Abstract
Rogues are relatively unstable taxa in phylogenetic analyses that are of concern if they obfuscate relationships, or support for relationships, of interest among more stable taxa. RogueNaRok is a recently developed heuristic solution to the problem of identifying rogue taxa. We illustrate the performance of RogueNaRok with simple examples designed to clarify its behaviour. We argue that the optimality criterion currently used by RogueNaRok may sometimes be poorly suited to the task of detecting rogues and that the scores reported by RogueNaRok should not be interpreted as measures of taxon instability. We suggest how RogueNaRok might be enhanced and recommend that it not be relied upon exclusively for detecting rogues.
Introduction
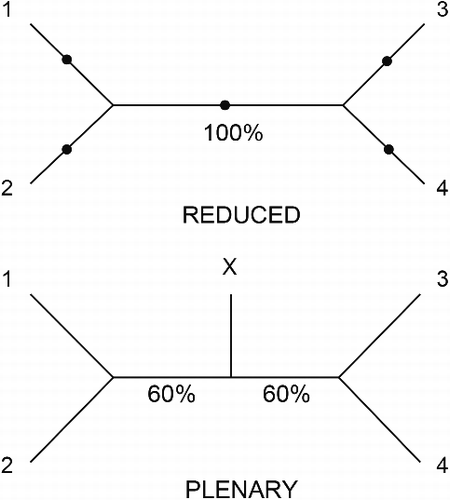
Some taxa may be relatively unstable in phylogenetic analyses either due to missing and/or conflicting data (Wilkinson, Citation1995a). By occupying different positions in a set of phylogenetic trees, such as those produced by bootstrap or Bayesian analyses, relatively unstable taxa may lead to unresolved or poorly supported relationships in consensus trees, and may thereby obscure the support for relationships among more stable taxa (e.g., Wilkinson, Citation1994, Citation2003). Relatively unstable taxa that are of concern because they have such undesirable effects are often called rogues (Wilkinson, Citation1996). gives a simple example of rogue behaviour. The tree for leaves 1–4 is maximally supported () but rogue taxon X lacks any relevant data such that all five possible placements of X on the underlying quartet tree for 1–4 so as to yield a binary (i.e., fully resolved) tree are equally probable. The strict consensus of these binary trees including X is unresolved and the majority rule consensus is resolved but with low support for the two internal branches (). The instability of rogue X obscures the impressive support for the relationship among 1–4.
Fig. 1. First example of the behaviour of RNR: ability to detect rogues. The upper quartet tree is a maximally supported (100%) unrooted tree for the quartet 1–4 of leaves. Dots indicate the five possible points at which a rogue (X) with no relevant data can be added to give a binary tree. The lower tree is the majority rule consensus of the five binary trees including X. Note that the quartet tree is also a reduced consensus of the five binary trees including X. ΣS is higher (120) for the plenary than for the reduced (100) consensus and thus RNR does not identify X as a rogue.
Countering the unhelpful effects of rogues in real datasets requires some means of identifying rogues. Several have been developed, including a priori methods such as safe taxonomic reduction (Wilkinson, Citation1995a) and concatabominations (Siu-Ting, Pisani, Creevey, & Wilkinson, Citation2015), but most involve a posteriori approaches, such as reduced consensus (Wilkinson, Citation1994, Citation1995b, Citation1996) and leaf stabilities (Thorley & Wilkinson, Citation1999; Wilkinson, Citation2006) in which rogues are identified from sets of trees. RogueNaRok (RNR) is a recently developed method for identifying rogues that has been shown to outperform alternatives on a range of real datasets and to yield more accurate trees in simulations (Aberer, Krompass, & Stamatakis, Citation2013). RNR is available via a flexible and user-friendly web-server (http://rnr.h-its.org).
RogueNaRok (RNR)
Here we use some simple examples to illustrate RNR and make some suggestions regarding its use and potential enhancement.
RNR casts rogue identification as an optimization problem: rogues are precisely those leaves that, given some set of trees and a consensus rule or threshold, are excluded from an optimal consensus tree. With this novel approach rogue taxa exist only when there is some ‘reduced’ consensus, i.e. a consensus of pruned trees, that, according to the RNR optimality criterion, is better than the corresponding plenary consensus, i.e., a consensus including all the leaves (Wilkinson, Thorley, Pisani, Lapointe, & McInerney, Citation2004).
RNR has two alternative optimality criteria. The relative information content (Pattengale, Aberer, Swenson, Stamatakis, & Moret, Citation2011) is simply the degree of resolution of the consensus (i.e., the number of internal branches), maximization of which is also what Wilkinson (Citation1995b) used to distinguish primary from other reduced consensus trees. Of more interest is the default optimality criterion, the relative bipartition information content (Aberer & Stamatakis Citation2011), which Aberer et al. (Citation2013) define as 'the sum of all support values divided by the maximum possible support in a fully bifurcating tree with the initial (i.e., before pruning any rogues) set of taxa'. Note that these optimality criteria are not strictly measures of information (see Thorley, Wilkinson, & Charleston, Citation1998; Wilkinson, Cotton, & Thorley, Citation2004) and that they are identical in the special case of strict consensus. We denote the sum of support values of tree as ΣS and, without concern for technical details of the algorithm, we can simply and equivalently characterize the RNR (in default mode) as seeking the consensus tree and smallest dropset (= set of leaves excluded from the consensus) with the highest ΣS. Following Pattengale et al. (Citation2011), Aberer et al. (Citation2013) call the optimal consensus a maximum information subtree consensus.
Excluding leaves reduces the number of internal branches that consensus trees can have, and can thereby contribute toward a reduction in ΣS. Only where this is offset by increased support for other internal branches will RNR detect rogues. Note that finding an optimal dropset is NP-hard (Akshay, Dong, & Fernández-Baca, Citation2012) and that there may not be a unique solution. Thus the RNR algorithm is a heuristic. We focus upon the behaviour of RNR with a majority-rule because of the importance of this consensus rule in summarizing support in bootstrap, jackknife and Bayesian analyses. Unless otherwise indicated reference to RNR is to its default implementation and optimality criterion.
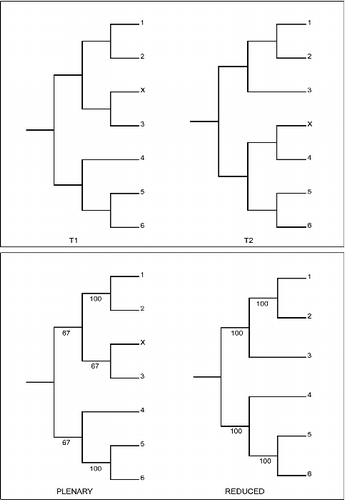
Consider the reduced and plenary consensus trees in . The plenary consensus including rogue X has a higher ΣS than does the reduced consensus (120 versus 100) and thus, perhaps surprisingly, RNR does not identify X as a rogue in this simple case. Note that RNR would identify X as a rogue using a strict consensus threshold in this case or if there were five stable and one unstable taxon. provides an illuminating example of the sensitivity of RNR rogue taxon identification to small differences in composition of the set of input trees.
Fig. 2. Second example of the behaviour of RNR: sensitivity. The alternative tree topologies (T1 & T2) differ only in the position of a rogue (X). The lower panel shows the plenary and a reduced majority-rule consensus for a set of 67 copies of T1 and 33 copies of T2. ΣS is higher (401) for the plenary than for the reduced (400) consensus and thus RNR does not identify X as a rogue. If instead there are 66 copies of T1 and 34 of T2 then ΣS is lower (398) for the plenary consensus and RNR identifies X as a rogue.
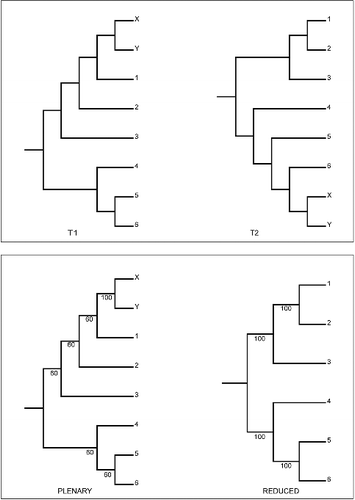
A group of taxa may constitute a rogue. illustrates such a case with a well-supported but unstable cherry (i.e., a pair of adjacent leaves). The RNR algorithm implements an upper bound on dropset size that by default is set to one. Using any bound means that RNR will be unable to detect any rogue group larger than the bound. Thus, with the default settings, RNR does not detect the roguish behaviour of the unstable cherry in . Only if dropset size is increased will RNR ever find the rogue cherry and, as in the previous example, RNR is sensitive to small differences in the set of input trees.
Fig. 3. Third example of the behaviour of RNR: dropset size. The alternative tree topologies (T1 & T2) differing only in the position of a rogue cherry comprising leaves X & Y. The lower panel shows the plenary and a reduced majority-rule consensus for a set of 60 copies of T1 and 40 copies of T2. ΣS is identical (400) for the plenary and the reduced consensus and thus RNR does not identify X and Y as rogue. If instead there are 59 copies of T1 and 41 of T2 then ΣS is lower (395) for the plenary consensus and RNR identifies X and Y as rogue if the dropset size is higher than the default. At the default setting X and Y are never identified as rogues.
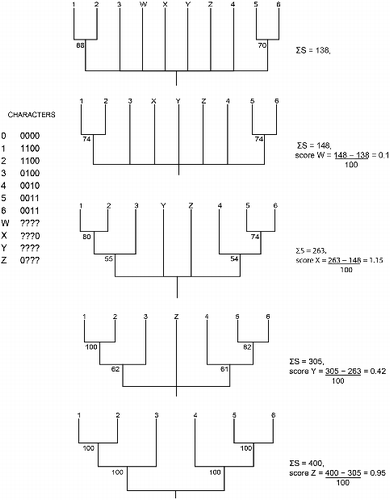
RNR outputs a list of any rogues found together with a score labelled as the 'sum of support value improvements for pruning this taxon' which merits some explanation. We used PAUP (Swofford, Citation2003) to generate 100 binary trees including four rogue taxa (W-Z) by resampling 1000 characters of the data in and applied RNR to these trees. illustrates the operation of RNR with the default dropset maximum of one and the calculation of RNR scores for the rogues. RNR iteratively identifies and prunes the dropset that leads to the largest increase in ΣS until no further increase is possible. The RNR score for each dropset is the difference between ΣS before and after pruning the dropset divided by 100. Thus the order in which dropsets are identified and pruned need not be given by the magnitude of the reported RNR scores and need not correspond to the relative instabilities of leaves. Consequently RNR scores should not be interpreted as measuring the impact of pruning just that dropset from the input trees and may not be useful as measures of taxon instability, contra to how these values have sometimes been used. For example, Agorreta et al. (Citation2013) and Selvatti, Gonzaga, Augusta, and Russo (Citation2015) considered only leaves with scores above 0.5 as rogues and Maronna, Miranda, Cantero, Barbeitos, and Marques (Citation2016) considered only the 10 leaves with the highest scores as rogues. In our example (), W and Y lack any data whereas X and Z have 75% missing data. Thus we expect (1) W to be similarly unstable to Y, (2) X to be similarly unstable to Z, and (3) W and Y to be more unstable than X and Z. Interpreted as a measure of stability, the RNR scores fulfil only the third of these expectations (differences between the scores of similarly unstable leaves are not insubstantial). In contrast, leaf stabilities (not shown) better fulfil all three expectations. Applied to our example, a 0.5 threshold for the RNR score would lead to the deletion of just X and Z, the two least unstable rogues, yielding a sub-optimal consensus tree with ΣS = 254 (not shown).
Fig. 4. Fourth example of the behaviour of RNR: scores. A data matrix containing four rogue taxa (W-Z) used to create 100 binary bootstrap trees and the corresponding plenary and four reduced majority rule consensus trees for successive deletions of W-Z showing changes in ΣS and calculation of associated RNR scores. The outgroup (0) is not shown in the tree.
There is no doubt that RNR has performed impressively in simulations and proven useful in some real cases. However, our first three simple examples highlight that RNR is sometimes unable to detect rogues and that small differences in sets of input trees and choice of settings can be the difference between success and failure in detecting rogues with this approach. Users need to be aware of the potentially profound impact of the default bound on dropset size and should consider exploring the impact of higher values. For example, Timmermans et al. (Citation2016) used RNR to identify 14 rogues with the default dropset size but 30 rogues with a dropset size of two. Otherwise, inability to detect rogues is attributable to the optimality criterion. Using the default settings, RNR favours a consensus with two internal branches with support of 50% over a reduced consensus with a single internal branch with maximal support, it favours a consensus with three internal branches with 67% support over a reduced consensus with two internal branches with maximal support, and it favours a consensus with four branches with 75% support over a reduced consensus with three maximally supported internal branches. This performance may not correspond well with how researchers value branch support. A simple alternative optimality criterion, the sum of squared support (ΣS2) gives greater weight to strong support and with it all the rogue taxa in our first examples would be detected as such. The RNR server already usefully enables specification of a minimum level of support and enables the default optimality criterion to be replaced with a simple count of the number of internal branches above some threshold, which may be more useful. For example, using this option with a threshold of 80% would identify all the rogue taxa in our simple examples. We suggest that there is scope for further improvement to the optimality criterion in RNR and that enabling users to specify or choose among criteria would be good. There may be other circumstances where we might seek an optimality criterion for choosing among consensus trees. For example, when comparing alternative codings (e.g., DNA, RY, Amino Acid) or in analyses of RADSeq data, where varying assembly parameters such as the depth of stacks and the amount of missing data, yield alternative datasets. We consider ΣS to be ill-suited to identifying the best codings or datasets in such comparisons.
RNR seeks dropsets associated with optimal ‘reduced consensus’ trees and is, perhaps unsurprisingly, closely related to reduced consensus methods. For example, the RNR method of determining minimal dropsets using set operations on pairs of bipartitions of taxa is analytically equivalent to the bipartition comparisons used in reduced consensus methods (Wilkinson, Citation1994, Citation1995b, Citation1996; Wilkinson & Thorley Citation2003). Reduced consensus methods were developed to produce comprehensive summaries of support but may require an unhelpfully large number of consensus trees to do so. Thus while reduced consensus can potentially provide more information relevant to the identification of rogues than does RNR, it will often be more efficient and sensible to focus, as does RNR, on one or a few of the most informative reduced consensus trees and the dropsets of rogue taxa that they identify. However, we recommend that researchers concerned about rogues should not rely only upon RNR's default classification of leaves as being rogues or not, but should be prepared to experiment with non-default setting and to also consider measures of how roguish is each taxon. In the simple cases considered here, where RNR fails to find rogue taxa the rogues nonetheless had the lowest leaf stability measures (which can also be calculated through the RNR server).
Acknowledgements
We thank David Gower and Jeff Streicher for discussions and Gabriela B. Bittencourt Silva for comments on the manuscript. This work was supported in part by the UK Biotechnology and Biological Sciences Research Council under Grant BB/K007440/1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aberer, A. J., Krompass, D., & Stamatakis, A. (2013). Pruning rogue taxa improves phylogenetic accuracy: An efficient algorithm and webservice. Systematic Biology, 62, 162–166. doi: 10.1093/sysbio/sys078
- Aberer, A. J., & Stamatakis, A. (2011). A simple and accurate method for rogue taxon identification. Atlanta, Georgia: IEEE International Conference on Bioinformatics and Biomedicine. doi: 10.1109/BIBM.2011.70
- Agorreta, A., Mauro, D. S., Schliewen, U., Van Tassell, J. L., Kovacic, M., Zardoya, R., & Ruber, L. (2013). Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Molecular Phylogenetics and Evolution, 69, 619–633. doi: 10.1016/j.ympev.2013.07.017.
- Akshay, D., Dong, J., & Fernández-Baca, D. (2012). Identifying rogue taxa through reduced consensus: NP-Hardness and exact algorithms. In L. Bleris, I. Măndoiu, R. Schwartz, & J. Wang (Eds.), International symposium on bioinformatics research and applications (pp. 87–98). Berlin Heidelberg (Germany): Springer-Verlag.
- Maronna, M. M., Miranda, T. P., Cantero, A. L. P., Barbeitos, M. S., & Marques, A. C. (2016). Towards a phylogenetic classification of Leptothecata (Cnidaria, Hydrozoa). Scientific Reports, 6. doi: 10.1038/srep18075
- Pattengale, N., Aberer, A. J., Swenson, K., Stamatakis, A., & Moret, B. (2011). Uncovering hidden phylogenetic consensus in large datasets. IEEM/ACM Transasctions on Computational Biology and Bioinformatics, 8, 902–911. doi: 10.1109/TCBB.2011.28
- Selvatti, A. P., Gonzaga, L. P., Augusta, C., & Russo, M. (2015). A Paleogene origin for crown passerines and the diversification of the Oscines in the New World. Molecular Phylogenetics and Evolution, 88, 1–15. doi: 10.1016/j.ympev.2015.03.018.
- Siu-Ting, K., Pisani, D., Creevey, C. J., & Wilkinson, M. (2015). Concatabominations: Identifying unstable taxa in morphological phylogenetics using a heuristic extension to safe taxonomic reduction. Systematic Biology, 64, 137–143. doi: 10.1093/sysbio/syu066
- Swofford, D. (2003). PAUP*: Phylogenetic analysis using parsimony. Version 4.0 b10. Sunderland, Massachusetts: Sinauer Associates.
- Timmermans, M. J. T.N., Barton, C., Haran, J., Ahrens, D., Culverwell, C. L., Ollikainen, A., … Vogler, A. P. (2016). Family-level sampling of mitochondrial genomes in Coleoptera: Compositional heterogeneity and phylogenetics. Genome Biology and Evolution, 8, 161–175. doi: 10.1093/gbe/evv241
- Thorley, J. L., & Wilkinson, M. (1999). Testing the phylogenetic stability of early tetrapods. Journal of Theoretical Biology, 200, 343–344. doi: 10.1006/jtbi.1999.0999
- Thorley, J. L., Wilkinson, M., & Charleston, M. A. (1998). The information content of consensus trees. In: A. Rizzi, M. Vichi, & H. H. Bock (Eds.), Advances in data science and classification (pp. 91–98). Berlin, Germany: Springer-Verlag.
- Wilkinson, M. (1994). Common cladistic information and its consensus representation: Reduced Adams and reduced cladistic consensus trees and profiles. Systematic Biology, 43, 343–368. doi: 10.1093/sysbio/43.3.343
- Wilkinson, M. (1995a). Coping with missing entries in phylogenetic inference using parsimony. Systematic Biology, 44, 501–514. doi: 10.1093/sysbio/44.4.501
- Wilkinson, M. (1995b). More on reduced consensus methods. Systematic Biology, 44, 435–439. doi: 10.2307/2413604
- Wilkinson, M. (1996). Majority-rule reduced consensus methods and their use in bootstrapping. Molecular Biology and Evolution, 13, 437–444. Retrieved from: http://mbe.oxfordjournals.org/ (accessed 19 October 2016).
- Wilkinson, M. (2003). Missing data and multiple trees: Instability, relationships, and support in parsimony analysis. Journal of Vertebrate Paleontology, 23, 311–323. doi: 10.1671/0272-4634
- Wilkinson, M. (2006). Identifying stable reference taxa for phylogenetic nomenclature. Zoologica Scripta, 35, 109–112. doi: 10.1111/j.1463-6409.2005.00213.x
- Wilkinson, M., Cotton, J. A., & Thorley, J. L. (2004). The information content of trees and their matrix representations. Systematic Biology, 53, 989–1001. doi: 10.1080/10635150490522737
- Wilkinson, M., & Thorley, J. L. (2003). Reduced consensus. In M. F. Janovitz, F. J. Lapointe, F. R. McMorris, B. Mirkin, & F. S. Roberts (Eds.), Bioconsensus, DIMCAS Series in Discrete Mathematics and Theoretical Computer Science (Vol. 61, pp. 195–204). Providence, RI: American Mathematical Society.
- Wilkinson, M., Thorley, J. L., Pisani, D., Lapointe, F. J., & McInerney, J. O. (2004). Some desiderata for liberal supertrees. In O. Bininda-Emonds (Ed.), Phylogenetic supertrees (pp. 227–246). Dordrecht, Netherlands: Kluwer Academic Publishers.
