Abstract
This study aimed to understand whether the geo-ecological segregation of native plant species affects the root-associated fungal community. Rhizoplane (RP) and rhizosphere (RS) fungal microbiota of Sedum takesimense native to three geographically segregated coastal regions (volcanic ocean islands) were analyzed using culture-independent methods: 568,507 quality sequences, 1399 operational taxonomic units, five phyla, and 181 genera were obtained. Across all regions, significant differences in the phyla distribution and ratio were confirmed. The Chao’s richness value was greater for RS than for RP, and this variance coincided with the number of genera. In contrast, the dominance of specific genera in the RS (Simpson value) was lower than the RP at all sites. The taxonomic identity of most fungal species (95%) closely interacting with the common host plant was different. Meanwhile, a considerable number of RP only residing fungal genera were thought to have close interdependency on their host halophyte. Among these, Metarhizium was the sole genus common to all sites. These suggest that the relationship between potential symbiotic fungi and their host halophyte species evolved with a regional dependency, in the same halophyte species, and of the same natural habitat (volcanic islands); further, the fungal community differenced in distinct geographical regions. Importantly, geographical segregation should be accounted for in national culture collections, based on taxonomical uniqueness.
1. Introduction
Since the Nagoya Protocol calls for strengthening the rights of the “management nation” toward biological resources, competition between countries to secure national microbial diversity and various culture collections has become fierce [Citation1]. As the nation’s right who secure microbial resources has been strengthened, it has become important to quickly secure various microbial culture collections. Therefore, the concept of “valuable national biodiversity” and “microbiological assets” was already introduced [Citation1]. Further, microbial culture collections enable sustainable development goals (SDGs), particularly regarding bio-economy [Citation2] and under such circumstances, scientific studies for securing microbial resources that show high diversity and applicability have been conducted vigorously [Citation3].
Microbial resources that closely interact with higher host organisms have been studied to assess their benefits in agriculture or other environmental aspects, such as stimulating host growth or boosting the immune system and helping humans cope with global warming or climate change [Citation4]. Especially, symbiotic interactions between root layer-associated soil microbes and higher host plant species have been consistently studied. Its functionality has been demonstrated using large-scale screening: increase crop potential via growth promotion, increasing their resistance to environmental stress, and protecting them from environments where diseases and natural changes are severe [Citation5–8].
Although these methods can theoretically reveal interactions between microorganisms and host plants, certain limitations have been noted, particularly concerning the diversity of bio-asset [Citation1,Citation3]. Due to an increasing number of studies focused on understanding functionalities, taxonomic positions of secured culture collection strains are gradually becoming nonspecific [Citation8]. In fact, many studies do not present taxonomical uniqueness. Importantly, it should be possible to determine which natural factors contribute most significantly to securing unique microbial resources and to identify how many unique resources are present within such unique natural factors of note, competition for microbial resources is intensifying among countries [Citation9,Citation10].
Meanwhile, although it has been reported that unique beneficial microorganisms can be discovered in special natural habitats, this will one day become challenged, as there is an inherent limit to the geographical diversity within any given nation [Citation8]. Thus, if the differentiation of a fungal community is identified within special environments (particularly geographically isolated ones), the concept of geographical isolation itself will increase the potential to secure unique microbial species or improve diversity, providing another strategy to ensure the identification of diverse and unique microbial resources [Citation3,Citation8]. However, geographic isolation does not refer to only physical segregation, but may also include temporal concepts. Hence, it must be clarified whether the persistence of isolation can increase the differentiation of symbiotic microorganisms because continuous differentiation and evolution of microbial species can occur within the isolated local ecosystem.
To ensure the competitive value of natural microbial resources, the uniqueness of the total microbiome and its species composition must be demonstrated [Citation3]; the specificity of microbial taxon composition does not directly predict the existence of uniqueness of microbial resources. However, clearly distinct, rare species or genera found in separate environments can be interpreted as minimal evidence of the presence of unique microbial species in those environments [Citation3]. By demonstrating the presence of microbial structures or unique species compositions, we can focus on securing such unique microbial species. Further, such results will help determine the extent to which geographical isolation influences the unique distribution of fungi.
Therefore, this study assessed the extent to how geographical isolation and the following ecological segregation of higher halophyte affects root layer-associated fungal diversity. For accurate analysis, we compared the fungal community of the RS (rhizosphere) and RP (rhizoplane) [Citation11,Citation12] using culture-independent analysis [Citation13,Citation14] of the same halophyte species distributed commonly in geographically isolated volcanic islands in the East Sea of the Korea peninsula. Further, using such comparative analysis, host dependency and potential symbiosis also can be proposed. If fungal genera are found commonly in geographically isolated areas, and closely interact (depend) with host plants, they are more likely to have a potential symbiotic relationship. This study proposes an example strategy for acquiring uniqueness in national fungal diversity or unique microbial collection.
2. Materials and methods
2.1. Sampling sites
The geomorphic traits of the Korean peninsula are derived from old landforms and young volcanic islands (). Such diverse natural topography induces geographical segregations, leading to ecological isolation of higher organisms and lack of genetic exchange [Citation15–18]. To assess how geographical isolation affects the characteristics and uniqueness of fungal microflora, three sites in two volcanic islands located offshore from the mainland Korean peninsula, including the Dongdo Islands (a constituent of Dokdo Islands) (), Sadong (), and Taeha () in the Ulleungdo Islands (), were selected. The Dokdo and Ulleungdo Islands are separated by 120 km from the mainland Korean peninsula () [Citation16]. The distance between the Ulleungdo and Dokdo Islands is 88 km, and they are separated by nearshore waters of the Pacific Ocean (). In addition, Sadong and Taeha are over 10 km apart () and are segregated by inland cliffs and valleys () [Citation19]. We selected Sedum takesimense Nakai, which is commonly distributed among these three sites as the target host halophyte species.
Figure 1. Geographical location and segregation of each sampling site. (A) Geographical location of the Ulleungdo and Dokdo Islands. The two volcanic islands are ecologically segregated from mainland and each other. (B) Site 1: Dongdo island of the Dokdo Islands. (C) Ulleungdo Islands; (C1) Site 2: Sadong, slope cliff near the seashore, (C2) Site 3: Primitive virgin forest in Taeha in the Ulleungdo Islands. The sampling sites are circled. The map images were downloaded from the Cultural Heritage Administration of Korea (CHA) Portal Site (http://www.heritage.go.kr/heri/idx/index.do., accessed on 21 October 2020) and modified for this study.
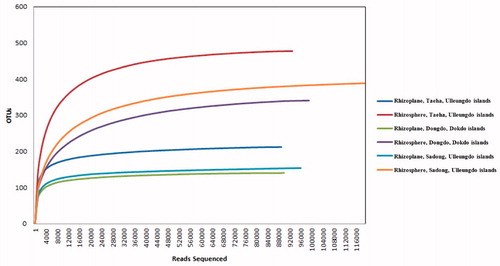
Figure 2. Rarefaction curves for operational taxonomic units (OTUs) from each site. OTUs were clustered at 3% dissimilarity using CD-HIT. The microbial community in the rhizoplane and the rhizosphere of Dongdo, Dokdo Islands (OTUs, 140/341; sequencing reads, 84,267/94,745), Sadong, Ulleungdo Islands (OTUs, 154/389; sequencing reads, 95,395/117,939), and Taeha (OTUs, 212/478; sequencing reads, 86,299/89,864) are shown.
2.2. Host plant candidate
Sedum takesimense was selected as the host plant for the comparison of associated soil fungi. Sedum takesimense is a representative native halophyte and has a long natural history of adaptation to ocean volcanoes [Citation20]. As perennial plants [Citation8,Citation21], it flourishes in an evergreen community and forms a shrubbery zone able to stably interact with root-associated soil for a long period. () [Citation8]. Further, it forms a shrubbery zone in the narrow saxicoline zone of the soil, as a saxicolous plant [Citation8]. These ecological traits avoid interference with other species in the island vegetation. The geographical information of the sampled shrubbery zone is listed in .
Table 1. Geographical information regarding the commonly identified halophyte species, Sedum takesimense, from the islands of the Korean peninsula.
2.3. Sampling of S. takesimense
RP and RS of host plants from each site were sampled; full-grown adult S. takesimense species were sampled as targets species. In the Dokdo Islands, soil loss because of steeply inclined planes is often observed [Citation21]. Host plant sites were selected such that they bore no marks of soil loss. Over 15 individual S. takesimense were sampled, and the sampled soil depth was above 30 cm. Specific permission was required for one location (Dokdo) no endangered or protected species live in the geographical regions disturbed. In each site, three soil samples (1 kg each) were collected in the vertices of 1 m side equilateral triangle and mixed in a unique representative analytical sample [Citation22]. Soil samples from each site were subdivided in two representative sub-samples: the first one was air-dried, 2 mm sieved, then chemically and physically analyzed, whereas the second one was stored at −80 °C and later processed for the pyrosequencing analysis.
RS represents soil areas affected by the plant physiological activity and physiologically interacting microorganisms [Citation23]. An RS soil core (depth: 30 cm, diameter: 2 mm) was sampled from each site. The RP region represents the region in contact with the soil of plant root surfaces containing fungal mycelia or microbial community [Citation24]. Three RP and RS samples from the three geographical regions were pooled per site. In each geographically isolated region, the RP and RS soil were obtained from three well-developed halophytic colonies and combined in one last sample for sequencing. In poor condition plant objects that make up, colonies were excluded in the sampling and pretreatment stages. For representative samples, the pooling was conducted by region, and each of the three S. takesimense colonies in a particular region was within the permitted population in an administrative manner, in the natural resources and natural cultural heritage protection zones. Soils strongly associated with plant roots were separated by vigorous vortex with 50 ml of sterile phosphate-buffered saline (PBS) solution. The roots were stirred vigorously with sterile forceps to remove all soil from the root surface. Suspensions were centrifuged (Sigma-Aldrich, St. Louis, MO, USA) at 5000 rpm for 10 min at 4 °C, and the sediment was considered RS soil. After removing all the soils, the roots were put into a tube with 50 ml sterile PBS and then sonicated for 10 s (three times) at 50 Hz in a sealed state (ultrasonic cleaner, Branson Ultrasonics, Danbury, CT, USA). The roots were then removed using sterile forceps, and the solution was centrifuged at 5000 rpm for 10 min at 4 °C. The resulting pellets were considered as RP soil.
2.4. Soil analysis
Air-dried and sieved soil sub-samples were analyzed for the following physical and chemical properties: soil texture classification; reaction (pH), and organic carbon (Corg). Soil texture was determined via the pipette method, without carbonate and organic matter removal, and after complete removal of soluble salts using distilled water [Citation25]. The variation of the pH was measured on 1:2.5 (w/v) soil to water mixtures; Corg was obtained using the Walkley and Black method [Citation26]. Total nitrogen (TN) was analyzed using the Kjeldahl method [Citation27].
2.5. DNA extraction and PCR amplification
All samples were transported to the laboratory for DNA extraction. All soil DNA was extracted using the Power Soil DNA extraction kit (MO BIO Laboratories, Carlsbad, CA, USA), following the protocol described by the manufacturer. The fungal internal transcribed spacer (ITS) region 1 was amplified using the ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) primers. The resulting ITS 1 amplicons were sequenced at Macrogen (Macrogen, Inc., Seoul, Korea), using the paired-end (2 × 300 nt) Illumina MiSeq sequencing system (Illumina, San Diego, CA, USA).
2.6. Sequence processing and statistical analyses
Paired-end sequences were assembled using the PANDAseq software [Citation28]. After assembly, all sequence data were processed using the Mothur pipeline [Citation29]. For fungal community analysis, the flanking gene fragments were removed from the ITS1 region using an ITSx version 1.0.9 [Citation30]. Putative chimeric sequences were detected and removed using the Chimera Uchime algorithm available within Mothur [Citation31], in de novo mode. QIIME2 implementation of UCLUST [Citation32,Citation33] was used to assign the OTUs, defined with a limit threshold of 97% sequence similarity. All singleton OTUs were removed from the datasets before analysis. The taxonomic classification was performed using Mothur’s version of the Naïve Bayesian classifier, using the UNITE database for fungi [Citation34]. All samples were standardized via random sub-sampling, using the “sub-sampling” command (http://www.Mothur.org/wiki/Sub.sample) in Mothur. Richness, diversity indices, and rarefaction values [Citation35] were also estimated using Mothur [Citation36,Citation37]. The raw sequences obtained from this study have been deposited at the NCBI Sequence Read Archive (SRA metadata) under the project PRJNA616037, and the sample accession numbers SAMN14480218 to SAMN14480223 (14480218-9: RS, RP from Dokdo Islands; 14480220-1: RS, RP from Sadong, Ulleungdo Islands; 14480222-3 RS, RP from Taeha, Ulleungdo Islands). The environmental sequence information released date was April 2, 2020.
3. Results and discussion
3.1. Pyrosequencing results and statistical analyses
Here, 568,507 high-quality fungal sequences were obtained from soil samples and classified into 1399 operational taxonomic units (OTUs) and were confirmed at the 97% similarity level (). Rarefaction curves for OTUs were deduced (). The number of fungal phyla did not vary based on the coastal location associated with the host plant (RS and RP). The five fungal phyla from all geographical locations (RP and RS) were Ascomycota, Basidiomycota, Chytridiomycota, Glomeromycota, and Zygomycota (). One-hundred eighty-one fungal genera were identified from the three sampling sites (six sampling soil groups) (). The ratios of confirmed fungal genera between RP and RS were 44:86 (Dongdo), 44:97 (Sadong), and 43:98 (Taeha) ().
Table 2. Pyrosequencing data and statistical analyses.
3.2. Soil texture classification, alkalinity, TN, and Corg
In previous studies, the type of rock that originates soil properties are derived from major trachyandesite, and trachyte, rhyolite, and tuff of the Dokdo islands [Citation38]. The soil properties of the Dokdo series were classified as different from the series of mainland Korean peninsula soil. The soil texture of the Dokdo sandy, compared to the coast and inland of the Ulleungdo (). Although the Dokdo and Ulleungdo soil share the same natural formation history (created by the erosion of volcanoes in the ocean), the small island areas of the Dokdo (18.7 ha) [Citation38] suffer fiercer sea-wind and extreme rainfall. Therefore, it can be interpreted that the proportion of sand in the soil has increased because of such effects (the ratio of sand in soil increased Dongdo of the Dokdo > coast of the Ulleungdo > inland of the Ulleungdo).
Table 3. Soil analysis results per geographical site.
The salt concentration of the Dongdo of the Dokdo was 0.019%. This may be associated with penetration of seawater caused by the stronger ocean breeze and sea fog. Although TN and Corg content can be affected by plant activity, the Dokdo root layer showed a relatively higher Corg and TN contents, than those in other sites. Like the inland of the Ulleungdo, which has mild conditions, higher contents of the Dokdo rhizosphere may be related to the well-established and flourishing native halophyte S. takesimense community. Although the salinity difference between the coast of the Ulleungdo and inland areas is not clear, the pH level of the coastal areas is close to the seawater. This indicates that the soil in the coastal areas of the Ulleungdo is suffering from severe seawater intrusion, similarly to the reported in previous studies [Citation20,Citation21]. Although seawater intrusion in the Dokdo is severe, strong acidity of the soil is thought to owing to the nutrient loss by extreme rainfall and sea-wind [Citation21].
Overall, the natural formation history of the three geographical regions is equal [Citation21], and an effect of strong marine condition attenuated by surrounding mountains and canopy forest in the natural habitat soil of S. takesimense community at the Ulleungdo inland might exist. Compared to such inland terrain, the S. takesimense habitat in the Dokdo Islands is viewed as being under the influence of extreme marine conditions. Meanwhile, native halophyte species include S. takesimense in such a diverse marine habitat have their own ecological function, which eliminates Na+ cations from their root layer soil [Citation39]. Therefore, the microbial community from such characteristic ocean islands needs to be discussed regarding the host halophyte ecological function and the habitat character. A recent meta-analysis of soil microbial communities reported that the global microbial composition in saline soil is influenced more by salinity than by any other extreme chemical factors such as temperature or pH [Citation40,Citation41]. Notably, the purpose of this study is not to discover how the microbial phase changes with salt concentration or soil texture, but to provide indicators for further future studies on how naturally isolated special marine terrains gave us advantageous unique microbial resources. Several studies have shown there is a clear association between the existence of such unique microbial resources and unique marine habitats [Citation42–45]. Thus, the association between two concepts (geological isolation of unique terrains) should be intensively studied amid intensifying competition to secure microbial resources.
3.3. Fungal diversity in coastal regions
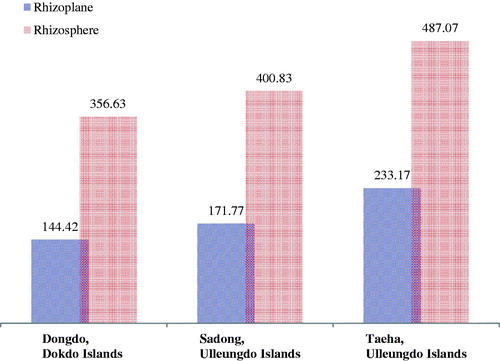
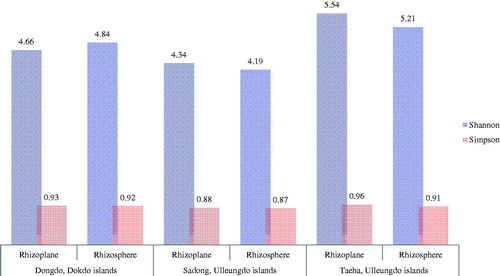
The Shannon, Chao richness and Simpson indices were used to analyze the fungal diversity [Citation46–48]. The evenness and diversity (Shannon index) of fungal genera exhibited higher values for the Taeha in the Ulleungdo (RP: 233.17, RS: 487.07) than those in other sites (RP/RS of Dongdo in the Dokdo: 144.42/356.63; Sadong in the Ulleungdo: 171.77/400.83) (). Sadong, which is located nearer to the ocean than Taeha in the Ulleungdo Islands, was thought to be affected by stronger selection pressure from the marine conditions. In the Dongdo, species richness showed the lowest values (RP: 144.42, RS: 356.63) among those in all coastal regions, regardless of RS or RP (). This is presumably because the Dokdo (Dongdo) Islands are affected by selective pressure from the maritime environment than the two Ulleungdo sites. In previous geographical studies, the extreme environment of the Dokdo Islands or their influences on the native halophyte community is already reported [Citation3,Citation21,Citation49]. Thus, we confirmed that the diversity of fungal biota, especially for halophyte-associated soils, reflects their environment. The variance of diversity values between geographically isolated sites regardless of RS or RP suggests that environmental influences fungal microbiota. Nevertheless, lower fungal richness was observed in RP (), as compared to that for RS in all geographically segregated sites across all coastal regions.
Figure 3. Fungal diversity variation from rhizoplane to rhizosphere in each island. The Simpson’s Diversity Index quantifies the biodiversity of a habitat, and considers the number of species present, and the abundance of each species. The Shannon’s diversity index (H) accounts for both abundance and evenness of the species.
3.4. Distribution and variation of the fungal phyla
The dominance of each fungal phylum was different (). The most significant differences were observed between locations or between RS and RP. The specific fungal phyla dominant in RP of Dongdo (of the Dokdo) were Ascomycota (23.23%) > Basidiomycota (21.57%) > Zygomycota (13.42%). In contrast, Zygomycota (30.14%) > Ascomycota (23.73%) > Basidiomycota (10.44%) (). In contrast, the most abundant fungal group in the RS was Zygomycota, whereas that in RP was Ascomycota.
Figure 5. Comparison of the variation in dominant fungal phyla between the rhizosphere and the rhizoplane. Differences are highlighted. The dominance ratio (%) is represented per fungal phyla. Comparing the sites, the rank, and ratio of dominance are similar in the rhizoplane of all sampling sites, but with the rhizosphere areas, more affected by the marine environment, a change in the order of dominance was observed for each sampling point.
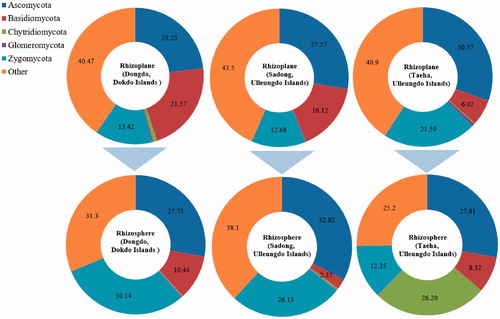
Table 4. Distribution of fungal phyla from the host halophyte soil (unit: %, RS: rhizosphere, RP: rhizoplane).
In contrast to observations in Dongdo (of the Dokdo), there were no marked phylum variations between the RP and RS in the Sadong or Taeha regions (of the Ulleungdo). Ascomycota was the most dominant phylum in all sampling sites, regardless of RS or RP.
The Dokdo Islands, especially the Dongdo, are geographically segregated from the inland Korean peninsula by over 120 km [Citation19,Citation50]. Macrophytic trees cannot survive there, but the community of herbaceous native halophyte species is well-developed in the Dongdo Islands [Citation19,Citation21]. Therefore, the soil fungal community associated with host plants may vary between RS and RP. This phenomenon agrees with the observed variation in fungal diversity (, and ) and the constituent fungal taxa between RS and RP, as described below.
3.5. Distribution and variation of fungal genera
Fungal microbiota structures were different in each of the coastal regions, as described above ( and ). The number of fungal genera varied markedly according to the coastal region () (RP/RS of Dongdo Islands: 44/86; Sadong: 44/97; and Taeha: 43/98). Higher genera diversity was observed in the RS, as compared to that in the RP, and comparable results were obtained for all coastal regions. Furthermore, OTUs (RP/RS of Dongdo Islands: 140/341; Sadong: 154/389; Taeha: 212/478) varied similarly with fungal genera diversity (). These patterns were like the results of statistical analyses examining total read counts, total bases, or the number of valid sequences (). This result is noteworthy, bacterial diversity is less in the rhizoplane compared to the rhizosphere [Citation14]. The same is true for fungi. In addition, the total number of fungal genera in RS was greater than RP (), which was also confirmed by Chao’s richness index in all coastal sites (). Thus, we confirmed that fungal biota in RP is more strongly affected by the host plant than that in the RS soil. This unique interaction between the host plant and RP may lead to the emergence or dominance of only closely related, specifically interacting, fungal genera. The dominance of specific fungal genera could limit the emergence of other fungal genera.
Figure 6. Unique fungal genera distribution in each coastal region. Fungal microbiota structures were different in each of the coastal regions. Only those fungal genera that accounted for 0.7% or more of each community were labeled. The ratio (%, dominance) of each fungal genus.
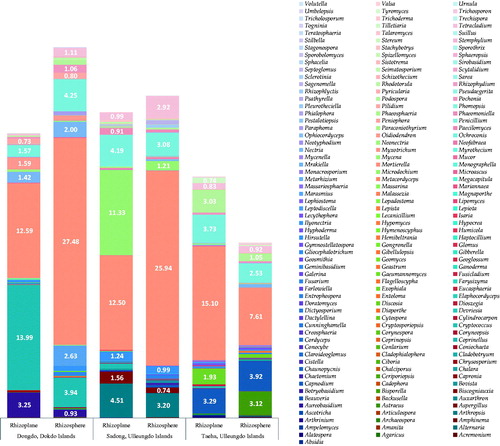
Contrary to what has been observed for RP, the RS fungal microbiome may be more affected by the maritime environment soil and less affected by S. takesimense. This assumption is supported by two results. (1) A higher ratio of the number of fungal genera (, Supplementary Tables 1 and 2) in RS versus RP in the Ulleungdo (Sadong: 120.5% increased, compared to their RP; Taeha: 127.9% increased, compared to their RP), which possess milder environmental (maritime) conditions, than those in the Dokdo Islands (95.5% increased, compared to their RP) (, Supplementary Table 1 and 2) [Citation21]. The harsh environment in the Dokdo Islands may limit the richness of genera in the S. takesimense RS soil. (2) Among newly appeared genera (134 genera across three sites) in the RS (which were not observed in RP) (Supplementary Table 1), a considerable number of fungal genera (38 genera, 28.4%) shared taxonomical locations between the three geographical sites (, Supplementary Tables 1 and 2). These commonly distributed genera are better adapted to coastal soil than to S. takesimense. Therefore, these differences in RP and RS demonstrate a powerful influence of the host plant on their RP. Further, the uniqueness of the closely interacting fungal biota with their host plants in RP was revealed. However, follow-up studies are needed to understand if other native plant species show the same pattern.
3.6. Uniqueness of fungal microbiomes in the RP
The uniqueness of fungal biota was assessed based on the identification and comparison of the fungal community () across three geographical sites. Fungal genera in RP and RS were compared to determine the influence of host halophyte community on their proximity soil. Using this method, uniquely interacting fungal genus groups could be identified. We compared RP and RS residing fungal genera and focused on fungal genera only found in RP or RS compartment. If a genus identified as only residing in RP (but not identified in RS) can be estimated to have close interactions and interdependency with host plants (symbiosis, mutualism, or other negative interaction such as parasitic based on strong dependency to host). In the Dongdo (of the Dokdo Islands), eight fungal genera among 44 (18.2%) (Supplementary Table 2, ) were found only in RP, and 50 new fungal genera were only found in RS (Supplementary Tables 1 and 5, ). In the Sadong region (Ulleungdo Islands) (Supplementary Tables 1 and 2, ) genera only found in RP were counted to nine genera out of a total 44 in RP (11.4%), but they were not found in RS, and 58 new genera identified in the RS part (Supplementary Table 1, ). In the Taeha region (in the Ulleungdo), eight fungal genera among a total of 43 were identified only in RP (18.6%) (Supplementary Table 2, ) and they were absent in RS compartment, whereas 63 new genera were identified in the RS part not found in RP (Supplementary Table 1 and 2, ).
Owing to these results, interactive fungal genera that reside only in RP potentially have a strong dependency on their host halophyte species, and these dependencies decrease with distance from their host plant in contrast to another fungal genus, which was not found in RP, but only in RS. We can obtain information about the interdependency between host species and microbes by comparing fungal genera in the RP and the RS. However, such dependencies cannot always be interpreted as symbiotic relationships [Citation51], because certain pathogens usually have strong host dependencies, as is well known in traditional pathobiology or plant pathophysiology [Citation52,Citation53]. Therefore, evaluating symbiosis using ecological principles should contemplate a meta-analysis of previous scientific reports of fungal species. If Alatospora and Dioszegia species occur, it is reported as a plant root endophyte [Citation54–57]. If genus Amanita occurs, it is reported as ectotrophic mycorrhiza [Citation58]. Several species of genus Arthrinium were reported with plant pathogenicity [Citation59]. Species belonging to the genus Entrophospora are reported as endo and ecto-mycorrhizae [Citation60,Citation61]. Aureobasidium are reported as endophyte or biocontrol strains [Citation62]. Mariannaea have been reported as fungal resources as entomopathogenic fungi, an antagonist for crop insect vector [Citation63]. Some species in the genus Cryptococcus are reported as arbuscular mycorrhizal [Citation57]. Species of genus Cladophialophora were reported as root-associated fungi [Citation64] and Stereum are saprobic [Citation65]. Species belongings to the genus Metarhizium have not been reported as pathogens, yet they were reported as root symbionts [Citation66,Citation67]. Pathogenicity of some species of genera Gaeumannomyces and Malassezia to plant species, was reported [Citation68–70]. Isaria and Ophiocordyceps species have been reported as endophyte or root layer species and often fungal resources as entomopathogenic fungi [Citation71–73]. Tyromyces species are often saprophytic mycorrhizae along with Penicillium and Mucor [Citation74].
Further, these closely interacting fungal genera are not common in geographically segregated regions, even if they reside in the same host plant species. Geographically isolated volcanic islands in this study induced the inevitable ecological segregation of plant species. Owing to this natural history, fungal species interacting with the host plants might adapt and evolve independently from other coastal environments. This interpretation implies that geographical isolation can be a clue for unique fungal resources, and this study demonstrates a useful strategy to cope with situations that accelerate competition for biological resources. According to a previous study, to secure the uniqueness of microbial resources, it must be proven that specific environment-derived microbial species are unique strains and are rarely found in other natural environments [Citation3]. To accomplish this, the microbial community database from the specific environment must show unique constituent taxa comparable to other sites.
Regardless, it is well known that microbial species that co-exist in geographically isolated regions with identical hosts are more likely to be strong symbiotic with dependency to host species [Citation75]. However, if the specific microbial taxon that exists only in the RP (and not found in RS) are found in common geographically isolated areas, there is a stronger possibility of close relations between fungal species and host plants. Thus, in this study, a comparison of the entire fungal community between geographically isolated regions was not performed, and we only compared RP residing species not found in the RS. Interestingly, most fungal genera that reside only in RP do not share taxonomical identity across three geographical locations in this study, except for one genus, Metarhizium (Supplementary Table 2). In this study, geographical isolation and uniqueness of plant-associated fungal genera was shown. Nevertheless, commonly identified fungal genera found in geographically isolated regions that overcome ecological segregations (such as Metarhizium in this study) could be another biological resource. Often, commonly identifiable microbial species from higher organisms are regarded as close symbionts [Citation75–80]. Metarhizium was confirmed in the Dokdo and Ulleungdo, which are over 80 km apart (). Metarhizium has been reported as a plant-associated fungal species, which has the benefit of entomo-pathogenicity and is resistant against plant diseases [Citation78–82]. Metarhizium anisopliae has been reported as a novel tool for “paratransgenesis” in vector-borne infectious disease. Of note, it is a close symbiont of a diverse host insect vector [Citation83–85]. There have been few reports confirming the functionality of Metarhizium from halophytes or surrounding coastal soil environments. Therefore, understanding the relationship between geographical isolation and fungal community can be an effective tactic for securing unique microbial resources that show good colonization and settlement in diverse or extreme agriculture applications.
3.7. Fungal microbiome in the RS
The percentages of newly emerged genera in RS are higher in the Ulleungdo Islands (Sadong: 31.8%, Taeha: 41.6%) than in the Dongdo Islands (13.6%) (Supplementary Tables 1 and 2). These newly identified fungal species, found only in RS, not in RP, share their taxonomic identity in all three sites with a high ratio (38 genera among 134, 28.4%) (Supplementary Table 1), compared to that of fungal genera that disappear upon moving from RP to RS (5.0%) (Supplementary Table 2). Likewise, the ratio of the number of fungal genera moving from RP to RS, in the Ulleungdo Islands (Sadong: increased 120.5%, Taeha: increased 127.9%) outnumbered that in the Dokdo Islands (Dongdo: increased 95.5%) (Supplementary Tables 1 and 2, ). This phenomenon could be interpreted as terrain of the Ulleungdo that suffering low stresses from marine condition [Citation21] compared to the Dokdo exert lower selective pressure on RS derived from marine terrains. These results also follow salinity analysis results in . Although RP conditions and the inhabiting fungal species are affected by host plants, RS is thought to be under another influence that originated from the marine environment. Fungal genera identified only in the RS showing common and uncommon distributions between the three coastal environments are listed in Supplementary Table 2.
A considerable number of fungal genera identified only in RS were revealed as belonging to the same fungal taxon (Supplementary Table 2). In the Dongdo 25 among 50 fungal genera (50.0%), 25 among 58 in the Sadong (43.1%), and 25 among 63 genera (39.7%) in the Taeha of the Ulleungdo shared similar taxonomic identity with fungal strains from other locations (Supplementary Table 2). Characteristically, identified only in the RS of the Taeha region, were the most diverse among the three tested coastal environments. These results were thought to low selective pressure in the Taeha of the Ulleungdo Islands, as compared to that in the Dokdo. Variation in Chytridiomycota in the three coastal environments supports these results. The ratio of specific phyla increased moving from RP to RS, Zygomycota in the Dongdo or the Sadong of the Ulleungdo (Dongdo: increased from 13.420 to 30.140%, Sadong: 12.680 to 26.130%), and Chytridiomycota in the Taeha (0.180 to 26.200%) (, ). In particular, the fungal phylum Chytridiomycota is evolutionary close to the kingdom Animalia [Citation86]. Therefore, it has a motility apparatus (zoospore) [Citation85] and is more adapted to a freshwater environment [Citation86], increasing its dominance ratio in the RS of the Taeha or Sadong of the Ulleungdo compared to halophyte habitat in the Dokdo RS soil (). The dominance of this phylum, not identified in the RS of the Dongdo of the Dokdo, decreases the ratio of the number of fungal genera and is scored as 54.3%. This result implies that severe salt intrusion occurred in the RS soil of the Dongdo as compared to that of the Ulleungdo, as has been reported in previous geographical or related ecological studies of Dokdo [Citation3,Citation20,Citation21]. Own ecological function of halophyte species, such as the salt elimination function of S. takesimense [Citation87] used as a study target in this study, was thought to be limited in RS due to severe salt intrusion, which may relatives to explain Chytridiomycota absence.
Specific fungal genera closely interacting with their host plants, showed geographical uniqueness, even for host plants belonging to the same plant taxon. However, as the distance from the host halophyte community in the associated soil increased, more diverse fungal genera appeared. A considerable number of common genera were confirmed in three geographical sites. These results imply several important strategies for securing fungal resources that interact with higher organisms.
4. Conclusions
To assess the effect of geographical isolation on the emergence of the uniqueness of fungal resources, S. takesimense was selected as a host halophyte species from the Dokdo and the Ulleungdo Islands. A culture-independent analysis was conducted to compare the fungal diversity between RP and RS.
Variations in the fungal profile patterns of RP versus RS were observed in all geographical sites. Soil analysis shows that although the geographical formation factors of each island are the same, the characteristics of the soil properties and physiochemistry were characteristically different depending on the size of the island and the distance from the coast (described in the section of soil texture classification, alkalinity, TN, Corg). Furthermore, such natural factors had a discriminatory effect on the formation of vegetation. Moreover, the comparison of RS and RP of three sites of volcanic islands also showed there was a difference in selective pressure on fungal microbiomes depending on the intensity of the marine environment.
However, the complex interpretation of these different conclusions requires attention. It is impossible to determine whether such climate differences affect soil characteristics and are differentiated in the formation of vegetation or fungal clusters, or whether climate differences directly affected soil and fungal clusters, respectively. However, the strength of the marine condition, the characteristics of vegetation, and the characteristics of soil have combined to differentiate the current climax fungal communities in each site. Therefore, characteristic analysis data were obtained in this study, which shows the differences between geographical isolation and local environmental characteristics affect the distribution of microorganisms (or symbiotic fungal species in them) associated halophyte species. This also allows that estimation of geographical isolation can be a strategy for securing unique fungal taxa. Notably, in the geographically isolated volcanic islands, a few fungal genera were commonly confirmed in RP but absent in the RS: Metarhizium. Results demonstrate that symbiotic fungi and their host plants evolved with regional independence, even if the host plant belonged to the same species. In contrast, a considerable number of commonly confirmed rhizosphere fungal genera, which were not present in the RP, were found in RS. Thus, unique fungi can be obtained from geographically isolated unique terrain. Repeated investigation and studies on various plant species are needed to establish other biological resource acquisition strategies because the competition to secure biological resources is fiercer than ever.
Supplemental Material
Download MS Word (133.5 KB)Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability statement
All data are available within the manuscript and its supplementary materials.
Additional information
Funding
References
- Ryan MJ, McCluskey K, Verkleij G, et al. Fungal biological resources to support international development: challenges and opportunities. World J Microbiol Biotechnol. 2019;35(9):139.
- Kevin M. A review of living collections with special emphasis on sustainability and its impact on research across multiple disciplines. Biopreserv Biobank. 2017;15(1):20–30.
- Park JM, Hong JW, Son JS, et al. A strategy for securing unique microbial resources—focusing on Dokdo islands-derived microbial resources. Israel J Ecol Evol. 2018;64(1–4):1–11.
- Bloemberg GV, Lugtenberg BJ. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol. 2001;4:343–350.
- Li HB, Singh RK, Singh P, et al. Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Front Microbiol. 2017;14(8):1268.
- Rohini S, Aswani R, Kannan M, et al. Culturable endophytic bacteria of ginger rhizome and their remarkable multi-trait plant growth-promoting features. Curr Microbiol. 2017;75:505–511.
- Wang Z, Li T, Wen X, et al. Fungal communities in rhizosphere soil under conservation tillage shift in response to plant growth. Front Microbiol. 2017;11(8):1301.
- Park JM, Hong JW, Lee W, et al. Fungal clusters and their uniqueness in geographically segregated wetlands: a step forward to marsh conservation for a wealth of future fungal resources. Mycobiology. 2020;48(5):351–363.
- Ministry of Environment, Republic of Korea. The Fifth national report to the convention on biological diversity [Internet]. Sejong (South Korea): MoE; 2014 [cited 2019 October 19]. Available from: https://www.cbd.int/nr5/
- Ministry of Environment, Republic of Korea. Report: conservation and sustainable use of biological resources [Internet]. Sejong (South Korea): MoE; 2015 [cited 2019 October 19]. Available from: http://hdl.handle.net/20.500.11822/9049
- Lucas M, Balbín-Suárez A, Smalla K, et al. Root growth, function, and rhizosphere microbiome analyses show local rather than systemic effects in apple plant response to replant disease soil. PLoS One. 2018;13(10):e0204922.
- Wu L, Chen J, Xiao Z, et al. Barcoded pyrosequencing reveals a shift in the bacterial community in the rhizosphere and rhizoplane of Rehmannia glutinosa under consecutive monoculture. Int J Mol Sci. 2018;19(3):850.
- Yang Y, Dou Y, Huang Y, et al. Links between soil fungal diversity and plant and soil properties on the Loess plateau. Front Microbiol. 2017;78:2198.
- Zhang Z, Zhou X, Tian L, et al. Fungal communities in ancient peatlands developed from different periods in the Sanjiang Plain, China. PLoS One. 2017;12(12):e0187575.
- Lee DS. Geology of Korea. Seoul (Republic of Korea): Kyohak-sa; 1987.
- Kong WS, David W. The plant geography of Korea with an emphasis on the alpine zones. Berlin (Germany): Springer; 1993.
- Thorpe RS, Surget-Groba Y, Johansson H. Genetic tests for ecological and allopatric speciation in anoles on an island archipelago. PLoS Genet. 2010;6(4):e1000929.
- Zhao C, Wang CB, Ma XG, et al. Phylogeographic analysis of a temperate-deciduous forest restricted plant (Bupleurum longiradiatum Turcz.) reveals two refuge areas in China with subsequent refugial isolation promoting speciation. Mol Phylogenet Evol. 2013;68(3):628–643.
- NGII (National Geographic Information Institute). The geography of Dokdo. Suwon (Republic of Korea): Press of National Geographic Information Institute; 2015.
- You YH, Park JM, Park JH, et al. Specific rhizobacterial resources: characterization and comparative analysis from contrasting coastal environments of Korea. J Basic Microbiol. 2016;56(1):92–101.
- CHA (Cultural Heritage Administration), Republic of Korea. Natural heritage of Korea, Dokdo, Cultural Heritage Administration of Korea. Daejeon (Republic of Korea): Cultural Heritage Administration; 2009.
- FAO. Guidelines for soil description. 4th ed. Rome (Italy): Food and Agriculture Organisation of the United States; 2006.
- Flores-Vargas RD, O’Hara GW. Isolation and characterization of rhizosphere bacterial with potential for biological control of weed in vineyards. J Appl Microbiol. 2006;100(5):946–954.
- Vega NOW. A review on beneficial effects of rhizosphere bacterial on soil nutrient availability and plant nutrient uptake. Rev Fac Nac Agron Medellín. 2007;60(1):3621–3643.
- U.S. Salinity Lab Staff. Diagnosis and improvement of saline and alkali soils. In: Richards LA, editor. Agriculture handbook 60. Washington (DC): USDA; 1954. p. 122–124.
- FAO, United Nations. Standard operating procedure for soil organic carbon, Walkley-Black method (Titration and colorimetric method). Rome (Italy): Global Soil Laboratory; 2019.
- Bremner JM, Mulvaney CS. Nitrogen total. In: Page AL, editor. Methods of soil analysis: part 2 chemical and microbiological properties, 9.2.2. 2nd ed. Madison (WI): American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America; 1983.
- Masella AP, Bartram AK, Truszkowski JM, et al. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 2012;13(1):31.
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541.
- Bengtsson Palme J, Ryberg M, Hartmann M, et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol. 2013;4(10):914–919.
- Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200.
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336.
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461.
- Abarenkov K, Nilsson RH, Larsson KH, et al. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol. 2010;186:281–285.
- Heck KL, van Belle G, Simberloff D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology. 1975;56(6):1459–1461.
- Lambshead PJD, Platt HM, Shaw KM. The detection of differences among assemblages of marine benthic species based on an assessment of dominance and diversity. J Nat Hist. 1983;17(6):859–874.
- Chao A, Shen TJ, Ma KH, et al. User's guide for program SPADE (Species Prediction And Diversity Estimation). Taiwan: National Tsing Hua University; 2016 [cited 2019 Dec 18]. Available from: http://chao.stat.nthu.edu
- Sonn YK, Park CW, Zhang YS, et al. Characteristics of soils distributed on the Dokdo Island in South Korea. Korean J Soil Sci. 2011;44(2):187–193.
- Chapman VJ. Salt marshes and salt deserts of the world. In: Polunin N, editor. Ecology of halophytes. New York (NY): Academic Press; 1974.
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440.
- Ma B, Gong J. A meta- analysis of the publicly available bacterial and archaeal sequence diversity in saline soils. World J Microbiol Biotechnol. 2013;29:2325–2334.
- Tazi L, Breakwell DP, Harker AR, et al. Life in extreme environments: microbial diversity in Great Salt Lake, Utah. Extremophiles. 2014;18(3):525–535.
- Zain Ul Arifeen M, Ma YN, Xue YR, et al. Deep-Sea fungi could be the new arsenal for bioactive molecules. Mar Drugs. 2019;18(1):9.
- Blaud A, Lerch TZ, Phoenix GK, et al. Arctic soil microbial diversity in a changing world. Res Microbiol. 2015;166(10):796–813.
- Otlewska A, Migliore M, Dybka-Stępień K, et al. When salt meddles between plant, soil, and microorganisms. Front Plant Sci. 2020;16(11):553087.
- Haegeman B, Hamelin J, Moriarty J, et al. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013;7:1092–1101.
- Magurran AE. Measuring biological diversity. Oxford (UK): Blackwell Science; 2004.
- Chao A, Gotelli NJ, Hsieh TC, et al. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr. 2014;84(1):45–67.
- Gong Y, Son SJ. A study of oceanic thermal fronts in the Southern Japan Sea. Bull Fish Res Dev Agency. 1982;28:25–54.
- Park B, Kang GW, Song GM, et al. Ten newly recorded species of insect on Dokdo Island, South Korea. J Species Res. 2017;6(3):280–290.
- Johnson NC, Graham JH, Smith FA. Functioning of mycorrhizal associations along the mutualism–parasitism continuum. New Phytol. 1997;135(4):575–585.
- Bäumler A, Fang FC. Host specificity of bacterial pathogens. Cold Spring Harb Perspect Med. 2013;3(12):a010041.
- Morris CE, Moury B. Revisiting the concept of host range of plant pathogens. Annu Rev Phytopathol. 2019;57:63–90.
- Sati SC, Belwal M. Aquatic hyphomycetes as endophytes of riparian plant roots. Mycologia. 2005;97(1):45–49.
- Quilliam RS, Jones DL. Fungal root endophytes of the carnivorous plant Drosera rotundifolia. Mycorrhiza. 2010;20(5):341–348.
- Sapkota R, Jørgensen LN, Nicolaisen M. Spatiotemporal variation and networks in the mycobiome of the wheat canopy. Front Plant Sci. 2017;8:1357.
- Renker C, Blanke V, Börstler B, et al. Diversity of Cryptococcus and Dioszegia yeasts (Basidiomycota) inhabiting arbuscular mycorrhizal roots or spores. FEMS Yeast Res. 2004;4(6):597–603.
- Nehls U. Review Mastering ectomycorrhizal symbiosis: the impact of carbohydrates. J Exp Bot. 2008;59(5):1097–1108.
- Chen K, Wu XQ, Huang MX, et al. First report of brown culm streak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Dis. 2014;98(9):1274.
- Palenzuela J, Barea JM, Ferrol N, et al. Entrophospora nevadensis, a new arbuscular mycorrhizal fungus from Sierra Nevada National Park (southeastern Spain). Mycologia. 2010;102(3):624–632.
- Amna T, Puri SC, Verma V, et al. Bioreactor studies on the endophytic fungus Entrophospora infrequens for the production of an anticancer alkaloid camptothecin. Can J Microbiol. 2006;52(3):189–196.
- Busby PE, Ridout M, Newcombe G. Fungal endophytes: modifiers of plant disease. Plant Mol Biol. 2016;90(6):645–655.
- Araújo CAS, Dias LP, Ferreira PC, et al. Responses of entomopathogenic fungi to the mutagen 4-nitroquinoline 1-oxide. Fungal Biol. 2018;122(6):621–628.
- Toju H, Tanabe AS, Sato H. Network hubs in root-associated fungal metacommunities. Microbiome. 2018;6(1):116.
- Jaber E, Xiao C, Asiegbu FO. Comparative pathobiology of Heterobasidion annosum during challenge on Pinus sylvestris and Arabidopsis roots: an analysis of defensin gene expression in two pathosystems. Planta. 2014;239(3):717–733.
- Barelli L, Waller AS, Behie SW, et al. Plant microbiome analysis after Metarhizium amendment reveals increases in abundance of plant growth-promoting organisms and maintenance of disease-suppressive soil. PLoS One. 2020;15(4):e0231150.
- Lovett B, St Leger RJ. Stress is the rule rather than the exception for Metarhizium. Curr Genet. 2015;1(3):253–261.
- Karakkat BB, Hockemeyer K, Franchett M, et al. Data for designing two isothermal amplification assays for the detection of root-infecting fungi on cool-season turfgrasses. Data Brief. 2018;20:471–479.
- Xu F, Yang G, Wang J, et al. Spatial distribution of root and crown rot fungi associated with winter wheat in the north china plain and its relationship with climate variables. Front Microbiol. 2018;25:9.
- Xu J, Saunders CW, Hu P, et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci USA. 2007;104(47):18730–18735.
- Kamel NM, Abdel-Motaal FF, El-Zayat SA. Endophytic fungi from the medicinal herb Euphorbia geniculata as a potential source for bioactive metabolites. Arch Microbiol. 2020;202(2):247–255.
- Zhong X, Peng QY, Li SS, et al. Detection of Ophiocordyceps sinensis in the roots of plants in alpine meadows by nested-touchdown polymerase chain reaction. Fungal Biol. 2014;118(4):359–363.
- Wang JB, St LR. Wang C. Advances in genomics of entomopathogenic fungi. Adv Genet. 2016;94:67–105.
- Cullings K, Makhija S. Ectomycorrhizal fungal associates of Pinus contorta in soils associated with a hot spring in Norris Geyser Basin, Yellowstone National Park, Wyoming. Appl Environ Microbiol. 2001;67(12):5538–5543.
- Barton LL, Northup DE. Microbial ecology. Hoboken (NJ): Wiley-Blackwell; 2011.
- Park JM, You YH, Back CG, et al. Fungal load in Bradysia agrestis, a phytopathogen-transmitting insect vector. Symbiosis. 2018;74(2):145–158.
- Park JM, You YH, Park JH, et al. Cutaneous microflora from geographically isolated groups of Bradysia agrestis, an insect vector of diverse plant pathogens. Mycobiology. 2017;45(3):160–171.
- Baker DK, Rice SJ, Leemon DM, et al. Horizontal transmission of Metarhizium anisopliae (Hypocreales: Clavicipitacea) and the effects of infection on oviposition rate in laboratory populations of Musca domestica (Diptera: Muscidae). Pest Manag Sci. 2017;74(4):987–991.
- Liao X, Lovett B, Fang W, et al. Metarhizium robertsii produces indole-3-acetic acid, which promotes root growth in Arabidopsis and enhances virulence to insects. Microbiology. 2017;163(7):980–991.
- Masoudi A, Koprowski JL, Bhattarai UR, et al. Elevational distribution and morphological attributes of the entomopathogenic fungi from forests of the Qinling mountains in China. Appl Microbiol Biotechnol. 2017;102(3):1483–1499.
- Mukherjee K, Vilcinskas A. The entomopathogenic fungus Metarhizium robertsii communicates with2the insect host Galleria mellonella during infection. Virulence. 2017;9(1):402–413.
- Pelizza SA, Schalamuk S, Simón MR, et al. Compatibility of chemical insecticides and entomopathogenic fungi for control of soybean defoliating pest, Rachiplusia nu. Rev Argent Microbiol. 2017;50(2):189–201.
- Wilke AB, Marrelli MT. Paratransgenesis: a promising new strategy for mosquito vector control. Parasit Vectors. 2015;8:342.
- Ashraf M, Farooq M, Shakeel M, et al. Influence of entomopathogenic fungus, Metarhizium anisopliae, alone and in combination with diatomaceous earth and thiamethoxam on mortality, progeny production, mycosis, and sporulation of the stored grain insect pests. Environ Sci Pollut Res Int. 2017;24(36):28165–28174.
- Kabaluk T, Li-Leger E, Nam S. Metarhizium brunneum - An enzootic wireworm disease and evidence for its suppression by bacterial symbionts. J Invertebr Pathol. 2017;150:82–87.
- Alexopoulos CJ, Mims CW, Blackwell M. Introductory mycology. 4th ed. Hoboken (NJ): John Wiley & Sons, Inc.; 1996.
- Hameed A, Gulzar S, Aziz I, et al. Effects of salinity and ascorbic acid on growth, water status, and antioxidant system in a perennial halophyte. AoB Plants. 2015;7:1–11.