Abstract
There exists a highly tumorigenic subset of esophageal squamous cell carcinoma (ESCC) cells defined by high expression of CD44. A novel therapy targeting these cancer stem-like cells (CSCs) is needed to improve prognosis of ESCC. CSCs of ESCC have a mesenchymal phenotype and epithelial-mesenchymal transition (EMT) is critical to enrich and maintain CSCs. EGFR, frequently overexpressed in ESCC, has pivotal roles in EMT induced by TGF-β in invasive fronts. Thus, EMT in invasive fronts of ESCC might be important for CSCs and EGFR could be a target of a novel therapy eliminating CSCs. However, effects of EGFR inhibitors on CSCs in ESCC have not been fully examined. EGFR inhibitors, erlotinib and cetuximab, significantly suppressed enrichment of CSCs via TGF-β1-mediated EMT. Importantly, EGFR inhibitors sharply suppressed ZEB1 that is essential for EMT in ESCC. Further, EGFR inhibitors activated Notch1 and Notch3, leading to squamous cell differentiation. EGFR inhibition may suppress expression of ZEB1 and induce differentiation, thereby blocking EMT-mediated enrichment of CSCs. In organotypic 3D culture, a form of human tissue engineering, tumor cells in invasive nests showed high expression of CD44. Erlotinib significantly blocked invasion into the matrix and CD44 high expressing CSCs were markedly suppressed by erlotinib in organotypic 3D culture. In conclusion, EMT is a critical process for generation of CSCs and the invasive front of ESCC, where EMT occurs, might form a CSC niche in ESCC. EGFR inhibitors could suppress EMT in invasive fronts and be one therapeutic option targeting against generation of CSCs in ESCC.
Abbreviations
| CK13 | = | cytokeratin 13; CSCs, cancer stem-like cells; DMSO, dimethyl sulfoxide; EMT, epithelial–mesenchymal transition; EGFR, epidermal growth factor receptor; ESCC, esophageal squamous cell carcinoma; FACS, fluorescence-activated cell sorting; IHC, immunohistochemistry; IVL, involucrin; TGF-β, transforming growth factor-β; ZEB, zinc finger E-box binding protein |
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the deadliest human cancers and ESCC is the sixth leading cause of cancer death and the eighth common cancer type worldwide.Citation1 Surgery and chemoradiotherapy are accepted treatment options for patients with locally advanced ESCCCitation2 and endoscopic treatments (mucosal resection / submucosal dissectionCitation3,4 and ablationCitation5,6) are also used for those with very early stage of ESCC. Although treatments for ESCC have greatly improved, prognosis is still poor in patients with advanced ESCC because of its malignant biological behavior and limited therapeutic options.
Cancer stem-like cells (CSCs) have been identified in many cancers and are thought to attribute to aggressive tumor behavior. CSCs can self-renew and have extremely high tumor initiating capacity. Furthermore, CSCs are highly invasive and resistant to conventional chemotherapy and radiation. Such characteristics of CSCs lead to recurrence, metastasis and inadequate response to the conventional therapies.Citation7,8 A novel therapy targeting CSCs is needed to improve prognosis of ESCC. CD44 has been used as a CSC marker in many cancers including ESCC, head and neck squamous cell carcinoma (SCC), cervical SCC and lung SCC.Citation9-12 We have confirmed that high CD44 expression correlates with enhanced ESCC cell tumorigenicity in vivo.Citation13 Interestingly, in some cancer types CSCs defined by high expression of CD44 have a mesenchymal phenotype and epithelial-mesenchymal transition (EMT) is critical to enrich and maintain CSCs.Citation14-16 Consistent with those reports, we have shown that TGF-β induces EMT, resulting in conversion of CD44 low expressing cells to CD44 high expressing CSCs. Furthermore, we have previously reported that tumor cells undergo EMT in invasive fronts of ESCC and EMT contributes to tumor progression and poor prognosis.
Epidermal growth factor receptor (EGFR), frequently overexpressed in ESCC,Citation17 has pivotal roles in EMT induced by TGF-β.Citation18 Thus, EMT in invasive fronts of ESCC might be important for CD44 high expressing CSCs and EGFR could be a target of a novel therapy eliminating CSCs in ESCC. However, effects of EGFR inhibitors on CSCs in ESCC have not been fully examined and remain to be elucidated. In the present study, we examined effects of EGFR inhibitors on CSCs in ESCC using organotypic 3D culture system, a form of human tissue engineering, and showed that EGFR inhibitors suppressed EMT and generation of CSCs.
Results
EGFR inhibitors suppress enrichment of CSCs induced by TGF-β1 in ESCC
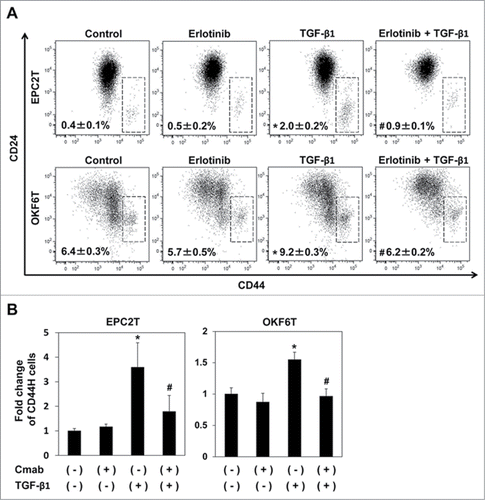
EPC2T and OKF6T cells are transformed keratinocyte cell lines overexpressing EGFR and p53R175H. EMT was induced by TGF-β1 in EPC2T and OKF6T cells as reported by us previously,Citation18,19 and CSCs defined by high expression of CD44 and low expression of CD24 (CD44High / CD24Low) were significantly enriched (). Erlotinib, one of EGFR inhibitors, significantly suppressed enrichment of CSCs via TGF-β1-mediated EMT as expected (). Cetuximab, another EGFR inhibitor, also suppressed enrichment of CSCs in both cell lines (); however, EGFR inhibitors per se had no effects on the preexisting CD44High / CD24Low CSC population (). Interestingly, treatment with EGFR inhibitors resulted in increased expression of CD24 in the non-CSC population (CD44Low / CD24High cells), suggesting that EGFR inhibition may induce differentiation in non-CSC population as CD24 is a keratinocyte differentiation marker that is expressed in post-mitotic, non-clonogenic suprabasal keratinocytes.Citation20 These results indicate that EGFR inhibitors may block EMT by inducing differentiation in non-CSC populations, leading to suppressed enrichment of CSCs via EMT; however, these EGFR inhibitors have no effect on preexisting CSCs.
Figure 1. EGFR inhibitors suppressed enrichment of CSCs induced by TGF-β1. (A) EPC2T cells and OKF6T cells were treated with or without erlotinib (2.5 μM) and TGF-β1 (5 ng/ml) for 72 hours. CD44 high expressing CSCs were enriched by TGF-β1 and the enrichment of CSCs by TGF-β1 was significantly suppressed by erlotinib. (*P < 0.05 vs. DMSO control, #P < 0.05 vs TGF-β1) (B) EPC2T cells and OKF6T cells were treated with or without cetuximab (10 μg/ml) and TGF-β1 (5 ng/ml) for 72 hours. Fold change of CD44 high expressing CSCs was shown. (*P < 0.05 vs. DMSO control, # P < 0.05 vs TGF-β1)
EGFR inhibitors suppress ZEBs and induce differentiation in ESCC
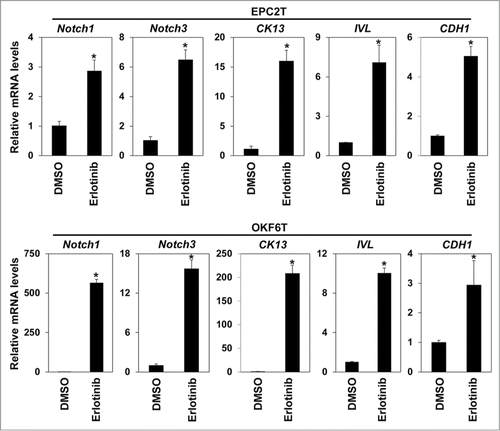
We have previously reported that Zinc finger E-box binding proteins (ZEBs) are essential for TGF-β1 mediated EMT.Citation18 Thus we examined the effects of EGFR inhibition on expression of ZEBs. Both erlotinib and cetuximab sharply suppressed expression of ZEB1 and ZEB2 (). Furthermore, Notch1 and Notch3, essential transcription factors in keratinocyte differentiation of esophagus,Citation21 as well as CK13 and involucrin, keratinocyte differentiation markers, were all up-regulated by erlotinib in EPC2T and OKF6T cells. These results are consistent with upregulation of CD24, a keratinocyte differentiation marker (). A major epithelial marker, CDH1, was also significantly increased by EGFR inhibition (). These findings suggest that EGFR inhibition may suppress expression of ZEBs and induce differentiation, thereby blocking EMT-mediated enrichment of CSCs.
Figure 2. EGFR inhibitors suppressed expression of ZEB1 and ZEB2. (A) EPC2T cells were treated with erlotinib for 72 hours and expression levels of ZEB1 and ZEB2 were determined by real-time RT-PCR. (*P < 0.05 vs. DMSO control) (B) EPC2T cells were treated with cetuximab (10 μg/ml) for 72 hours and expression levels of ZEB1 and ZEB2 were determined by real-time RT-PCR. (* P < 0.05 vs. vehicle control)
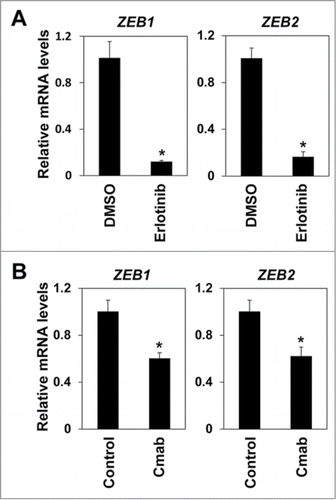
Figure 3. Erlotinib upregulated Notch transcriptional factors and induced differentiation. EPC2T cells and OKF6T cells were treated with erlotinib (2.5 μM) for 72 hours and expression levels of indicated genes were determined by real-time RT-PCR. Notch1 and Notch3 are critical transcriptional factors in keratinocyte differentiation. CK13 and involucrin (IVL) are differentiation markers of keratinocytes. (* P < 0.05 vs. DMSO control)
Effects of EGFR inhibition in organotypic 3-D culture
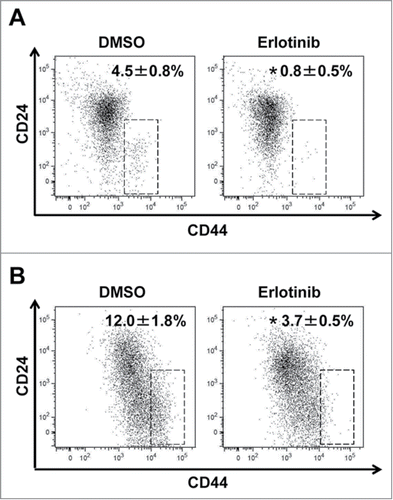
We then carried out experiments with organotypic 3D culture to assess effects of EGFR inhibition in a more physiologically relevant context. In organotypic 3D culture, EPC2T and OKF6T cells formed epithelial tumor compartments upon a matrix consisting of fibroblasts, type I collagen and matrigel, and also showed invasion into the matrix (). EGFR was activated in most cells and erlotinib sufficiently blocked EGFR activation, confirmed by IHC with the anti-phospho EGFR antibody (). Erlotinib sharply suppressed growth of tumor cells in the epithelial compartments and also blocked invasion into the matrix (). Expression of E-cadherin was increased by erlotinib in organotypic 3D culture (). Another EGFR inhibitor, cetuximab, had the same effects and suppressed growth and invasion of tumor cells in organotypic 3D culture (Fig. S1). Tumor cells at the basal layer of the epithelial compartments and in invasive nests actively proliferated and were positive for Ki67. Consistent with suppression of tumor cell growth, Ki67 positive tumor cells were significantly decreased by erlotinib (). Tumor cells at the basal layer in the epithelial compartments and in invasive nests showed high expression of CD44 in EPC2T cells. In OKF6T cells, most tumor cells highly expressed CD44 in organotypic 3D culture. Expression of CD44 was markedly suppressed by erlotinib in both EPC2T and OKF6T cells in organotypic 3D culture (). Further, we isolated tumor cells from the whole tissue obtained by organotypic 3D culture and evaluated expression of CD44 by FACS. Consistent with the results of IHC, erlotinib significantly suppressed expression of CD44 in EPC2T cells () and OKF6T cells () cultured in 3D organotypic culture. We have previously reported that EMT occurs in invasive nests of ESCC in this organotypic 3D culture system as well as in surgically resected clinical samples.Citation18,19,22 The present findings suggest that EGFR inhibitors have the potential to suppress EMT and generation of CD44 high expressing CSCs in response to cues from the local microenvironment in the context of ESCC.
Figure 4. Erlotinib suppressed tumor growth and invasion as well as enrichment of CSCs in organotypic 3D culture. EPC2T cells and OKF6T cells were cultured in organotypic 3D culture system with or without erlotinib (5 μM). (A) Tissues were stained by hematoxylin and eosin (H&E) (upper panels), anti-phospho EGFR antibody (middle panels) and anti-E-cadherin antibody (lower panels). Scale bar indicates 100 μm. (B) Cell growth was evaluated by Ki-67 staining. Histograms show percentage of Ki-67 positive cells at the basal layer. Scale bar indicates 100 μm. (*P < 0.05 vs. DMSO control) (C) Tissues were stained by anti-CD44 antibody. Scale bar indicates 100 μm.
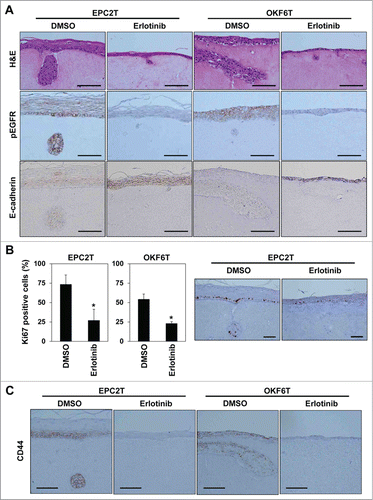
Figure 5. CD44 high expressing CSCs were reduced by erlotinib in organotypic 3D culture. EPC2T cells and OKF6T cells were cultured in organotypic 3D culture system with or without erlotinib (5 μM). Then, tumor cells were isolated from the whole organotypic 3D culture tissue and expression levels of CD24 and CD44 were analyzed by FACS. (*P < 0.05 vs. DMSO control)
Discussion
We have previously reported that EMT is induced by TGF-β in invasive fronts of ESCC and that EMT contributes to tumor progression.Citation18,19,22 EGFR is frequently overexpressed in ESCCCitation17 and EGFR is critical in the process of EMT. EGFR is necessary for EMT to negate senescence induced by TGF-β, which is also the most important inducer of EMT in ESCC.Citation18 Furthermore, mesenchymal cancer cells that have undergone EMT have been reported to be CSCs in several types of cancer. Mani SA et al. have reported that CD44High / CD24Low cancer stem-like cells are dramatically enriched by EMT induced by TGF-β in transformed human mammary epithelial cells.Citation14 Consistent with this report, CSCs were significantly increased after EMT induced by TGF-β in the present study. EMT is also a critical step for generation and maintenance of CSCs in ESCC. These findings led us to hypothesize that EGFR may serve as an effective therapeutic target against CSCs in ESCC. EGFR inhibitors sufficiently suppressed EMT and generation of CSCs as expected (). However, EGFR inhibitors did not impact the preexisting CSCs pool in the present study, suggesting that established mesenchymal CSCs are refractory to inhibition of EGFR. Indeed, we confirmed that sorted CD44High / CD24Low CSCs are significantly more resistant to the EGFR inhibitors compared to CD44Low / CD24High non-CSCs (Fig. S2). Basu D et al. have also reported that mesenchymal CSCs are EGFR independent and resistant to EGFR inhibitors.Citation23 Thus, while EGFR inhibition may prove to be effective in the prevention of CSC expansion in response to cues from the local tumor microenvironment, development of a further therapeutic strategy that concomitantly targets preexisting mesenchymal CSCs may be necessary for complete eradication of advanced ESCCs.
EGFR inhibitors sharply suppressed expression of ZEB1 and ZEB2 in the present study (). ZEBs are transcriptional factors that play a critical role in promoting EMT. ZEBs can directly bind to the promoter region of E-cadherin and suppress its transcription and also induce miR-200 family to regulate several EMT related genes.Citation24,25 ZEB1 is especially critical for EMT in ESCC and we have previously shown EGFR promotes EMT in ESCC via up-regulation of ZEB1 and that knock-down of ZEB1 is sufficient to block EMT induced by TGF-β in ESCC.Citation19 Consistent with previous findings, EGFR inhibitors can block EMT and suppress generation of CD44 high expressing CSCs by down-regulation of ZEB1. Furthermore, EMT and differentiation represent opposing cell fates in keratinocyte-derived cells. Recently, Lee B et al. have reported that Ovol1/Ovol2-deficient epidermal cells show an EMT-like phenotype and fail to undergo terminal differentiation and that knock-down of ZEB1 is sufficient to restore differentiation.Citation26 We also have shown that mesenchymal ESCC cell lines overexpressing ZEB1 downregulate expression of keratinocyte-differentiation markers, such as CK13 and involucrin.Citation19 Thus, EGFR inhibitors not only suppress CSCs but also induce keratinocyte-differentiation () potentially via downregulation of ZEB1, a novel mechanistic finding.
In the present study we utilized an organotypic 3D culture system of ESCC which allows for recapitulation of a complex tumor tissue closer that is more similar to that found in vivo than cells grown in conventional 2D culture. Moreover, 3D organotypic culture enables us to analyze heterogeneous tumor cells more precisely.Citation27 ZEB1, one of the most important regulators of EMT in ESCC, is strongly expressed in invasive compartments of surgically resected human ESCC samples.Citation19 Consistent with this observation, CD44 is highly expressed in invasive fronts where EMT occurs in the 3D culture system ().Citation19,22 Our previous and present data indicate that a niche may form at ESCC invasive fronts, presenting a hospitable environment for generation and maintenance of CSCs. Furthermore, this CSC niche may be exploited to therapeutically target CSCs. In the present study, we focused on EGFR, one of the tumor-promoting factors that is present on tumor cells; however, ESCC malignant potential is also influenced by the tumor stromal environment. In the organotypic -D culture system, tumor cell invasion is dramatically altered by the background of the fibroblasts in the matrix.Citation28 As secretion of hepatocyte growth factor (HGF) by fibroblasts influences tumor cell invasion in 3D organotypic culture, it is possible that HGF and other unknown factors secreted by various cells forming CSC niche may represent novel therapeutic target against CSCs. One limitation of organotypic 3D culture is that this system does not contain inflammatory cells, endothelial cells etc., all of which are also critical for tumor progression, malignant biological behavior and development of a CSC niche.Citation29,30 Thus, further studies utilizing in vivo ESCC models are necessary for a more precise understanding of CSCs and the CSC niche.
In conclusion, EMT is a critical process for generation and maintenance of CSCs and the invasive front of ESCC, where EMT occurs, might form a CSC niche in ESCC. EGFR inhibitors could suppress EMT in invasive fronts and be one therapeutic option targeting against generation of CSCs in ESCC.
Materials and Methods
Cell lines and reagents
EPC2T cells, transformed esophageal epithelial cells, were cultured in Keratinocyte-SFM serum-free medium (KSFM) (Lifetechnologies) as described previously.Citation18 Another immortalized keratinocytes, OKF6-TERT2 cellsCitation31 were transformed by p53R175H and EGFR by retroviral-mediated gene transfer as described previously,Citation32 and named OKF6T. OKF6T cells were grown in KSFM. Recombinant human TGF-β1 was purchased from Lifetechnologies. EGFR inhibitors, erlotinib and cetuximab were purchased from Santa Cruz Biotechnologies and Bristol-Meyers Squibb, respectively.
Real-time RT-PCR
RNA was extracted by RNeasy mini kit (QIAGEN) and cDNA was synthesized by Super Script II (Lifetechnologies) according to the manufacturer's instruction. Real-time RT-PCR was done with SYBR® Green (QIAGEN) and ABI 7300 real time PCR system (Lifetechnologies). β-actin was used as an internal control. Primers used for real-time RT-PCR were listed in .
Table 1 Primers used for real-time RT-PCR
Drug sensitivity assay
Five thousand cells per well were seeded in a 96 well plate. Twenty-four hours after seeding, drugs were added at indicated concentrations. After 72 hours incubation, MTS assay was carried out using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer's instruction. Absorbance (A490 – A630) was measured by Sunrise™ plate reader (F039302, TECAN) and survival rate was determined.
Flow cytometry and Fluorescence Activated Cell Sorting (FACS)
Cells were suspended in Hank's balanced salt solution (Invitrogen) containing 1% BSA (Sigma-Aldrich) and stained with PE/Cy7-anti-CD24 (BioLegend) and APC-anti-CD44 at 1:20 (BD Biosciences) on ice for 30 min. FACSCantoII (BD Biosciences) and FlowJo (Tree Star) were used for analysis.
Organotypic 3D culture of ESCC
Neoplastic squamous epithelium was reconstituted in organotypic 3D culture as described previously.Citation27 In brief, 0.5 × 106 of tumor cells were seeded on top of the type I collagen and Matrigel (BD Biosciences) matrices containing FEF3 human fetal esophageal fibroblasts and grown in submerged conditions for 4 d Cultures were then raised to the air–liquid interface for additional 4 d and harvested for hematoxylin and eosin staining or immunofluorescence. Each organotypic 3D culture experiment was performed in triplicate.
FACS with organotypic-3D culture samples
The whole organotypic 3D culture tissue including the matrix was minced into 1 mm3 pieces and incubated in DMEM containing 1mg/ml collagenase I (Sigma-Aldrich) for 90 min at 37°C. After centrifugation and removal of supernatant, the cells were incubated in 0.05% trypsin/EDTA (Life Technologies) for 10 min at 37°C and then incubated in dispase (1U/ml, BD Bioscience) and DNase I (100 μg/ml, Roche, Indianapolis) for 10 min at 37°C. Cells were passed through 40 μm filter and washed with DMEM. For FACS, cells were resuspended in FACS buffer and stained with 7-AAD (Life Technologies), PE/Cy7-anti-CD24 (BioLegend) and APC-anti-CD44 (BD Biosciences). Dead cells were excluded by 7-AAD and designated populations were analyzed by FACSCantoII (BD Biosciences).
Immunohistochemistry
Hematoxylin and Eosin (H&E) staining and immunohistochemistry (IHC) were performed as described previously.Citation33 The deparaffinized sections were heat-treated with antigen retrieval solution (Target Retrieval Solution, pH 9.0; Dako) at 95°C for 20 min using the Dako PT Link system. Then sections were incubated with anti-Ki-67 polyclonal antibody (Novocastra), anti-phospho EGFR monoclonal antibody (D7A5; Cell signaling technologies), anti-E-cadherin monoclonal antibody (4A2C7; Lifetechnologies), anti-CD44 monoclonal antibody (Abcam) overnight at 4°C. Detection was performed using a standard polymer method according to the manufacturer's instructions (EnVision Flex system, Dako).
Statistical analysis
Data from triplicate experiments are presented as mean ± SD and were analyzed by 2-tailed Student's t test. P < 0.05 was considered significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Figures
Download Zip (562.1 KB)Additional information
Funding
References
- Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet 2013; 381:400-12; PMID:23374478; http://dx.doi.org/10.1016/S0140-6736(12)60643-6
- Fokas E, Weiss C, Rodel C. The role of radiotherapy in the multimodal management of esophageal cancer. Dig Dis 2013; 31:30-7; PMID:23797120; http://dx.doi.org/10.1159/000347170
- Oyama T, Tomori A, Hotta K, Morita S, Kominato K, Tanaka M, Miyata Y. Endoscopic submucosal dissection of early esophageal cancer. Clin Gastroenterol Hepatol 2005; 3:S67-70; PMID:16013002; http://dx.doi.org/10.1016/S1542-3565(05)00291-0
- Shimizu Y, Tsukagoshi H, Fujita M, Hosokawa M, Kato M, Asaka M. Long-term outcome after endoscopic mucosal resection in patients with esophageal squamous cell carcinoma invading the muscularis mucosae or deeper. Gastrointest Endosc 2002; 56:387-90; PMID:12196777; http://dx.doi.org/10.1016/S0016-5107(02)70043-6
- Shaheen NJ, Sharma P, Overholt BF, Wolfsen HC, Sampliner RE, Wang KK, Galanko JA, Bronner MP, Goldblum JR, Bennett AE, et al. Radiofrequency ablation in Barrett's esophagus with dysplasia. N Engl J Med 2009; 360:2277-88; PMID:19474425; http://dx.doi.org/10.1056/NEJMoa0808145
- Bergman JJ, Zhang YM, He S, Weusten B, Xue L, Fleischer DE, Lu N, Dawsey SM, Wang GQ. Outcomes from a prospective trial of endoscopic radiofrequency ablation of early squamous cell neoplasia of the esophagus. Gastrointest Endosc 2011; 74:1181-90; PMID:21839994; http://dx.doi.org/10.1016/j.gie.2011.05.024
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 2011; 17:313-9; PMID:21386835; http://dx.doi.org/10.1038/nm.2304
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10:717-28; PMID:22704512; http://dx.doi.org/10.1016/j.stem.2012.05.007
- Zhao JS, Li WJ, Ge D, Zhang PJ, Li JJ, Lu CL, Ji XD, Guan DX, Gao H, Xu LY, et al. Tumor initiating cells in esophageal squamous cell carcinomas express high levels of CD44. PloS One 2011; 6:e21419; PMID:21731740; http://dx.doi.org/10.1371/journal.pone.0021419
- Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 2007; 104:973-8; PMID:17210912; http://dx.doi.org/10.1073/pnas.0610117104
- Feng D, Peng C, Li C, Zhou Y, Li M, Ling B, Wei H, Tian Z. Identification and characterization of cancer stem-like cells from primary carcinoma of the cervix uteri. Oncol Rep 2009; 22:1129-34; PMID:19787230
- Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 2011; 11:254-67; PMID:21390059; http://dx.doi.org/10.1038/nrc3023
- Natsuizaka M, Kinugasa H, Kagawa S, Whelan KA, Naganuma S, Subramanian H, Chang S, Nakagawa KJ, Rustgi NL, Kita Y, et al. IGFBP3 promotes esophageal cancer growth by suppressing oxidative stress in hypoxic tumor microenvironment. Am J Cancer Res 2014; 4:29-41; PMID:24482736
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133:704-15; PMID:18485877; http://dx.doi.org/10.1016/j.cell.2008.03.027
- Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad A, Banerjee S, Azmi AS, Miele L, Sarkar FH. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett 2011; 307:26-36; PMID:21463919; http://dx.doi.org/10.1016/j.canlet.2011.03.012
- Chen C, Wei Y, Hummel M, Hoffmann TK, Gross M, Kaufmann AM, Albers AE. Evidence for epithelial-mesenchymal transition in cancer stem cells of head and neck squamous cell carcinoma. PloS One 2011; 6:e16466; PMID:21304586; http://dx.doi.org/10.1371/journal.pone.0016466
- Itakura Y, Sasano H, Shiga C, Furukawa Y, Shiga K, Mori S, Nagura H. Epidermal growth factor receptor overexpression in esophageal carcinoma. An immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer 1994; 74:795-804; PMID:8039107; http://dx.doi.org/10.1002/1097-0142(19940801)74:3%3c795::AID-CNCR2820740303%3e3.0.CO;2-I
- Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, Kalabis J, Vega ME, Kalman RA, Nakagawa M, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res 2010; 70:4174-84; PMID:20424117; http://dx.doi.org/10.1158/0008-5472.CAN-09-4614
- Ohashi S, Natsuizaka M, Naganuma S, Kagawa S, Kimura S, Itoh H, Kalman RA, Nakagawa M, Darling DS, Basu D, et al. A NOTCH3-mediated squamous cell differentiation program limits expansion of EMT-competent cells that express the ZEB transcription factors. Cancer Res 2011; 71:6836-47; PMID:21890822; http://dx.doi.org/10.1158/0008-5472.CAN-11-0846
- Bergoglio V, Larcher F, Chevallier-Lagente O, Bernheim A, Danos O, Sarasin A, Rio MD, Magnaldo T. Safe selection of genetically manipulated human primary keratinocytes with very high growth potential using CD24. Mol Ther 2007; 15:2186-93; PMID:17712330; http://dx.doi.org/10.1038/sj.mt.6300292
- Ohashi S, Natsuizaka M, Yashiro-Ohtani Y, Kalman RA, Nakagawa M, Wu L, Klein-Szanto AJ, Herlyn M, Diehl JA, Katz JP, et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology 2010; 139:2113-23; PMID:20801121; http://dx.doi.org/10.1053/j.gastro.2010.08.040
- Natsuizaka M, Ohashi S, Wong GS, Ahmadi A, Kalman RA, Budo D, Klein-Szanto AJ, Herlyn M, Diehl JA, Nakagawa H. Insulin-like growth factor-binding protein-3 promotes transforming growth factor-{β}1-mediated epithelial-to-mesenchymal transition and motility in transformed human esophageal cells. Carcinogenesis 2010; 31:1344-53; PMID:20513670; http://dx.doi.org/10.1093/carcin/bgq108
- Basu D, Bewley AF, Sperry SM, Montone KT, Gimotty PA, Rasanen K, Facompre ND, Weinstein GS, Nakagawa H, Diehl JA, et al. EGFR inhibition promotes an aggressive invasion pattern mediated by mesenchymal-like tumor cells within squamous cell carcinomas. Mol Cancer Ther 2013; 12:2176-86; PMID:23939378; http://dx.doi.org/10.1158/1535-7163.MCT-12-1210
- Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J 2011; 30:770-82; PMID:21224848; http://dx.doi.org/10.1038/emboj.2010.349
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci 2009; 66:773-87; PMID:19011757; http://dx.doi.org/10.1007/s00018-008-8465-8
- Lee B, Villarreal-Ponce A, Fallahi M, Ovadia J, Sun P, Yu QC, Ito S, Sinha S, Nie Q, Dai X. Transcriptional mechanisms link epithelial plasticity to adhesion and differentiation of epidermal progenitor cells. Dev Cell 2014; 29:47-58; PMID:24735878; http://dx.doi.org/10.1016/j.devcel.2014.03.005
- Kalabis J, Wong GS, Vega ME, Natsuizaka M, Robertson ES, Herlyn M, Nakagawa H, Rustgi AK. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc 2012; 7:235-46; PMID:22240585; http://dx.doi.org/10.1038/nprot.2011.437
- Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, Diehl JA, Herlyn M, Han M, Nakagawa H, et al. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc Natl Acad Sci U S A 2010; 107:11026-31; PMID:20534479; http://dx.doi.org/10.1073/pnas.0914295107
- Ghiabi P, Jiang J, Pasquier J, Maleki M, Abu-Kaoud N, Rafii S, Rafii A. Endothelial Cells Provide a Notch-Dependent Pro-Tumoral Niche for Enhancing Breast Cancer Survival, Stemness and Pro-Metastatic Properties. PloS One 2014; 9:e112424; PMID:25380486; http://dx.doi.org/10.1371/journal.pone.0112424
- Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: attacking cancer's inflammatory roots. Clin Cancer Res 2011; 17:6125-9; PMID:21685479; http://dx.doi.org/10.1158/1078-0432.CCR-10-2743
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 2000; 20:1436-47; PMID:10648628; http://dx.doi.org/10.1128/MCB.20.4.1436-1447.2000
- Okawa T, Michaylira CZ, Kalabis J, Stairs DB, Nakagawa H, Andl CD, Johnstone CN, Klein-Szanto AJ, El-Deiry WS, Cukierman E, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev 2007; 21:2788-803; PMID:17974918; http://dx.doi.org/10.1101/gad.1544507
- Naganuma S, Whelan KA, Natsuizaka M, Kagawa S, Kinugasa H, Chang S, Subramanian H, Rhoades B, Ohashi S, Itoh H, et al. Notch receptor inhibition reveals the importance of cyclin D1 and Wnt signaling in invasive esophageal squamous cell carcinoma. Am J Cancer Res 2012; 2:459-75; PMID:22860235
