Tewari BB. (2010). Studies on complexation in solution with a paper electrophoretic technique [the system copper(II)/cobalt(II)−methionine − penicillamine. J Chem Eng Data 55:1779–83.
- Thomas CE, Ehrhardt A, Kay MA. (2003). Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet 4:346–58.
- Verma IM, Somia N. (1997). Gene therapy – promises, problems and prospects. Nature 389:239–42.
- Wang F, Hu K, Cheng Y. (2016). Structure–activity relationship of dendrimers engineered with twenty common amino acids in gene delivery. Acta Biomater 29:94–102.
- Wang J, Wang Y, Wang Z, et al. (2019). A thermosensitive gel based on w1/o/w2 multiple microemulsions for the vaginal delivery of small nucleic acid. Drug Deliv 26:168–78.
- Wang S, Wang F, Zhang Q, Cheng Y. (2017). A core–shell structured polyplex for efficient and non-toxic gene delivery. J Mater Chem B 5:5101–5108.
- Wu P, Zhao T, Wang S, Hou X. (2014). Semicondutor quantum dots-based metal ion probes. Nanoscale 6:43–64.
- Yang C, Yin X, Huan SY, et al. (2018). A Two-photon DNAzyme-Gold Nanoparticle Probe for Imaging Intracellular Metal Ions. Anal Chem 90:3118–23.
- Yang J, Zhang Q, Chang H, Cheng Y. (2015). Surface-engineered dendrimers in gene delivery. Chem Rev 115:5274–5300.
- Yi WJ, Zhang QF, Zhang J, et al. (2014). Cyclen-based lipidic oligomers as potential gene delivery vehicles. Acta Biomater 10:1412–22.
- You S, Cai Q, Zheng Y, et al. (2014). Perylene-cored star-shaped polycations for fluorescent gene vectors and bioimaging. ACS Appl Mater Interfaces 6:16327–34.
- Yue Y, Huo F, Lee S, et al. (2017). A review: the trend of progress about pH probes in cell application in recent years. Analyst 142:30–41.
- Yue P, Zhang Y, Guo ZF, et al. (2015). Synthesis of bifunctional molecules containing [12]aneN3 and coumarin moieties as effective DNA condensation agents and new non-viral gene vectors. Org Biomol Chem 13:4494–4505.
- Zhang Y, Gao YG, Shi YD, et al. (2015). [12]aneN3-based BODIPY as a selective and sensitive off–on sensor for the sequential recognition of Cu2+ ions and ADP. Chin Chem Lett 26:894–98.
- Zhang L, Lichtmannegger J, Summer KH, et al. (2009). Tracing copper − thiomolybdate complexes in a prospective treatment for Wilson’s disease. Biochemistry 48:891–7.
- Zhong L, Xing F, Bai Y, et al. (2013). Aspartic acid functionalized water-soluble perylene diimide as “Off–On” fluorescent sensor for selective detection Cu2+ and ATP. Spectrochim Acta A 115:370–5.
- Zhu L, Shuqi W, Jiahuai H, Shoufa H. (2011). Imaging of intracellular acidic compartments with a sensitive rhodamine based fluorogenic pH sensor. Analyst 136:3698–706.