ABSTRACT
Soy and maize are the most cultivated genetically modified (GM) crops in the world, which are commonly used in many food products. Establishing regulations for food products containing transgenic materials are obligatory in many countries in order to provide consumers with information. For this purpose, an accurate qualitative polymerase chain reaction (PCR) method was applied to investigate GM food products from Iran. In present study, a total of 90 non-labeled soy and maize samples were collected from Tehran’s market and their DNA were extracted by a kit. All samples were analyzed to screen the 35 S promoter and (nopaline synthase) NOS terminator elements. Having screened the GM positive samples, they were subjected to identification of specific transgenic events RR soy, Bt11 and MON810 maize with PCR. According to our results, 95% of soy and 60% of maize analyzed samples were positive for the 35 S promoter and NOS terminator. Furthermore, event-specific analysis indicated the presence of RR soy, Bt11 and MON810 maize in tested samples, while none of these food-derived samples demonstrated any GM label. The obtained results revealed that the necessity of monitoring system to provide a good reliable control of GM materials in food products and subsequently on their labeling.
Introduction
Gene modification techniques have been applied to a number of agricultural crops in order to insert new appropriate traits such as herbicide tolerance, insect and disease resistance, improve the nutritional content of food and feed products and increasing of various desirable traits.[Citation1,Citation2] Among the genetically modified (GM) plants soybean and maize are the two main cultivated GM crops in the globe, which covered 47% and 32% of the transgenic plants area respectively, and commonly represent as ingredients in many foods.[Citation3,Citation4] About 12 percent of global farming area were planted with these products according to the reports of 2014.[Citation5] Although several events have been approved for commercialization and consumption worldwide, public concerns increase about the human health risk and environmental effects of GM crops.[Citation6] On the other side, it has been shown that more than 81% of transgenic crops containing herbicide-tolerant genes for weed control.[Citation7] The majority of those known as the glyphosate-resistant plants, such as Roundup Ready crops of which several reports indicated that accumulation and widespread use of glyphosate in foods and environment can have an adverse effect on human health.[Citation8] Therefore, because of existing concerns about them, in several countries to provide consumers with freedom of choice, regulation for labeling of food containing GM ingredients above the threshold level was established.[Citation9]
Currently various genetically modified organism (GMO) detection methods based on protein and DNA are available to realize these legislations. Taking into account that DNA is more stable than protein, DNA-based methods are more sensitive to detection of GM ingredients in raw and processed food products. Hence, qualitative polymerase chain reaction (PCR) is a valid amplification technique to detect such materials from GMOs.[Citation10] Qualitative analysis can be divided into screening and event-specific methods. In the screening methods the common sequences in many various GMOs such as CaMV 35 S promoter from cauliflower mosaic virus and nopaline synthase (nos) terminator are monitored and totally are the first point of detection. Whereas, the purpose of event-specific PCR method is to identify the transgenic events to detect the differences between the inserted trait and genomic DNA. However, the screening is a common strategy for detection of transgenic products, prior to the identification of unique sequences by event-specific method.[Citation11]
Detection and identification of GMO products using PCR method have been reported worldwide. In Turkey, Arun et al. monitored the food containing soy and maize ingredients for detection of CaMV 35 S promoter and NOS terminator elements by PCR. Their results showed the presence of GM maize and soy in a number of samples.[Citation12] To date, two small qualitative GMO studies were conducted in Iran. In the second study which was carried out in 2016 on five soy samples it is indicated that two samples were positive for CaMV 35 S promoter.[Citation13] The aim of this study was detection of genetically modified soy and maize products, from Tehran’s market using reliable PCR method. Thus, present study focuses on screening and event-specific methods to detection and identification of raw and middle process samples (include maize and soy), using qualitative PCR.
Materials and methods
Sampling
A total of 90 commercially middle processed soy and maize samples, including different brands (51 soy proteins, 9 soy flours, 24 maize grains, 6 maize flours), were purchased randomly from Tehran markets in 2018. The origin of collected samples were countries with various GMO legislations including Iran, Brazil and Argentina. All the samples were grounded and homogenized by a laboratory electric grinder, then the acquired flour is preserved at −20°C before DNA extraction.
Certified reference materials
Certified reference materials (CRMs) were purchased from the Institute of Reference Materials and Measurements (JRC-IRMM, Geel, Belgium) for performance appraisal method. The following CRMs of GM lines were used as a positive control to evaluation of soy and maize events: GM powders containing 0.1, 0.5, 1 and 5% of RR soy, Bt-11 and MON810 maize. Additionally, the GM-free lines of soy and maize powders were used as a negative control in present work. It has to be mentioned the obtained CRMs cover the CaMV 35 S promoter and NOS terminator lines in order to GMO screening and also provide specific genes to detect the endogenous targets (soy lectin and maize invertase).
DNA extraction and purification
DNA extraction was performed from collected samples and provided CRMs using DNA extraction kit from plant materials (MBST, Iran). Based on the manufacturer’s instructions of kit, 100 mg of material taken from a previously ground sample was weighed in a 1.5 ml tube and mixed with the 300 μl lysis buffer, 20 μl proteinase K and incubated for 15 min at 60°c. After incubation, 580 μl binding buffer was added, mixed by vortexing and incubated for 10 min at 70°c. The mixture was centrifuged for 1 min by 8000 x g and the supernatant transferred into the new 1.5 ml tube. Then 440 μl ethanol (100%) was added into the mixture, applied into a spin column and centrifuged at 8000 x g for 1 min. Following centrifugation, 500 μl wash buffer was added to column and centrifuged at 8000 x g for 3 min to remove the ethanol completely. The DNA was eluted with 35–50 ml of preheated (72°C) elution buffer, then incubated for 3 min at room temperature. After 1 min of centrifugation the obtained DNA was stored at −20°C. The concentration and purity of acquired DNA were determined by UV light absorption at 260 nm and 260/280 nm using a NanoDrop spectrophotometer, respectively.
Oligonucleotide primers
To quality determination of extracted DNA, oligonucleotide primers GM03/GM04 and IVR1-F/IVR1-R were used for soy lectin and maize invertase endogenous genes, respectively. Because approximately all of the transgenic soy and maize products containing at least one of the P-35 S and T-NOS elements, suitable primer pairs 35 S-cf3/35 S-cf4 and HA-nos118-f/HA-nos118- r were used for screening of the products, respectively.[Citation12] Additionly, the event-specific lines of RR soy, Bt11 and MON810 maize were identified by primers 35 s-f2/petu-r1, IVS2-2/PAT-B and VW01/VW03, respectively. Roundup Ready (RR) soybean, maize MON 810, maize Bt-11 are the most common transgenic elements in different types of foods in the market. The principle trait of RR soy and MON810 maize are herbicide and insect resistant, respectively. Bt11 includes the combination of these traits.[Citation14,Citation15] The final concentration of primers varied: 0.6 μM for 35 S and nos, 0.2 μM for RR, 0.5 μM for Bt11 and MON810. Selected primers were purchased and maintained at –20°C prior to use. The oligonucleotide primers are presented in .
Table 1. Primer pairs used to detection of endogenous genes and transgenic DNA sequences of food samples by PCR method
Qualitative PCR set-up
All PCR reactions were carried out by a thermal cycler (96 universals, PEQStar, Germany), in a final reaction mixture volume of 25 μl containing 2 μl of isolated DNA, 1 μl of each forward and reverse primer, 12 μl of ready to use master mix (Ampliqon, Denmark) (including Tris-Hcl pH 8.5, (NH4)2SO4, 3 mM Mgcl2, 0.2% Tween 20, 0.4 mM dNTPs, 0.2 units/μl Ampliqon Taq DNA polymerase, Inert red dye and stabilizer), and 9 μl of ultrapure water. In order to achieve valid results, for each reaction, positive and negative relevant controls were used. On the other side, we used the ultrapure water instead of DNA as a master mix control during PCR analysis. PCR thermal cycling condition for each set of primers were as follows: denaturation for 5 min at 94°C; amplification for 30 s at 94°C; annealing for 45 s at 58 for lectin, 54°C for invertase, 54°C for CaMV35 s, 54°C for nos, 54°C for Bt11, 54°C for MON 810, extension at 72°C for 75 s, final elongation for 8 min at 72°C.
Agarose gel electrophoresis
The obtained PCR products from food materials and CRMs were electrophoresed through a 2% agarose at 80 V for 1 h to separate DNA fragments. Agarose gel was prepared using Tris Borate EDTA (TBE) 1x (50 ml) and DNA Safe Stain (2 μl). To size determination of separated bands a 100 bp DNA ladder was used as a standard. Finally, DNA fragments were visualized by a UV light source.
Results and discussion
In the present research, nighty commercially raw or middle processed soy and maize samples were collected and analyzed during 2018. Food materials were grouped into soy protein (51 samples of 20 various brands), soy flour (nine samples of three various brands), maize grains (24 samples of 10 various brands) and maize flour (six samples of two various brands). All samples were assessed for detection and identification of transgenic sequences (P-35 S, T-NOS, RR soy, Bt11 and MON810) by a reliable qualitative PCR.
Quality assurance
In order to reduce false positive or false negative risk related to the contamination during DNA extraction procedure and PCR analysis, both appropriate CRMs and sterile ultra-pure water were used as control in a parallel process for each sample. Based on our CRM testing the sensitivity of qualitative analyzing method for GMO detection of soy and maize in each PCR reaction was 0.1% (). A Similar method was applied to achieve an accurate GMO detection method by Arun ÖÖ et al. (2013) who found that 25 out of 100 analyzed samples were positive for transgenic sequences.[Citation12]
DNA extraction and purification
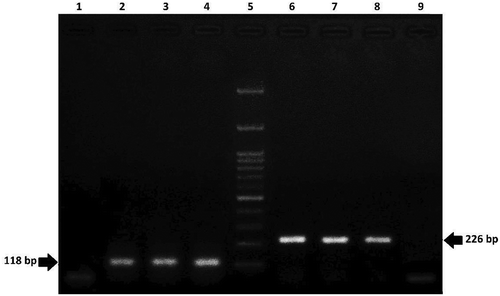
Extracting an adequate amount of DNA with an appropriate quality is required for reliable GMO testing.[Citation17] The absorbance ratios of obtained DNA at 260 nm ranged from 1.7 to 1.9, and concentration of DNA based on the kit extraction method was 20 to 100 ng/μl. Similarly, other studies showed that the possibility of extracting a high quality DNA from raw and middle processed food products by good extraction procedures.[Citation18,Citation19] On the other side, Vijayakumar et al. (2009) reported that increasing amount of sample can improve the DNA yield.[Citation20] However, the purity of obtained DNA can be significantly affected by degree of processing, extraction procedure and different components of sample matrix.[Citation1] Therefore, to reduce the false negative results quality confirmation of genomic DNA for all samples and CRMs were carried out by agarose gel electrophoresis.[Citation21,Citation22] It was observed that the specific lectin and invertase endogenous genes are available for further investigation of GM sequences from soy and maize, respectively (). This method has been used as quality confirmation of DNA yield to check and verify the DNA integrity by other researchers.[Citation23,Citation24]
Figure 2. Agarose gel electrophoresis of PCR products from DNA of food samples containing lectin (118 bp) and invertase (226 bp) genes. Lane 1: no template control; Lanes2-3: soy positive samples; Lane 4: soy positive control; Lane 5: 100 bp DNA ladder; Lane 6: maize positive control; Lanes 7–8: maize positive samples; Lane 9: no template control
Screening and event specific detection
For the screening and event specific detection of GM sequences, PCR analyzes indicated that the detection limit of 0.1% using soy and maize CRMs containing various GM levels (0.1, 0.5, 1 and 5%). A total of 60 soy and 30 maize non-labeled samples were collected from different regions of Tehran to detection of CaMV 35 S, NOS, RR soy, Bt11 and MON810 genes by qualitative PCR. The results of present study indicated that 95% of soy samples were positive for screening targets (CaMV 35 S and NOS) (). Also, CaMV 35 S and NOS were detected in 60% of maize samples ().
Table 2. Detection of endogenous genes and transgenic DNA sequences of soy product samples during 2018 by qualitative PCR method
Table 3. Detection of endogenous genes and transgenic DNA sequences of maize product samples during 2018 by qualitative PCR method
Our findings of collected soy products are in accordance with the results of Sieradzki et al. (2006), who screened soybean samples from Poland and proved that only 1 sample out of 45 was GM negative.[Citation25] Similarly, the evaluation of food products by Ujhelyi et al. (2007) demonstrated that 91.2% of non-labeled soy products from the Hungarian market contained the 35 S-promoter or NOS-terminator elements in their genome.[Citation14] CaMV 35 S promoter and NOS terminator are the two important screening elements for qualitative PCR analyzes. These elements are presented in most of the commercialized transgenic plants.[Citation15] In 2009, Holden et al. (2010) found that in 95% of GM foods in Europe the CaMV 35 S are present.[Citation26] Our results of GM soy screening were also close to the finding of Greiner and Konietzny (2007) those revealed that among 100 collected samples of soy in 2005, 35 S-promoter or NOS-terminator elements were detected in 78 (78%) samples.[Citation2] In Turkey, it was found that 11 out of the 57 (19.3) non-labeled tested soy containing samples were positive for GM targets.[Citation12] On the other hand, previous research which was carried out in Iran showed that of all the 5 imported soybean from different country, 3 of which were negative for mentioned GM elements, although, in that study the sensitivity of method did not determine during the study by using the appropriate CRMs.[Citation13]
The observation of maize food products was consistent with the study conducted in Malaysia which have indicated that of all 20 non-labeled maize samples tested, 13 (65%) were determined to be positive for GM materials.[Citation1] GMO screening study in Kingdom of Saudi Arabia indicated that out of 64 maize containing products, 28 were positive for CaMV 35 S or NOS terminator.[Citation27] In the other study, Gürakan and Aydın (2011), examined 31 maize sold samples for GMO detection by targeting screening materials. In that study, it has been stated that in 11 and 7 of them CaMV 35 S and NOS terminator were found, respectively.[Citation28] The obtained PCR data were also close to the results of Fernandes et al. (2013) since they showed that among 21 assessed commercialized maize samples in 2007, 48 (44%) were positive for GM elements (CaMV 35 S or NOS terminator).[Citation18] In contrast to these findings, Zdjelar et al. (2013) did not detect GM maize in 100 non-labeled samples from Serbia.[Citation19] Negative results of that study can be due to the selection of a special type of maize for analysis. The obtained data from local and imported processed maize sold commercially in Brazilian markets in 2000 to 2005 indicated that 8 to 11% of analyzed samples were transgenic.[Citation2] Compared to the results of screening GM elements in soy samples, maize food samples reflected a lower presence of such elements.
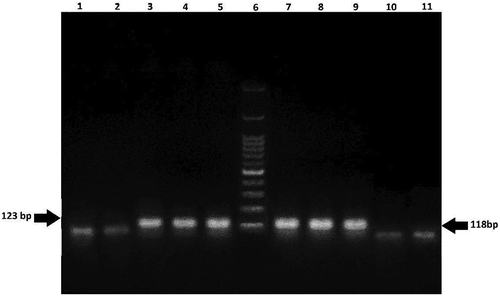
As mentioned above CaMV35 S promoter and NOS terminator are the most current elements in approved transgenic products worldwide. On the other side, these elements also occur naturally in various plants and soil microorganism, therefore a positive result will indicate the presence of GM sequences are is probable in food products.[Citation15,Citation29] However, the probability of positive result is higher when both the elements are present in a sample. In order to the confirmation of obtained results of screening PCR, and also identification of transgenic events, positive samples were further analyzed by event-specific method. As shown in (), further evaluation indicated that all of the 57 GM positive samples contained the RR soy event, which proved the presence of GM sequences in their genome. An example of positive signals of RR soy is displayed in . Our findings of soy-containing products were similar to the results of Arun et al. (2013) who showed that all of the 11 GM positive soy-based products, both local and imported, were contained RR soy line.[Citation12] Similarly, Zdjelar et al. (2013) showed that 8 out of 32 non-labeled soy samples originally from EU countries, USA, Argentina, Brazil, and Thailand were positive for RR line.[Citation19] RR soy is the only transgene plant variety approved for consumption in the EU market, but it’s not allowed to be cultivated. Branquinho et al. (2010) examined soy-containing products during 2004to 2007 in Brazil for identification of RR event, of 240 samples 68 were determined as RR soy.[Citation30] Another study conducted in Hungary also declared that 38% of analyzed samples gave positive amplification signal for RR specific event.[Citation14] Roundup Ready is the most widely cultivated soy event across the world. This event has been modified to be glyphosate-resistant during the cultivation.[Citation31] Glyphosate is a nonselective chemical substance commonly used in RR herbicides to weed control, hence cannot disrupt the grows of target plants that are modified to tolerate, although the accumulation of this chemical herbicide in soil and plants may lead to unintended effects on environment and human health.[Citation32] For instance, Mesnage et al. (2015) revealed that glyphosate has adverse effects under regulatory limits such as neurotoxicity, carcinogenicity, hepatic and kidney toxicity.[Citation33] On the other side, A study performed on RR soy grown in USA indicated the high residues of glyphosate (3.3 mg/kg) in collected samples.[Citation34] Therefore, because of the food safety issues it is necessary to determine the residue concentration of it in food products especially in glyphosate-resistant crops.
Figure 3. Agarose gel electrophoresis of PCR products from DNA of 3 soy samples for analysis of Roundup Ready (172 bp) line. Lanes 1–3: positive samples; Lane 4: 100 bp DNA ladder; Lane 5: negative control; Lane 6: positive control
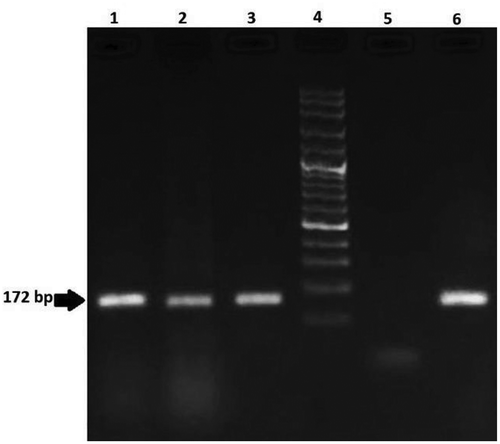
In the case of maize food samples, simplex PCR was used to detection of Bt11 and MON810 events. From , it is indicated that Bt11 maize was found in 40% and MON810 maize was found in 13.3% of samples. Moreover, both the Bt11- MON810 events were found in 10% of maize products, while two out of 18 positive samples for CaMV35 S promoter and nos terminator genes did not show any positive results for respective events. It has to be mentioned, in this work other events were not examined. A summary of electrophoresis results is displayed in and . This observation is consistent with those reported by Branquinho et al (2013), examined Bt11 and MON810 lines in incorrect labeled Brazilian maize samples in 2011 and 2012. The results of their study demonstrated that 27 out of 30 and 29 out of 30 samples were positive for Bt11 and MON810, respectively.[Citation35] Arun et al. (2013), showed that among 14 tested samples containing maize ingredients, 8 contained Bt11 maize transgenic variety.[Citation12] Similarly, previous study conducted in Iran proved that of all 5 non-labeled positive maize samples, 2 and 3 containing GM lines, that is CaMV35 S promoter were Bt11 and MON810, respectively.[Citation36] Our results also are in agreement with the findings of Greiner, Konietzny, & Villavicencio (2005), who informed that both the MON810 and Bt11 are the main GMO events in analyzes of maize.[Citation37] In contrast to our results, Dinon, De Melo and Arisi (2008), collected 83 labeled commercialized maize products in order to GMO detection during 2005 to 2007. Their findings reported that of all the 81 samples originally from Brazil gave negative signal for MON810 transgenic line, but 2 from Argentina were positive for MON810 maize line..[Citation38]
Figure 4. Agarose gel electrophoresis of PCR products from maize samples for analysis of Bt11 (189 bp) line. Lanes 1–3: positive samples; Lane 4: 100 bp DNA ladder; Lane 5: negative control; Lane 6: positive control
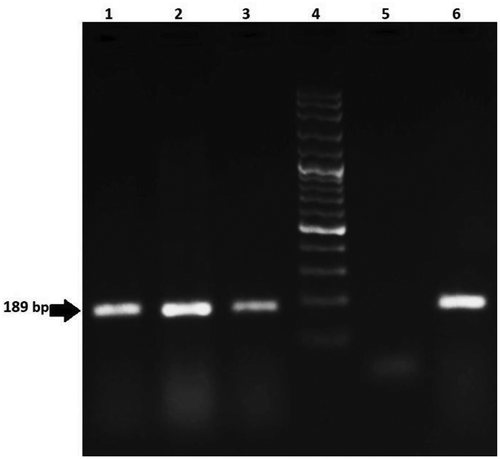
Figure 5. Agarose gel electrophoresis of PCR products from maize samples for analysis of MON810 (170 bp) line. Lanes 1–3: positive samples; Lane 4: 100 bp DNA ladder; Lane 5: negative control; Lane 6: positive control
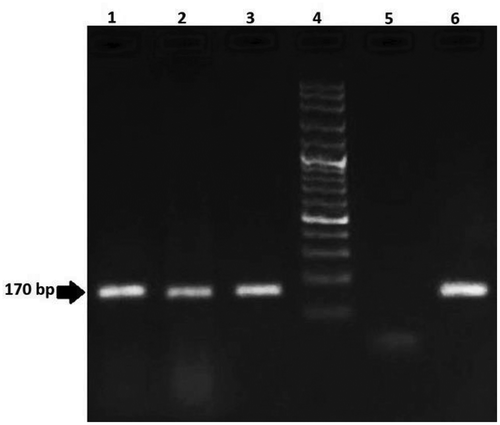
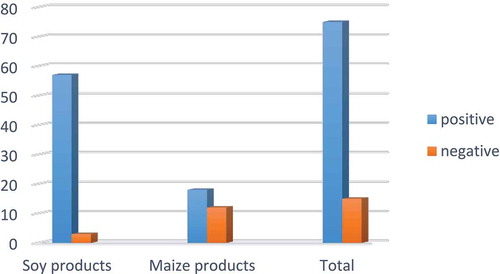
Maize transgenic event MON810 contains the genes from the bacterium Bacillus thuringiensis for increased resistance to some insects. Maize transgenic event Bt11 was designed to herbicide tolerance and insect resistance.[Citation39] As a result, these crops increasing the concerns regarding accumulation of herbicide in foods or environment and novel gene function. The overall results of this study demonstrate the presence of transgenic ingredients in 95% of soy and 60% of maize food products obtained from Tehran’s market. Most of the GM positive samples were imported into Iran. Distribution of positive and negative samples is shown in . In spite of the Iranian legislation regarding labeling of food materials derived from GM products, none of the collected samples in 2018 were appropriately labeled. To date, Iran has imported different types of transgenic crops such as soy and maize, but cultivation of these plants is prohibited. However, Food and Drug Administration of Iran is responsible for monitoring of GMO products. Additionally, in order to control of these products and protect the consumers concern about their biosafety establishing regulation provides a reliable monitoring program is recommended.
Conclusion
Monitoring of GMOs in food products are subjected to ensure the free choice of consumers and their protection. Based on our results, it could be concluded that DNA extraction procedure and qualitative PCR were reliable methods to DNA isolation and detection of GMOs in food products, respectively. The obtained data clearly showed the frequency of transgenic sequences in soy and maize food samples with construct-specific genes of RR soybean, Bt11 and MON810 maize. However, none of the samples containing GMOs sequences had label in order to provide information for consumers. Therefore, establishing an accurate chain traceability system to control and labeling of GM foods are advised in Iran.
Additional information
Funding
References
- Kaur, J.; Radu, S.; Ghazali, F. M.; Kqueen, C. Y. Real-time PCR-based Detection and Quantification of Genetically Modified Maize in Processed Feeds Commercialised in Malaysia. Food Control. 2010, 21(11), 1536–1544.
- Greiner, R.; Konietzny, U. Presence of Genetically Modified Maize and Soy in Food Products Sold Commercially in Brazil from 2000 to 2005. Food Control. 2008, 19(5), 499–505. DOI: 10.1016/j.foodcont.2007.05.016.
- James, C.;, Brief 43: Global Status of Commercialized biotech/GM Crops: 2011. ISAAA Brief, 2011 ( 44).
- Forte, V.; Di Pinto, A.; Martino, C.; Tantillo, G. M.; Grasso, G.; Schena, F. P. A General multiplex-PCR Assay for the General Detection of Genetically Modified Soya and Maize. Food Control. 2005, 16(6), 535–539.
- James, C. Global Status of Commercialized biotech/GM Crops: 2014. ISAAA brief, 2015. 49.
- Domingo, J. L.; Bordonaba, J. G. A Literature Review on the Safety Assessment of Genetically Modified Plants. Environ. Int. 2011, 37(4), 734–742. DOI: 10.1016/j.envint.2011.01.003.
- Bonny, S. Herbicide-tolerant Transgenic Soybean over 15 Years of Cultivation: Pesticide Use, Weed Resistance, and Some Economic Issues. The Case of the USA. Sustainability. 2011, 3(9), 1302–1322. DOI: 10.3390/su3091302.
- Cuhra, M. Review of GMO Safety Assessment Studies: Glyphosate Residues in Roundup Ready Crops Is an Ignored Issue. Environ. Sci. Eur. 2015, 27(1), 20. DOI: 10.1186/s12302-015-0052-7.
- Gruère, G. P.; Rao, S. A Review of International Labeling Policies of Genetically Modified Food to Evaluate India’s Proposed Rule. 2007. 10.1094/PDIS-91-4-0467B
- Gryson, N.; Effect of Food Processing on Plant DNA Degradation and PCR-based GMO Analysis: A Review. Anal. Bioanal. Chem. 2010, 396(6), 2003–2022. DOI: 10.1007/s00216-009-3343-2.
- Novak, P. K.; Gruden, K.; Morisset, D.; Lavrač, N.; Štebih, D.; Rotter, A.; Žel, J. GMOtrack: Generator of Cost-effective GMO Testing Strategies. J. AOAC Int. 2009, 92(6), 1739–1746.
- Arun, Ö. Ö.; Yılmaz, F.; Muratoğlu, K. PCR Detection of Genetically Modified Maize and Soy in Mildly and Highly Processed Foods. Food Control. 2013, 32(2), 525–531. DOI: 10.1016/j.foodcont.2013.01.023.
- Ghareyazie, B.; Sarmadi, L.; Alemzadeh, A., PCR-based Detection of Genetically Modified Soybean at a Grain Receiving Port in Iran. 2018.
- Ujhelyi, G.; Vajda, B.; Béki, E.; Neszlényi, K.; Jakab, J.; Jánosi, A.; Némedi, E.; Gelencsér, É. Surveying the RR Soy Content of Commercially Available Food Products in Hungary. Food Control. 2008, 19(10), 967–973.
- Herzallah, S. M. Detection of Genetically Modified Material in Feed and Foodstuffs Containing Soy and Maize in Jordan. J. Food Compost. Anal. 2012, 26(1–2), 169–172. DOI: 10.1016/j.jfca.2012.01.007.
- ISO 21569. (2005). Foodstuffs E Methods of Analysis for the Detection of Genetically Modified Organisms and Derived Products E Qualitative Nucleic Acid Based Methods. Switzerland.
- Safaei, P.; Aghaee, E.M.; Khaniki, G.J.; Afshari, S.A.; Rezaie, S.; A simple and accurate PCR method for detection of genetically modified rice. J. Environ. Health Sci. Eng. 2019 Dec 10:1–5.
- T. J. R.; Amaral, J. S.; Oliveira, M. B. P. P.; Mafra, I. A Survey on Genetically Modified Maize in Foods Commercialised in Portugal. Food Control. 2014, 35(1), 338–344.
- Zdjelar, G.; Nikolić, Z.; Vasiljević, I.; Bajić, B.; Jovičić, D.; Ignjatov, M.; Milošević, D. Detection of Genetically Modified Soya, Maize, and Rice in Vegetarian and Healthy Food Products in Serbia. Czech J. Food Sci. 2013, 31(1), 43–48.
- Vijayakumar, K.; Martin, A.; Gowda, L. R.; Prakash, V. Detection of Genetically Modified Soya and Maize: Impact of Heat Processing. Food Chem. 2009, 117(3), 514–521.
- Taski-Ajdukovic, K.; Nikolic, Z.; Vujakovic, M.; Milosevic, M.; Ignjatov, M.; Petrovic, D. Detection of Genetically Modified Organisms in Processed Meat Products on the Serbian Food Market. Meat Sci. 2009, 81(1), 230–232.
- Cardarelli, P.; Branquinho, M. R.; Ferreira, R. T. B.; da Cruz, F. P.; Gemal, A. L. Detection of GMO in Food Products in Brazil: The INCQS Experience. Food Control. 2005, 16(10), 859–866.
- Smith, D. S.; Maxwell, P. W.; De Boer, S. H. Comparison of Several Methods for the Extraction of DNA from Potatoes and Potato-derived Products. J. Agric. Food Chem. 2005, 53(26), 9848–9859. DOI: 10.1021/jf051201v.
- Abdullah, T.; Radu, S.; Hassan, Z.; Hashim, J. K. Detection of Genetically Modified Soy in Processed Foods Sold Commercially in Malaysia by PCR-based Method. Food Chem. 2006, 98(3), 575–579.
- Sieradzki, Z.; Walczak, M.; Kwiatek, K. Occurrence of Genetically Modified Maize and Soybean in Animal Feedingstuffs. Bull. Vet. Inst. Pulawy. 2006, 50(4), 567.
- Holden, M. J. The Use of 35S and Tnos Expression Elements in the Measurement of Genetically Engineered Plant Materials. Anal. Bioanal. Chem. 2010, 396(6), 2175–2187.
- Elsanhoty, R. M.; Al-Turki, A.; Ramadan, M. F. Prevalence of Genetically Modified Rice, Maize, and Soy in Saudi Food Products. Appl. Biochem. Biotechnol. 2013, 171(4), 883–899. DOI: 10.1007/s12010-013-0405-x.
- Gürakan, G. C.; Aydin, G., Qualitative Detection of GM Maize (Bt11) in Food and Feed Sold Commercially in Turkey by PCR Based Methods. 2011.
- Jinxia, A.; Qingzhang, L.; Xuejun, G.; Yanbo, Y.; Lu, L.; Minghui, Z. A Multiplex Nested PCR Assay for the Simultaneous Detection of Genetically Modified Soybean, Maize and Rice in Highly Processed Products. Food Control. 2011, 22(10), 1617–1623.
- Branquinho, M. R.; Ferreira, R. T.; Cardarelli-Leite, P. Survey of Compliance with Labeling Legislation in Food Containing GMOs in Brazil. J. Food Compost. Anal. 2010, 23(3), 220–225. DOI: 10.1016/j.jfca.2009.09.004.
- Bonny, S.; Genetically Modified Glyphosate-tolerant Soybean in the USA: Adoption Factors, Impacts and Prospects. A Review. Agron Sustain Dev. 2008, 28(1), 21–32. DOI: 10.1051/agro:2007044.
- Bai, S. H.; Ogbourne, S. M. Glyphosate: Environmental Contamination, Toxicity and Potential Risks to Human Health via Food Contamination. Environ. Sci. Pollut. Res. 2016, 23(19), 18988–19001. DOI: 10.1007/s11356-016-7425-3.
- Mesnage, R.; Defarge, N.; Spiroux de Vendômois, J.; Séralini, G. E. Potential Toxic Effects of Glyphosate and Its Commercial Formulations below Regulatory Limits. Food Chem. Toxicol. 2015, 84, 133–153. DOI: 10.1016/j.fct.2015.08.012.
- Bøhn, T.; Cuhra, M.; Traavik, T.; Sanden, M.; Fagan, J.; Primicerio, R. Compositional Differences in Soybeans on the Market: Glyphosate Accumulates in Roundup Ready GM Soybeans. Food Chem. 2014, 153, 207–215. DOI: 10.1016/j.foodchem.2013.12.054.
- Branquinho, M. R.; Gomes, D. M. V.; Ferreira, R. T. B.; Lawson-Ferreira, R.; Cardarelli-Leite, P. Detection of Genetically Modified Maize Events in Brazilian Maize-derived Food Products. Food Sci. Technol. 2013, 33(3), 399–403.
- Rabiei, M.; Mehdizadeh, M.; Rastegar, H.; Vahidi, H.; Alebouyeh, M. Detection of genetically modified maize in processed foods sold commercially in Iran by qualitative PCR. Iran. J. Pharm. Res. 2013, 12(1): 25.
- Greiner, R.; Konietzny, U.; Villavicencio, A. L. Qualitative and Quantitative Detection of Genetically Modified Maize and Soy in Processed Foods Sold Commercially in Brazil by PCR-based Methods. Food Control. 2005, 16(8), 753–759. DOI: 10.1016/j.foodcont.2004.06.015.
- Dinon, A. Z.; de Melo, J. E.; Arisi, A. C. M. Monitoring of MON810 Genetically Modified Maize in Foods in Brazil from 2005 to 2007. J. Food Compost. Anal. 2008, 21(6), 515–518. DOI: 10.1016/j.jfca.2008.04.008.
- Querci, M.; Jermini, M.; Van den Eede, G., The Analysis of Food Samples for the Presence of Genetically Modified Organisms. TRAINING COURSE ON, 2006: p. 33.