ABSTRACT
Identification of meat and meat products is significant for economic, religious, or public health reasons. This study focused on the detection and differentiation of meat animal species. A total of sixty-four meat samples (10 samples of each like beef, buffen, chevon, and mutton; twenty-four samples of pork) were analyzed for these purposes. PCR-RFLP of 12S rRNA gene with four restriction enzymes was performed on individual meat samples as well as mixture of different meat. Species specific primers as well as sequencing of DNA were also used to detect and verify the RFLP results. PCR amplification yielded a 456bp fragments from 100% samples. AluI digested beef DNA into 359 and 97bp, chevon and mutton DNA into 246 and 210bp fragments and pork DNA into 290 and 166bp fragments. However, it cannot digest buffen DNA. On the other hand HhaI digested only buffen DNA into 247 and 209bp fragments but not others. ApoI restricts only mutton DNA and yielded 329 and 127bp fragments but not others. Nevertheless, BspTI yielded 323 and 133bp fragments only from chevon. Besides, mixed samples (1:1) of beef-buffen and chevon-mutton can be differentiated by AluI & HhaI and BspTI & ApoI, respectively. Chord analysis based on our sequence and respective sequences of studied animals from GenBank indicated that the cutting sites are conserved in these species of animals. Phylogenetically each of the species was clustered separately. This technique may be useful for meat animal species detection and differentiation either from individual meat and mixed meat samples.
Introduction
Cattle meat (beef), buffalo meat (buffen), goat meat (chevon), and sheep meat (mutton) account for the largest share within the total meat consumption in Bangladesh. A major portion of meat is used as fresh meat for immediate consumption, while some portion is processed in different ways into a wide variety of processed meat products. Adulteration of meat products has become a serious issue in the past three decades in different countries of the world including Bangladesh.[Citation1] Adulteration of meat may take in the form of substitution of costly meat with cheaper one.[Citation2,Citation3] Mixing of buffen with beef is a very old and common practice in many countries of the world.[Citation4,Citation5] In some countries, since the pork is cheaper than beef, it was often found that producers mix pork into beef, mainly in processed meat products such as meatballs for economic benefits.[Citation6,Citation7]
Beef is the most popular red meat in the world preferred for its taste, flavor, and texture. Therefore, it is the most expensive meat, and consumers are ready to pay more prices for its delicacy. Whereas buffalo meat is relatively coarse and less popular compared to beef. Although a considerable number (1.49 million) of buffalo population is available in Bangladesh[Citation8] and transportation of these species to the metropolitan cities is a common scenario of the country, but it is hard to find butcher shop or slaughterhouse in the market who sells buffalo meat. On the other hand, goat meat or chevon is also popular red meat in Bangladesh and stands second in position (goat population in Bangladesh is 26.21 million)[Citation8] among the produced red meat. Sheep meat is generally not popular with consumers in Bangladesh due to its characteristic odor. Similar to buffalo, it is very hard to find sheep meat in the shop though 3.51 million sheep are available in Bangladesh[Citation8] and they are also transported to the city areas. Bangladesh is now planning for large-scale exports of cattle meat (http://theindependentbd.com/post/212545). Hence, to support the livestock industry and protect from adulteration, meat species identification is significant.[Citation9] Furthermore, from the perspective of religions, pork and diet or foods containing pork or pork derivatives are prohibited for Muslims. Besides, beef and diet or foods containing beef or beef derivatives are prohibited for Hindus. Besides consumer satisfaction, certain social and religious concerns, and possible health hazards associated with particular type of meat warrants honest labeling of the source of meat and meat products. The malpractice of fraudulent meat substitution or mislabeling is more common in countries with rather poor economy and high population with ever-increasing demand for meat and meat products and their high cost. In addition to socio-religious factors, food allergy due to consumption of particular type of meat or meat products has emerged as another major health concern implicating the beef (73%), pork (58%), and chicken (41%) as the most common cause.[Citation10] Therefore, precise differentiation of the origin of meat has become a vital element in food quality control procedures.
Protection of consumers and producers from mislabeled meat products, fraudulent actions, and bad practices of meat adulterations through processing and marketing and the prevention of illegal sale of protected species were always critical concerns that enforce legal authorities as well as many researchers to develop different techniques and analytical methods for species identification present in meat or their products including a wide range of degraded and processed materials that were broadly based on measuring either DNA or protein.[Citation11–17] DNA based techniques developed in the recent past have been found to be of potential value in the identification of meat animal species overcoming the disadvantages of conventional methods. DNA characterized by greater stability under intensive heating, pressure, and chemical processing, has conserved structure in whole body cells, has a great identification power since they are relied on the recognition of specific DNA segments sequence of a particular tissue or animal.[Citation12,Citation18–25] Some of the techniques employed for this purpose include dot blot,[Citation26] PCR,[Citation27] RAPD-PCR,[Citation28,Citation29] and PCR-RFLP,[Citation30] nucleotide sequencing.[Citation31]
The PCR-RFLP technique employed for differentiation of meat animal species consists of amplification of a conserved region of DNA sequence by PCR and subsequent digestion of the PCR product employing restriction enzymes that reveal the differences between species Partis et al.[Citation32] PCR-RFLP has been reported as a trustworthy technique for DNA sequencing[Citation33] Henceforward, it can be used as an alternative to DNA sequencing for meat species identification. This technique requires a low amount of DNA but produces co-dominant genetic markers.[Citation34] The technique is simple, easy to perform, less time consuming, and indeed cheaper than DNA sequencing.[Citation35] Additionally, it does not require sophisticated equipment or high technical skills. The technique has been successfully applied targeting different regions viz. cytochrome b gene,[Citation36] 16S rRNA gene,[Citation37] mitochondrial 12S rRNA gene.[Citation22] PCR-RFLP is also used in wild and domestic small herbivores as an aid to wildlife forensic.[Citation38] In the present study PCR-RFLP is used targeting the mitochondrial 12S rRNA gene for identification of beef, buffen, chevon, mutton, and pork from individual as well as from mixed meat samples.
Materials and method
Instruments and reagents
Different instruments were used in this study such as centrifuge machine (Hitachi, CF16RX70, Japan), NanoDrop 2000 spectrophotometer (Thermo scientific, USA), Thermocycler (GeneAtlas, G-02, Japan), Gel Electrophoresis chamber with comb (MGU-402 T, CBS Scientific Company, USA), UV trans-illuminator (WUV-L50, Korea), high-performance gel documentation chamber (Major Science, UVDI-254.), and −20°C freezer (Sanyo, MDF-U5411, Japan). Reagents such as dNTPs, DNA marker, PCR grade water, agarose, and Taq polymerase were purchased from Bio Basic, Inc., Canada. Ethidium bromide was purchased from Sigma Aldrich, Germany. Restriction endonuclease (AluI, HhaI, ApoI, and BspTI) was acquired from New England Biolabs (NEB).
Collection of meat samples
A total of 40 meat samples, 10 from each species of cattle, buffalo, goat, and sheep were collected from butcher shops of different regions of the country. Besides, 24 meat samples of pig (12 from Jessore and 12 derived from pig meat shop at Farmgate, Dhaka) were included in this study. Samples were transported to the laboratory of Animal Biotechnology Division (ABD), National Institute of Biotechnology (NIB) and stored at −20°C until processed. For every sample about 250 g meat was collected.
Isolation of genomic DNA
The genomic DNA of the meat samples was extracted as per the method described earlier [Citation39] with some modifications. Briefly, about 0.5 g of meat was minced with pestle and mortar and taken into a microcentrifuge tube. About 600 μl of TNES buffer (50.0 mM Tris, 400 mM NaCl, 100.0 mM EDTA, 0.5% SDS) and 35 μl of Proteinase-K (20 mg/ml) were added and mixed several times by inverting the tube. The mixture was incubated overnight at 50°C. About 166.7 μl of 6 M NaCl, 225 µl of chloroform was added and mixed well for 10 minutes by shaking the tube followed by centrifugation at 15,000 rpm for 10 minutes. The supernatant was collected in to a new microcentrifuge tube, and equal volume (~800 μl) of ice-cold 100% ethanol was added, gently mixed by inverting the tube and centrifuged at 15,000 rpm for 15 minutes at 4°C. The supernatant was poured off, and DNA pellets were washed with 700 μl of 100% ethanol. Then, 200 µl TE buffer was added and incubated at 37°C for overnight. The mixture was homogenized by pipetting about 30 times and stored at −20°C for future use.
Primers
Primers used in this study were reported earlier[Citation40–42] and depicted in . All primers were obtained from IDTT, USA.
Table 1. List of primes used in the present study
Polymerase chain reaction
Polymerase chain reaction was carried out in 25 µl volume containing 2.5 µl of 10x PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-HCl [pH8.0], 0.1% Triton X-100), 1.8 µL of 25 mmol/L MgCl2, 2.5 µl of 2 mmol/L of each dNTP, 2.0 µl of 50 ng/µl genomic DNA, 2.5 µl of 5 µmol/L each primer, 1.0 U Taq DNA polymerase & the rest was ddH2O. Primers used in this study were reported earlier (). Thermal condition for universal primers were as follows: initial denaturation at 94°C for 5 minutes followed by 30 cycles of denaturation at 94°C for 45 seconds, annealing at 60°C for 45 seconds and extension at 72°C for 1 minute, and final extension at 72°C for 10 minutes as described earlier.[Citation22,Citation40] Thermal cycling was performed using GeneAtlas (Model: G02, Japan). The PCR product was analyzed by electrophoresis in 1.0% agarose gel staining with ethidium bromide.
Restriction fragment length polymorphism analysis
PCR amplicons of the mitochondrial 12S rRNA gene was subjected to restricted digestion as reported previously.[Citation22,Citation40] Four restriction enzymes viz. AluI, HhaI, ApoI, and BspTI were used. Digestion reaction consisted of 10X Buffer Tango 1 µl, respective restriction enzyme 0.5 µl, the sample (PCR product) 5 µl and nuclease free water 3.5 µl. Digested product was visualized by electrophoresis in 2% agarose gel.
Preparation of meat mixture, isolation of DNA and PCR-RFLP
Meat mixtures were prepared by mixing the meat samples of two different species. For this purpose, equal volume of beef & buffen (50:50) and chevon & mutton (50:50) was mixed. Then, DNA extraction and PCR-RFLP were performed as mentioned above.
Detection of meat samples by species specific primers
Only two sets of primers () for detection of mutton and beef were employed. The reaction mixture was the same as mentioned above. The thermal conditions for specific primers consisted of 30 cycles and each cycle included holding at 94°C for 45seconds, at 58°C for 45seconds, and at 72°C for 90seconds.[Citation41,Citation42] The PCR product was analyzed by electrophoresis in 1.5% agarose gel staining with ethidium bromide.
Sequencing of genes and restriction site analysis
DNA from a total of nine samples (5 beef, 1 buffen, 1 mutton, 1 chevon, and 1 pork) were sequenced using Genetic Analyzer 3130 (Applied Biosystems) using dideoxy chain termination method.[Citation43] Sequences were edited and analyzed by Molecular Evolutionary Genetics Analysis (MEGA-X) software.[Citation44] Restriction sites for endonucleases those are used in this study were searched for in the obtained sequences. Phylogenetic trees were made using the neighbor joining method with 1000 bootstrap replications.[Citation45] The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site.[Citation46]
R plot: chord analysis
The best BLAST hit 500 sequences from GenBank were obtained, analyzed, and visualized by R using Circlize package.[Citation47] Here, we developed a chord diagram, where the data were arranged radially around a circle to find out the relationships of tested restriction enzymes with the presence or absence of their cutting sites within the amplified 12S rRNA gene region of examined meat samples of five different animals.
GenBank accession number
The nucleotide sequences were submitted to GenBank under the accession number MT253537, MT253538, MT253539, MT253540, and MT253541 for beef, MT253542 for buffen, MT253543 for mutton, MT253544 for chevon and MT253545 for pork.
Results
Amplification of genomic DNA
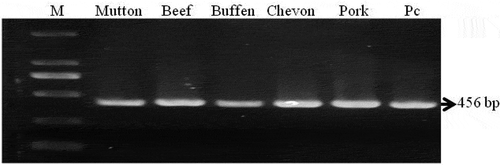
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis of 12S rRNA gene was performed to determine and differentiate the animal species of meat. A total of 64 samples were used in this analysis. About 456bp fragment from mitochondrial 12S rRNA gene of all samples (100%) of five species, viz. cattle, buffalo, sheep, goat, and pig meat samples were amplified (). A representative image of amplified 12S rRNA gene is presented in .
Table 2. Identification of meat of different animal species by PCR-RFLP of 12s rRNA gene
Identification of different meats by RFLP
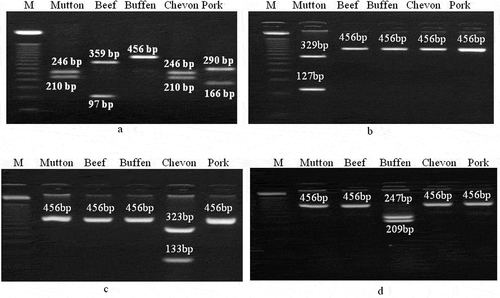
PCR amplicons obtained from meat samples of different animal species were digested with restriction enzymes. This digestion resulting in a pattern that could identify and differentiate each of the species employed in the study ( and ). The procedure was also employed for mixed meat samples (beef & buffen mixture and chevon & mutton mixture) to identify and differentiate meat species. It was clear that only AluI has a cutting site in amplified DNA of four species viz. mutton, beef, chevon, and pork. Each of the other three enzymes has a cutting site only in DNA from one species. Amplicons obtained from meat samples of different animal species were digested with AluI and the enzyme cuts the 456bp products of mutton DNA into two fragments of 246 and 210bp, beef DNA into 359 and 97bp, chevon DNA into 246 and 210 bp, pork DNA into 290 and 166 bp (). However, AluI could not cut the DNA from buffen. Digestion of PCR amplicons with ApoI restriction enzyme generated two fragments of 329 and 127bp from sheep meat sample. ApoI has no cutting site within the tested 456bp fragments of DNA from beef, buffen, chevon, and pork (). PCR products digested with BspTI restriction enzyme produced 323 and 133bp fragments from chevon but remain undigested DNA from mutton, beef, buffen and pork. () HhaI cut buffen DNA into two fragments of 247 and 209bp (). However, HhaI could not cut the DNA from beef, mutton, chevon, and pork ().
Figure 2. RFLP of 456bp fragment of mitochondrial 12S rRNA gene for differentiation of meat from different species of animals. Digestion pattern of 456bp DNA fragments of different species by a) AluI, b) ApoI, c) BspTI, and d) HhaI. Different fragment sizes are indicated in the figure. Lane M: DNA marker
PCR-RFLP of DNA from mixed meat samples
Results of detection and differentiation of mixed meat samples by PCR-RFLP are presented in . PCR amplicons derived from DNA from mixed meat samples of beef and buffen were digested with AluI and HhaI. Three types of bands were found in both cases. PCR amlplicons were digested with AluI that yielded fragments of 456, 359, and 97 bp, while 456, 247 and 209 bp fragments were found in the case of HhaI digestion. On the other hand, PCR amplicons of DNA from mutton and chevon mixed samples were digested with BspTI and ApoI. BspTI generated fragments of 456, 323 and 133 bp and ApoI generated 456, 329, and 127 bp fragments.
Table 3. Detection and differentiation of mixed meat samples by PCR-RFLP
Identification of mutton and beef by species specific primers
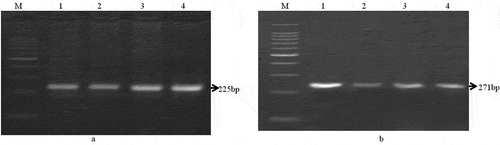
For further identification of mutton and beef PCR was performed with species specific primers, where a 225bp fragment of mitochondrial 12S rRNA gene from mutton () and 271bp fragment from beef () were amplified, respectively.
Restriction site analysis
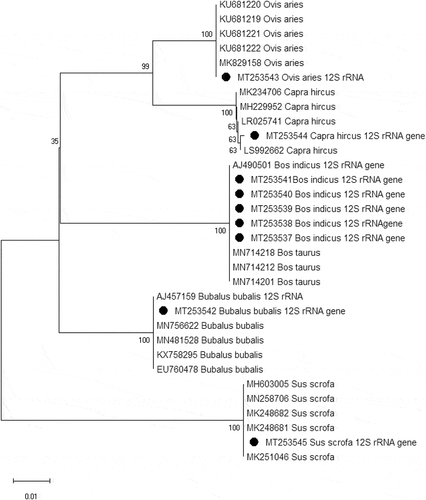
To confirm the digestion results of DNA, it was sequenced from all the species tested. Obtained sequences were analyzed for restriction sites for the enzymes used in this study and found restriction sites and patterns are consistent for each animal with the digestion result of RFLP banding pattern of 12S rRNA gene in the agarose gel. Phylogenetic analysis revealed the animal species of meat samples used in this study. Specifically, the sequences were clustered species wise and well separated from each species ()
Figure 4. Phylogenetic analysis of mitochondrial 12S rRNA gene of Cattle (Bos indicus), Buffalo (Bubalus bubalis), Sheep (Ovis aries), Goat (Capra hircus) and Pig (Sus scrofa). The tree was inferred using Neighbor-Joining method with 1000 bootstrap replication. Sequences generated in this study are marked with black circle in the tree. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site
Chord analysis
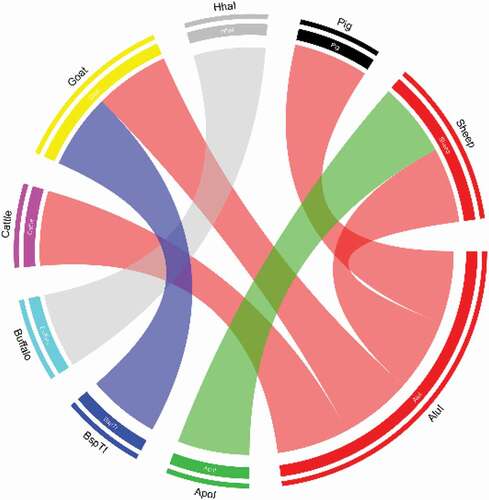
In addition to analysis of sequences of samples from this study, the best BLAST hit 500 sequences from GenBank were analyzed and found to have identical restriction sites and positions for each animal type. From restriction digestion analysis and studying the chord diagram () we found that restriction enzyme AluI have the capability to cut the amplified 12S rRNA gene fragment of ship, pig, goat, and cattle but not buffalo DNA. In addition, ApoI, BspTI and HhaI have the cutting in amplified sheep, goat, and buffalo DNA, respectively.
Figure 5. Chord analysis of restriction enzymes in portion of 12S rRNA gene of cattle, buffalo, goat, sheep and pig. In the diagram all four different types of colored arcs are originates from four different restriction enzymes. Color of each type of arcs indicates their cutting or digestion capacity against amplified 12S rRNA DNA fragment of meat sample of five different animals tested. Here, red color indicates restriction enzyme AluI, green indicates ApoI, blue indicates BspTI, and gray indicates HhaI
Discussion
Identification of meat species is necessary for various purposes including the consumer preferences, religious beliefs, public health matters, regulatory purposes, and so on. Meat adulteration has become a hot topic for researchers as it is becoming a common illegal practice especially in under-developed countries.[Citation48–50] Import of bone and meat meal in Bangladesh also requires laboratory confirmation regarding the content source that is the product is not preparing from swine origin materials. Although meat species identification is a burdensome and intricate process, nevertheless many DNA-based methods have been emerged as a potential tool in precise identification of animal meat species targeting highly conserved mitochondrial DNA sequence.[Citation35,Citation51,Citation52] This method has been empowered by exploring universal primers design for 12S rRNA gene,[Citation40] and the difference in its sequences is sufficient to use it as a target for species identification.[Citation22,Citation53,Citation54] In this study employed PCR-RFLP technique was used targeting mitochondrial 12S rRNA gene isolated from five meat species viz. beef, buffen, chevon, mutton, and pork that had been collected from different local markets in Bangladesh. The PCR products of all meat samples were produced 456bp fragment that suggested the reliability of universal primers made from 12S rRNA gene. Girish et al.[Citation22] reported the identification and differentiation of beef, buffen, chevon, and mutton meat. However, the present study added pig meat in addition with previously published study.
The restriction mapping of mt12S rRNA gene exhibited a significant variation and produced distinct fragment size followed by digesting with AluI, HhaI, BspTI and ApoI to the PCR amplicons of different meat samples, thus, enabling PCR-RFLP to distinguish specific meat in the mixture. In a similar report, Partis et al.[Citation32] generated DNA fingerprints for 22 animal species by amplifying 359bp regions within the cytochrome b gene and digesting the amplified product with HaeIII and HinfI. Our study used PCR-RFLP to differentiate between closely related meat species such as cattle-buffalo and goat-sheep. Here, AluI restriction enzyme produced two fragments 359 and 97bp from beef DNA, whereas 246 and 210bp fragment for mutton and chevon (). Therefore, AluI restriction enzyme can differentiate mutton and chevon from beef as per their generated fragment size. Furthermore, HhaI restriction enzyme has specific cutting site for buffen PCR amplicons and produce two fragments of 247 and 209bp (). Hence, the findings of this study are in agreement with those of Girish et al.[Citation22] Furthermore, this study explored the specific cutting site of AluI in the pork and generated 290 and 166bp fragments (). Moreover, the analyses of meat species were executed in the mixture of meats and found that this technique is suitable for detection and differentiation of meat in mixture (). However, the limitation of this experiment is that only two meats were mixed in a ratio/proportion of 50:50. Variation in animal species and mixture ratio/proportion may yield different results. In this connection, Mahajan et al.[Citation55] conducted an experiment with deliberately adulterated 54 meat samples. They used six combinations of cattle, buffalo, goat, and sheep meats in three proportions viz. 50:50, 75:25, and 90:10 of mixture. They identified and differentiated all samples of 50:50 and 75:25 proportions. But in mixtures with proportion of 90:10 they could not confirm the identity of the species from all combination except cattle and buffalo meat mixture. Thus, PCR-RFLP analysis of mt12S rRNA gene is a technique of potential value in identification and differentiation of meat species. Besides, both sequences of our sample and chord analysis with the sequences from GenBank reveal the potentials of this technique for differentiation of meat animal species.
Conclusion
Identification of meat is important due to religious belief, social, forensic, and public health reasons. Here, we showed that PCR-RFLP is suitable for identification and differentiation of individual mutton, beef, buffen, chevon, and pork, as well as mixed meat samples. Therefore, this study can be applicable for both individual and mixture of meats for their authentication that could help in the investigation and protection of meat adulteration.
Acknowledgments
The study was supported by the core fund of the National Institute of Biotechnology (NIB), Savar, Dhaka.
References
- Khatun, M. A.; Hossain, A.; Hossain, M. S.; Munshi, M. K.; Huque, R. Detection of Species Adulteration in Meat Products and Mozzarella-type Cheeses Using Duplex PCR of Mitochondrial Cyt B Gene: A Food Safety Concern in Bangladesh. Food Chem. Mol. Sci. 2021, 100017. DOI: https://doi.org/10.1016/j.fochms.2021.100017.
- Bhat, J. H.; Para, P. A.; Bukhari, S. A.; Ganguly, S.; Bhat, A. A.; Wakchaure, R.; Qadri, K. Fraudulent Adulteration/substitution of Meat. Int. J. Recent Res. Appl. Stud. 2015, 2(12), 22–33.
- Yman, I. M.; Emanuelsson, R. New Technology for Faster Disclosure of Meat Adulteration. VarFada (In Swedish). 1998, 3, 6–7.
- Cheng, J.; Chou, H.; Lee, M.; Sheu, S. Development of Qualitative and Quantitative PCR Analysis for Meat Adulteration from RNA Samples. Food Chem. 2016, 192, 336–342. DOI: https://doi.org/10.1016/j.foodchem.2015.06.094.
- Munira, S.; Jahura, F. T.; Hossain, M. M.; Bhuiyan, M. S. A. Molecular Detection of Cattle and Buffalo Species Meat Origin Using Mitochondrial Cytochrome B (Cyt B) Gene. Asian J. Med. Biol. Res. 2016, 2(2), 177–182. DOI: https://doi.org/10.3329/ajmbr.v2i2.29008.
- Al-Jowder, O.; Kemsley, E. K.; Wilson, R. H. Mid-infrared Spectroscopy and Authenticity Problems in Selected Meats: A Feasibility Study. Food Chem. 1997, 59(2), 195–201. DOI: https://doi.org/10.1016/S0308-8146(96)00289-0.
- Ballin, N. Z.; Vogensen, F. K.; Karlsson., A. H. Species Determination – Can We Detect and Quantify Meat Adulteration? Meat Sci. 2009, 83, 165–174. DOI: https://doi.org/10.1016/j.meatsci.2009.06.003.
- BER. 2019. Bangladesh Economic Review. https://mof.portal.gov.bd/sites/default/files/files/mof.portal.gov.bd/page/f2d8fabb_29c1_423a_9d37_cdb500260002/Ch-07%20%28english-2019%29.pdf (Accessed March 20, 2020).
- Bangladesh Now Plans Large-scale Meat Export. 2021. http://theindependentbd.com/post/212545 (Accessed June 23 2021).
- Ayuso, R.; Lehrer, S. B.; Tanaka, L.; Ibanez, M. D.; Pascual, C.; Burks, A. W.; Sussman, G. L.; Goldberg, B.; Lopez, M.; Reese, G. Ig-E Antibody Response to Vertebrate Meat Proteins Including Tropomyosin. Ann Allergy Asthma Immunol. 1999, 83, 399–405. DOI: https://doi.org/10.1016/S1081-1206(10)62837-2.
- Aida, A. A.; Che Man, Y. B.; Wong, C. M. V. L.; Raha, A. R.; Son, R. Analysis of Raw Meats and Fats of Pigs Using Polymerase Chain Reaction for Halal Authentication. Meat Sci. 2005, 69, 47–52. DOI: https://doi.org/10.1016/j.meatsci.2004.06.020.
- Calvo, J. H.; Zaragoza, P.; Osta, R. Random Amplified Polymorphic DNA Fingerprints for Identification of Species in Poultry Pate. Poult. Sci. 2001, 80(4), 522–524. DOI: https://doi.org/10.1093/ps/80.4.522.
- Herman, B. L.;. Determination of the Animal Origin of Raw Food By-species-specific PCR. J. Dairy Res. 2001, 68, 429–436. DOI: https://doi.org/10.1017/S0022029901004940.
- Matsunaga, T.; Chikuni, K.; Tanabe, R.; Muroya, S.; Shibata, K.; Yamada, J.; Shinmura, Y. A Quick and Simple Method for the Identification of Meat Species and Meat Products by PCR Assay. Meat Sci. 1999, 51, 143–148. DOI: https://doi.org/10.1016/s0309-1740(98)00112-0.
- Meyer, R.; Hoefelein, C.; Luethy, J.; Candrian, U. Polymerase Chain Reaction-restriction Fragment Length Polymorphism Analysis: A Simple Method for Species Identification in Food. J. AOAC Int. 1995, 78, 1542–1551. DOI: https://doi.org/10.1093/jaoac/78.6.1542.
- Myers, M. J.; Yancy, H. F.; Farrell, D. E. Characterization of a Polymerase Chain Reaction-based Approach for the Simultaneous Detection of Multiple Animal-derived Materials in Animal Feed. J. Food Prot. 2003, 66, 1085–1089. DOI: https://doi.org/10.4315/0362-028x-66.6.1085.
- Peter, C.; Brunen-Nieweler, C.; Cammann, K.; Borchers, T. Differentiation of Animal Species in Food by Oligonucleotide Microarray Hybridization. Eur. Food Res.Technol. 2004, 21, 286–293. DOI: https://doi.org/10.1007/s00217-004-0958-6.
- Akasaki, T.; Yanagimoro, T.; Yamakami, K.; Tomonaga, H.; Sato, S. Species Identification and PCR-RFLP Analysis of Cytochrome B Gene in Cod Fish (Order Gadiformes) Products. J. Food Sci. 2006, 71(3), 190–195. DOI: https://doi.org/10.1111/j.1365-2621.2006.tb15616.x.
- Arslan, A.; Ilhak, O. I.; Calicioglu, M. Effect of Method of Cooking on Identification of Heat Processed Beef Using Polymerase Chain Reaction (PCR) Technique. Meat Sci. 2006, 72, 326–330. DOI: https://doi.org/10.1016/j.meatsci.2005.08.001.
- Frezza, D.; Favaro, M.; Vaccari, G.; Von-Holst, C.; Giambra, V.; Anklam, E. A Competitive Polymerase Chain Reaction based Approach for the Identification and Semiquantification of Mitochondrial DNA in Different Heat-treated Bovine Meat and Bone Meal. J. Food Prot. 2003, 66, 103–109. DOI: https://doi.org/10.4315/0362-028x-66.1.103.
- Girish, P. S.; Anjaneyulu, A. S. R.; Viswas, K. N.; Anand, M.; Rajkumar, N.; Shivakumar, B. M. Sequence Analysis of Mitochondrial 12S rRNA Gene Can Identify Meat Species. Meat Sci. 2004, 66, 551–556. DOI: https://doi.org/10.1016/S0309-1740(03)00158-X.
- Girish, P. S.; Anjaneyulu, A. S. R.; Viswas, K. N. Meat Species Identification by Polymerase Chain Reaction-restriction Fragment Length Polymorphism (PCR-RFLP) of Mitochondrial 12S rRNA Gene. Meat Sci. 2005, 70(1), 107–112. DOI: https://doi.org/10.1016/j.meatsci.2004.12.004.
- Iwobi, A.; Sebah, D.; Kraemer, I.; Losher, C.; Fischer, G.; Busch, U.; Huber, I. A Multiplex Real-time PCR Method for the Quantification of Beef and Pork Fractions in Minced Meat. Food Chem. 2015, 169, 305–313. DOI: https://doi.org/10.1016/j.foodchem.2014.07.139.
- Lanzilao, I.; Burgalassi, F.; Fancell, S.; Settimelli, M.; Fani, R. Polymerase Chain Reaction-restriction Fragment Length Polymorphism of Mitochondrial Cyt B Gene from Species of Dairy Interest. J. AOAC Int. 2005, 88, 128–135. DOI: https://doi.org/10.1093/jaoac/88.1.128.
- Rashid, P. M. A.; Babashekh, M. O.; Marouf, A. S.; Amin, K. M. Identification of Animal Species in Meat Broth by Simplex and Multiplex PCR. J. Zankoy Sulaimani-Part A. 2014, 16(1), 97–102. DOI: https://doi.org/10.17656/jzs.10288.
- Ebbehoj, K. F.; Thomson, P. D. Differentiation of Closely Related Species by DNA Hybridization. Meat Sci. 1991, 30, 359–366. DOI: https://doi.org/10.1016/0309-1740(91)90044-Q.
- Cespedes, A.; Garcia, T.; Carrera, E.; Gonzalez, I.; Fernandez, A.; Hernandez, P. E.; Martin, R. Identification of Sole (Solea Solea) and Greenland Halibut (Reinhardtius Hippoglossoides) by PCR Amplification of the 5S rDNA Gene. J. Agric. Food Chem. 1999, 47, 1046. DOI: https://doi.org/10.1021/jf9810970.
- Ganai, T. A. S.; Singh, R. K.; Butchaiah, G. DNA Amplification Fingerprinting of Cattle and Buffalo Genome by RAPD-PCR Utilizing Arbitrary Oligonucleotide Primers. Buffalo J. 2000, 3, 331–339.
- Koh, M. C.; Lim, C. H.; Chua, S. B.; Chew, S. T.; Phang, S. T. W. Random Amplified Polymorphic DNA (RAPD) Fingerprints for Identification of Red Meat Animal Species. Meat Sci. 1998, 48, 275–285. DOI: https://doi.org/10.1016/s0309-1740(97)00104-6.
- Chung, E. R.; Kim, W. T.; Kim, Y. S.; Han, S. K. Identification of Hanwoo Meat Using PCR-RFLP Marker of MC1R Gene Associated with Bovine Coat Color. Korean J. Anim. Sci. Technol. 2000, 42(4), 379–390.
- Colombo, F.; Marchisio, E.; Pizzini, A.; Cantoni, C. Identification of the Goose Species (Anser Anser) in Italian “Mortara” Salami by DNA Sequencing and a Polymerase Chain Reaction with an Original Primer Pair. Meat Sci. 2002, 61(3), 291–294. DOI: https://doi.org/10.1016/s0309-1740(01)00195-4.
- Partis, L.; Croan, D.; Guo, Z.; Clark, R.; Coldham, T.; Murby, J. Evaluation of a DNA Fingerprinting Method for Determining the Species Origin of Meats. Meat Sci. 2000, 54(4), 369–376. DOI: https://doi.org/10.1016/s0309-1740(99)00112-6.
- Haider, N. Development and Use of Universal Primers in Plants. PhD thesis, University of Reading, UK. 2003.
- Graner, A.; Streng, S.; Kellermann, A.; Schiemann, A.; Bauer, E.; Waugh, R.; Pellio, B.; Ordon, F. Molecular Mapping and Genetic Fine-structure of the Rym5 Locus Encoding Resistance to Different Strains of the Barley Yellow Mosaic Virus Complex. Theor. Appl. Genet. 1999, 98(2), 285–290. DOI: https://doi.org/10.1007/s001220051070.
- Haider, N.; Nabulsi, I.; Al-Safadi, B. Identification of Meat Species by PCR-RFLP of the Mitochondrial COI Gene. Meat Sci. 2012, 90(2), 490–493. DOI: https://doi.org/10.1016/j.meatsci.2011.09.013.
- Branciari, R.; Avellini, P.; Sukasi, S. R.; Antonio, E.; Rea, S. PCR-RFLP Analysis (Polymerase Chain Reaction Restriction Fragment Length Polymorphism) for Species Determination in Heat Treated Meat Products. Industrie-Alimentarie. 2000, 39, 313–318.
- Borgo, R.; Souty-Grosset, C.; Bouchon, D.; Gomot, L. PCR-RFLP Analysis of Mitochondrial DNA for Identification of Snail Meat Species. J. Food Sci. 1996, 61, 1–4. DOI: https://doi.org/10.1111/j.1365-2621.1996.tb14712.x.
- Rajput., N.; Shrivastav, A. B.; Parmar, S. N. S.; Ranjan, R. Characterization of 12S rRNA Gene for Meat Identification of Common Wild and Domestic Small Herbivores as an Aid to Wildlife Forensic. Vet. World. 2013, 6(5), 254–259. DOI: https://doi.org/10.5455/vetworld.2013.254-259.
- Saderi, M.; Saderi, A. H.; Rahimi, G. Identification of Bovine, Ovine and Caprine Pure and Binary Mixtures of Raw and Heat Processed Meats Using Species Specific Size Markers Targeting Mitochondrial Genome. Iranian J. Vet. Res. 2013, 14(1), 1, 29–34.
- Kocher, T. D.; Thomas, W. K.; Meyer, A.; Edwards, S. V.; Pääbo, S.; Villablanca, F. X.; Wilson, A. C. Dynamics of Mitochondrial DNA Evolution in Animals: Amplification and Sequencing of Conserved Primers. PNAS. 1989, 86(16), 6196–6200. DOI: https://doi.org/10.1073/pnas.86.16.6196.
- Lahiff, S.; Glennon, M.; O’Brien, L.; Lyng, J.; Smith, T.; Maher, M.; Shilton, N. Species-specific PCR for the Identification of Bovine, Porcine, and Chicken Species in Meat and Bone Meal (MBM). Mol. Cell.Probes. 2001, 15, 27–35. DOI: https://doi.org/10.1006/mcpr.2000.0336.
- Tartaglia, M.; Saulle, E.; Pestalozza, S.; Morelli, L.; Antonucci, G.; Battaglia, P. A. Detection of Bovine Mitochondrial DNA in Ruminant Feeds: A Molecular Approach to Test for the Presence of Bovine Derived Materials. J. Food Prot. 1998, 61(5), 513–518. DOI: https://doi.org/10.4315/0362-028x-61.5.513.
- Sanger, F.; Coulson, A. R. A Rapid Method for Determining Sequences in DNA by Primed Synthesis with DNA Polymerase. J. Mol. Biol. 1975, 94(3), 441–448. DOI: https://doi.org/10.1016/0022-2836(75)90213-2.
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35(6), 1547–1549. DOI: https://doi.org/10.1093/molbev/msy096.
- Felsenstein, J.;. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985, 39(4), 783–791. DOI: https://doi.org/10.2307/2408678.
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-joining Method. PNAS. 2004, 101(30), 11030–11035. DOI: https://doi.org/10.1073/pnas.0404206101.
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and Enhances Circular Visualization in R. Bioinformatics. 2014, 30(19), 2811–2812. DOI: https://doi.org/10.1093/bioinformatics/btu393.
- Singh, V.; Neelam, S. Meat Species Specifications to Ensure the Quality of Meat: A Review. Int. J. Meat Sci. 2011, 1, 15–26. DOI: https://doi.org/10.3923/ijmeat.2011.15.26.
- Pascoal, A.; Prado, M.; Castro, J.; Cepeda, A.; Barros-Velazquez, J. Survey of Authenticity of Meat Species in Food Products Subjected to Different Technological Processes, by Means of PCR-RFLP Analysis. Eur. Food Res.Technol. 2004, 218, 306–312. DOI: https://doi.org/10.1007/s00217-003-0846-5.
- Waqas, M.; Hussain, Z.; Ihsan, A. Meat Species Identification: Amplification Refractory Mutation System-polymerase Chain Reaction–based Assay. Food Anal. Methods. 2019, 12(12), 2813–2822. DOI: https://doi.org/10.1007/s12161-019-01640-2.
- Farag, M. R.; Alagawany, M.; Abd El-Hack, M. E.; Tiwari, R.; Dhama, K. Identification of Different Animal Species in Meat and Meat Products: Trends and Advances. Adv.Anim. Vet. Sci. 2015, 3(6), 334–346. DOI: https://doi.org/10.14737/journal.aavs/2015/3.6.334.346.
- Van Der Kuyl, A. C.; Kuiken, C. L.; Dekker, J. T.; Goudsmit, J. Phylogeny of African Monkeys Based upon Mitochondrial 12S rRNA Sequences. J. Mol. Evol. 1995, 40(2), 173–180. DOI: https://doi.org/10.1007/BF00167111.
- Girish, P. S.; Haunshi, S.; Vaithiyanathan, S.; Rajitha, R.; Ramakrishna, C. A Rapid Method for Authentication of Buffalo (Bubalusbubalis) Meat by Alkaline Lysis Method of DNA Extraction and Species Specific Polymerase Chain Reaction. J.Food Sci. Technol. 2013, 50(1), 141–146. DOI: https://doi.org/10.1007/s13197-011-0230-6.
- Prakash, S.; Patole, M. S.; Ghumatkar, S. V.; Nandode, S. K.; Yogesh, S.; Shouche, Y. S. Mitochondrial 12S rRNA Sequence Analysis in Wild Life Forensics. Current Sci. 2000, 78, 1239–1241.
- Mahajan, M. V.; Gadekar, Y. P.; Dighe, V. D.; Kokane, R. D.; Bannalikar, A. S. Molecular Detection of Meat Animal Species Targeting MT 12S rRNA Gene. Meat Sci. 2011, 88(1), 23–27. DOI: https://doi.org/10.1016/j.meatsci.2010.11.026.