ABSTRACT
Glycolysis contributes to cell metabolism and facilitates cell proliferation of oral squamous cell carcinoma (OSCC), the most common type of oral cancer. Understanding the regulatory mechanisms involved in the glycolysis of OSCC cells may provide important therapeutic inspirations. Immunohistochemistry was used to examine protein localization patterns in human OSCC tissues and Western blot was conducted to gauge protein level. Lentivirus transduction was used to overexpress or silence genes of interest. Cell proliferation was assessed by Cell Counting Kit (CCK)-8 assay while glycolysis was examined via measurement of extracellular acidification rate, oxygen consumption rate, and lactate and ATP production. In vivo cancer development was evaluated with a mouse tumor growth model. OSCC tissues displayed reduced expression of NEDD4L compared with normal tissues. NEDD4L expression positively correlated with 5-year patient survival rate, indicating that NEDD4L may be a prognosis marker for OSCC. NEDD4L overexpression suppressed proliferation, cell cycle transition, and glycolysis in OSCC cells, and inhibited in vivo tumor growth. UbiBrowser identified ENO1, an enzyme that catalyzes glycolysis, as a substrate of NEDD4L. Overexpression of NEDD4L resulted in the ubiquitination and subsequent degradation of ENO1 whereas overexpression of ENO1 reversed the functional effects of NEDD4L overexpression, restoring proliferation, cell cycle transition, and glycolysis in OSCC cells. NEDD4L elicits tumor-suppressive functions via inhibition of OSCC cell proliferation, cell cycle transition, and glycolysis by stimulating ENO1 ubiquitination and degradation. Our results unraveled a signaling axis important for OSCC cell survival and metabolism, which can serve as a potential therapeutic target.
KEYWORDS:
Introduction
Oral squamous cell carcinoma (OSCC) accounts for >90% of all oral neoplasm cases,Citation1 which can be triggered by several factors: tobacco and alcohol use, vitamin A, E, or C deficiency, immune defects, and more. Despite advances in medical approaches and technology in recent decades, OSCC morbidity and mortality remain relatively high, and current treatment options for OSCC are limited. Understanding the molecular mechanisms that contribute to OSCC progression is therefore critical for developing effective treatment strategies for OSCC.
Neural precursor cell expressed developmentally downregulated 4-like (NEDD4L)Citation2 is an E3 ubiquitin ligase whose substrate targets are mostly membrane proteins (e.g., ion channels and transporters).Citation3 NEDD4L is involved in the development of many types of cancer and is believed to be a promising target for anti-cancer therapy.Citation4 For instance, NEDD4L expression is downregulated in hepatocellular carcinoma and glioma, as well as prostate, endometrial, ovarian, and gastric cancers. Moreover, the extent of NEDD4L downregulation correlated with cancer severity and prognosis.Citation5–11 In hepatocellular carcinoma, NEDD4L inhibits the MAPK/ERK signaling pathway to stimulate cancer cell apoptosis,Citation11 suggestive of a tumor suppressive role. NEDD4L was also found to ubiquitinate STK35 and inhibit glycolysis in colorectal cancer.Citation12 Moreover, NEDD4L was shown to inhibit WNT signaling, a central pathway for the pathogenesis of colorectal cancer.Citation13 It was later reported that NEDD4L induces the degradation of LGR5, a receptor for R-spondin that promotes Wnt signaling, which might explain the underlying mechanism.Citation14 Nonetheless, whether NEDD4L is involved in the regulation and development of OSCC has not been investigated.
Accelerated metabolism is a critical feature of cancer cells, which mediates many activities relevant to cell survival and fate (apoptosis, ferroptosis, differentiation, etc.).Citation15–17 The metabolism of cancer cells largely depends on glycolysis, a process that digests glucose to derive energy independent of oxygen.Citation18 The glycolysis of cancer cells involves multiple enzymes, including hexokinase 2 (HKII), glucose transporter 1 (GLUT1), and phosphoglucose isomerase.Citation19 Among these, alpha-enolase (ENO1), a metalloenzyme that dehydrates 2-phospho-D-glycerate in the glycolytic pathway, is upregulated in multiple types of cancer.Citation20,Citation21 In lung cancer, for instance, ENO1 knockdown inhibited cancer cell proliferation and invasiveness, while ENO1 overexpression facilitated these processes.Citation22 Besides its function in glycolysis, ENO1 is also expressed as a cell surface plasminogen receptor and facilitates cancer cell adhesion and metastasis.Citation23,Citation24 Interestingly, a previous work found that ENO1 expression was elevated in OSCC and increased cell proliferation, with circ-angiomotin like 1 acting as the upstream mediator.Citation25 However, whether other mediators and signaling pathways are involved in regulating the expression and activity of ENO1 in OSCC is not clear.
In this work, by exploring the involvement of NEDD4L in OSCC pathogenesis, we found that NEDD4L was downregulated as OSCC progressed, whereas overexpression of NEDD4L in OSCC cells displayed anti-cancer functions. We further demonstrated that the anti-cancer function of NEDD4L was dependent on its function as an ubiquitin ligase for ENO1. By inducing ENO1 ubiquitination and degradation, NEDD4L inhibits the functions of ENO1 in cancer cell glycolysis and proliferation. Our work thus shed light on the molecular mechanisms of OSCC, and identified the NEDD4L/ENO1 signaling axis as a potential target in OSCC treatment.
Results
NEDD4L expression level is low in OSCC tissues and has prognostic correlation
To investigate the function of NEDD4L in OSCC, NEDD4L expression was examined in five different OSCC cell lines (HSC4, SSC4, SSC9, SSC15, and CAL27) and compared with a non-tumor control cell line (HOEC). Compared to control, the mRNA level of NEDD4L was decreased in all OSCC cell lines (Figure S1). In addition, NEDD4L protein levels were also slightly reduced, with SCC15 showing the most noticeable difference (Figure S1).
RT-PCR showed that the RNA level of NEDD4L was significantly reduced in human patient OSCC tissues than in normal tissues ()). The RNA level was also correlated with the severity of OSCC: tissues from Stage III/VI patients showed lower expression of NEDD4L than those from Stage I/II patients. Consistently, Western blot showed that the protein level of NEDD4L was reduced in OSCC tissues, and was almost eliminated in tissue samples from Stage III/VI patients ()).
Figure 1. NEDD4L expression level was low in OSCC tissues and had prognostic correlation. Paraffin sections of Stage 1 or 2 OSCC tissues (I/II), Stage 3 or 4 OSCC tissues (III/IV), and adjacent non-tumor tissues (Normal) were assessed. (a) Mean ± S.D. and individual points of NEDD4L mRNA levels. **p < .01; ***p < .001, compared to Normal; ##p < .01, compared to I/II; assessed by ANOVA followed by Tukey’s test. (b) Representative Western blot images showing protein expression levels of NEDD4L in tissues. (c) Representative immunohistochemistry (IHC) images showing the distribution of NEDD4L in NEDD4L-low and -high tumor tissues. (d) Comparison of probability of patient survival with high or low NEDD4L expression level using Log-rank test.
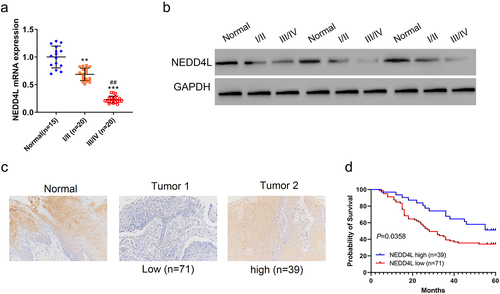
Using immunohistochemistry (IHC) analysis, we then quantitatively measured NEDD4L protein expression in 110 OSCC tissues, and segregated them into two groups that had high or low NEDD4L expression levels ()). We found that NEDD4L protein level strongly correlated with the tumor stage of the patients: most Stage I/II patients had high NEDD4L expression, whereas more Stage III/VI patients shifted to have low NEDD4L expression (). On the other hand, NEDD4L expression level did not correlate with gender, age, or pathologic differentiation of these patients (). More importantly, NEDD4L expression level turned out to be a strong predictor of prognosis: patients with high NEDD4L expression had a much higher 5-year survival rate than patients expressing low-level NEDD4L (~60% vs. <40%, P < .05) ()). Overall, our data showed that NEDD4L expression is downregulated in OSCC, and NEDD4L expression level serves as a good prognostic parameter to predict the lethality of the disease.
Table 1. Correlation of NEDD4L expression in OSCC tissues with different clinicopathological features (n = 110)
Overexpressing NEDD4L in OSCC cells inhibits cell proliferation and triggers cell cycle arrest
It is intriguing that NEDD4L expression level could predict OSCC prognosis. One straightforward hypothesis is that NEDD4L can affect OSCC disease progression. To test this hypothesis, we evaluated how NEDD4L overexpression affects OSCC cell growth, and therefore tumor growth.Citation26 Using NEDD4LOE, a lentivirus that induces NEDD4L overexpression. We then selected two OSCC cell lines that had relatively low NEDD4L expression, SCC15 and CAL27 (Figure S1), and transduced these cells with NEDD4LOE. Transduction of cells with NEDD4LOE lentivirus resulted in elevated NEDD4L expression compared with cells transduced with a control vector virus (), Figure S2A), confirming efficiency of overexpression. Importantly, after 24 h of cell culture, NEDD4L overexpression significantly inhibited proliferation of both SCC15 and CAL27 cells, and these inhibitory effects continued after 48 h ()).
Figure 2. Overexpressing NEDD4L in OSCC cells inhibited cell proliferation and triggered cell cycle arrest, suppressed glycolysis. (a) Western blot results showing NEDD4L expression levels in OSCC cell lines (SCC15 and CAL27) transduced with either control vector or NEDD4LOE. (b) Cell proliferation assay results of OSCC cells with or without NEDD4LOE transduction. (c) Cell cycle analysis results showing flow cytometry assessment of DNA content levels (left; green, yellow and cyan respectively represent cells in G0/G1, S and G2/M phases) and the calculated fraction of cell subpopulations in different phases (right). (d) Western blot results showing p21 and p27 expression levels in OSCC cells with or without NEDD4LOE transduction. (e-f) Mean ± S.E.M. of extracellular acidification rate (ECAR) (e) and oxygen consumption rate (OCR) (f) in OSCC cells with or without NEDD4LOE transduction. (g-h) Mean ± S.E.M. of lactate (g) and ATP (h) production of OSCC cells with or without NEDD4LOE transduction. **p < .01; ***p < .001, assessed by two-tailed Student’s t-test.
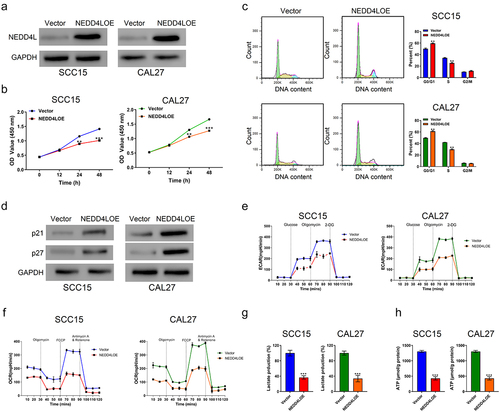
Further, our cell cycle assay revealed that NEDD4L overexpression resulted in an increased number of cells in the G0/G1 phase and less in the S phase, indicating cell cycle arrest ()). On the contrary, decreased expression of NEDD4L was observed when lentivirus mediated RNA interference was performed (Figure S2B, Figure S3A). Downregulation of NEDD4L promoted proliferation as well as cell cycle transition of OSCC cells (Figure S3B, C). p21 and p27 are two G1-checkpoint cyclin-dependent kinase inhibitors that mediate multiple human malignancies.Citation27–29 Here, we found that NEDD4L overexpression resulted in the overexpression of p21 and p27 as well ()), while downregulation of NEDD4L led to the reduced expression of p21 and p27 (Figure S3D), suggesting a potential molecular mechanism through which NEDD4L induces cell cycle arrest and in turn suppresses OSCC cell proliferation.
NEDD4L overexpression suppresses glycolysis in OSCC cells
Accelerated glycolysis is an important attribute of tumor cells and provides extra ATP and glucose to support the fast division and production of tumor cells.Citation30 Interestingly, as an ubiquitin ligase, NEDD4L regulates glycolysis by ubiquitinating serine/threonine kinase 3512 and PIK3CACitation31 inside cells. Notably, the effect of NEDD4L on glycolysis was found to be inhibitory in the former case, but facilitating in the latter, suggesting that when involved in different signaling pathways, the regulatory role of NEDD4L on glycolysis can be varied.
Here, we show that in SCC15 and CAL27 cells with NEDD4L overexpression, the extracellular acidification rate (ECAR)Citation32 ()) and oxygen consumption rate (OCR)Citation33 ()) were both significantly lower, which indicated a slower glycolytic rate. In agreement with these results, lactate ()) and ATP ()) production in these cells also became much lower. These results demonstrated that NEDD4L overexpression suppresses glycolysis in OSCC cells, consistent with its function in inhibiting cell proliferation. On the other hand, the ECAR, OCR, and lactate and ATP production rates were all increased by NEDD4L downregulation, indicating that NEDD4L interference facilitates glycolysis of OSCC cells (Figure S3E-H).
NEDD4L overexpression suppresses mouse tumor growth
Our above in vitro results clearly demonstrated anti-tumor functions of NEDD4L via inhibition of proliferation and metabolism of OSCC cells. We thus hypothesized that NEDD4L should inhibit the growth of tumor in vivo. To validate our hypothesis, we established an in vivo tumor growth model by subcutaneously injecting SCC15 cells into nude mice. Tumor formation and development were then tracked over 33 days. Although SCC15 cells with and without NEDD4LOE transduction both induced tumor development starting from Day 12, overexpression of NEDD4L clearly slowed down tumor growth ()). By the end of the observation time window, tumors in mice injected with NEDD4LOE-transduced SCC15 cells had an average size and weight of only one-third of the tumors developed in mice injected with normal SCC15 cells ()).
Figure 3. NEDD4L overexpression suppressed mouse tumor growth. Nude mice were injected subcutaneously with SCC15 cells with or without NEDD4LOE transduction. (a) Tumor volume was tracked from Day 12 to Day 33. (b) Tumor pictures (top) and weight (bottom; individual points and mean ± S.D.) on Day 33. (c) Representative IHC images showing Ki67 distribution. (d) Western blot results showing NEDD4L, p21, and p27 expression levels in representative tumor tissues. (e) Comparison of the probability of survival of mice with or without NEDD4L overexpression. *p < .05; ***p < .001, assessed by two-tailed Student’s t-test.
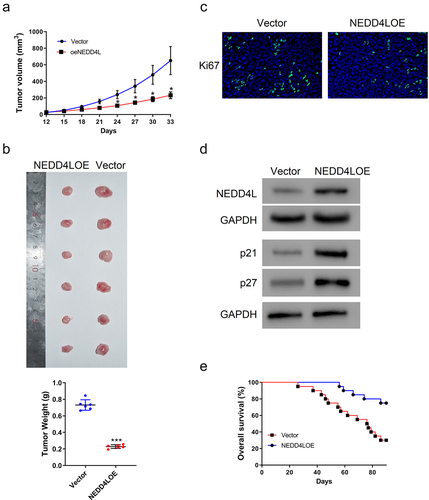
In agreement with these results, IHC staining of Ki67, a strong biomarker of tumor cell proliferation and growth,Citation34 showed much less signal in the tumor tissues of mice injected with NEDD4LOE-transduced SCC15 cells ()). Using Western blot, we further confirmed that these tumor tissues indeed expressed a higher level of NEDD4L; consistent with our in vitro results, these tissues also had an overexpression of p21 and p27 ()).
Our analysis on human patient samples has indicated that the expression level of NEDD4L is a good prognostic parameter to predict the lethality of cancer ()). Considering the results we collected so far, we argue that this good prognosis is in fact due to the protective function of NEDD4L that inhibits the development of cancer, thereby extending the patient’s lifetime. Indeed, we also found that mice injected with NEDD4LOE-trasnduced SCC15 cells could survive much longer than those injected with normal SCC15 cells, with the survival rate increased from 30% to >70% by the end of the 90-day observation ()).
NEDD4L binds to ENO1 and induces ENO1 ubiquitination
We next investigated the molecular mechanism through which NEDD4L inhibits proliferation and metabolism of OSCC cells. Because ENO1 is induced in many types of cancers and contributes to cancer progression,Citation15 and, more importantly, mediates glycolysis of cells,Citation35 we suspected that the functional effects of NEDD4L on OSCC might have some functional relevance to ENO1. Certain structural modifications of proteins, like phosphorylation and ubiquitination, can regulate protein degradation by marking the protein for further clearance mechanisms.Citation36 Considering that NEDD4L is a well-known E3-ubiquitin ligase,Citation37 we speculated that NEDD4L can induce ubiquitination of ENO1 and thereby accelerate ENO1 degradation. Supporting this, UbiBrowser, an online platform that predicts ubiquitin ligase-substrate interactions, predicts ENO1 as a likely target substrate of NEDD4L for ubiquitination (Figure S4).
In order to validate that ENO1 is indeed a substrate of NEDD4L, we first conducted co-immunoprecipitation assay and checked whether the two molecules can bind with each other. From cell lysates, a large amount of ENO1 was co-precipitated with NEDD4L; reciprocally, NEDD4L was also co-precipitated with ENO1 ()). These results confirmed interaction between NEDD4L and ENO1.
Figure 4. NEDD4L bound to ENO1 and induced its ubiquitination. (a) Co-immunoprecipitation results showing direct association between NEDD4L and ENO1. (b) ENO1 protein (top) and mRNA (bottom) levels in OSCC cells with or without NEDD4LOE transduction. (c) ENO1 protein (left) and mRNA (right) levels in OSCC cells with or without NEDD4L interference. (d) Western blot results showing ENO1 protein level in OSCC cells with or without NEDD4LOE transduction and with or without MG132 treatment. (e) Western blot results of ENO1 from OSCC cells with or without NEDD4LOE. (f) IHC analysis of paraffin sections from patients, staining ENO1 and NEDD4L. Case 1 represents high ENO1 and low NEDDL4, while Case 2 represents low ENO1 and high NEDDL4 (upper). A negative correlation between ENO1 and NEDD4L expression was identified using Fisher’s exact test (bottom).
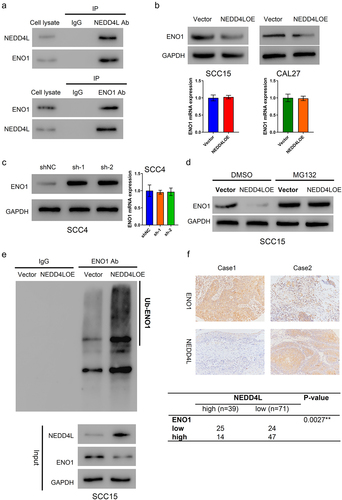
We then assessed whether and how NEDD4L regulates ENO1 in cells. In SCC15 and CAL27 cells that had NEDD4LOE transduction, the mRNA level of ENO1 was not affected; however, the protein level of ENO1 was greatly decreased ()). On the contrary, in cells that had sh-1 or sh-2 transduction, the protein level (but not the mRNA level) of ENO1 was greatly enhanced ()). These results indicated that NEDD4L negatively regulates the protein level of ENO1. Notably, if the cells were pre-treated with MG132, a cell-permeable proteasome inhibitor, then NEDD4L overexpression would have completely no inhibitory effect on ENO1 expression, suggesting that NEDD4L negatively regulates ENO1 by facilitating ENO1 degradation ()). Lastly, we identified that from the lysates of SCC15 cells with NEDD4LOE transduction, a large amount of ENO1 was ubiquitinated, whereas in cells with control vector transduction, the level of ubiquitination was much lower, confirming that NEDD4L induces ENO1 ubiquitination ()).
Importantly, the negative regulation of NEDD4L on ENO1 expression was also identified in patient samples. In IHC analysis of human patient paraffin sections, samples bearing high NEDD4L level would generally have low ENO1 level, and vice versa (representative images in ), top panel). Statistical analysis confirmed the negative correlation between NEDD4L and ENO1 expression levels to be highly significant (), bottom panel).
ENO1 overexpression reverses the functional effects of NEDD4L overexpression
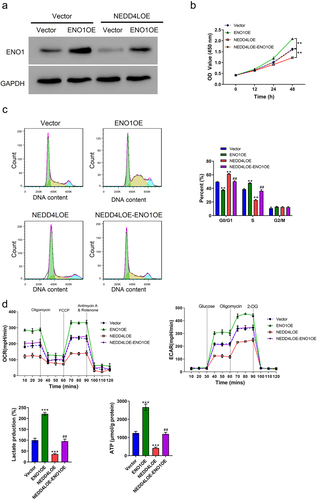
Although the above results identified ENO1 to be a target of NEDD4L for ubiquitination, it is still not certain whether this regulatory mechanism was underlying the function of NEDD4L in inhibiting proliferation and glycolysis of OSCC cells. Therefore, we used lentivirus transduction to artificially induce the overexpression of ENO1 and see if it could bypass the regulatory effects of NEDD4L. A vector, ENO1OE, was generated, and ENO1 overexpression in OSCC cells was confirmed by Western blot (), Figure S2C). Interestingly, the sole transduction of ENO1OE significantly enhanced cell proliferation and cycle transition, ()), and also increased cell glycolysis rate as reflected by enhanced OCR, ECAR, and lactate and ATP production rate ()). More interestingly, in cells that already had NEDD4LOE transduction, which inhibited cell proliferation and cycle transition, further introduction of ENO1OE restored the proliferative activity and glycolysis rate back to normal ()). Altogether, these results demonstrated that ENO1 overexpression reversed the functional effects of NEDD4L overexpression, and verified ENO1 to be the downstream target of NEDD4L in regulating OSCC cell proliferation and glycolysis.
Figure 5. ENO1 overexpression reversed the functional effects of NEDD4L overexpression. (a) Western blot results showing ENO1 protein level in OSCC cells with only ENO1OE transduction or with both NEDD4LOE and ENO1OE transduction. (b-c) Cell proliferation assay (b) and cell cycle analysis (c) of OSCC cells with NEDD4LOE and/or ENO1OE transduction. (d) Mean ± S.E.M. of ECAR and OCR (top) and lactate and ATP production (bottom) of OSCC cells with NEDD4LOE and/or ENO1OE transduction. **p < .01; ***p < .001, compared with Vector group; ##p < .01, compared with NEDD4LOE group; assessed by ANOVA followed by Tukey’s test.
Discussion
Like most other types of cancer, OSCC currently lacks a cure and accounts for a high rate of mortality worldwide, despite all currently available therapeutic approaches.Citation38 To develop novel strategies for treating OSCC requires in-depth understanding of the underlying molecular mechanisms. Recently, several molecular targets responsible for different aspects of cancer development were highlighted, including 1) epidermal growth factor receptor and 2) mammalian target of rapamycin, both of which mediate multiple-signaling pathways that lead to tumor cell proliferation, metastasis, and angiogenesis; 3) programmed cell death receptor 1, which results in the insensitivity and/or apoptosis of effector T cells; 4) vascular endothelial growth factor, which mainly contributes to angiogenesis;Citation38 and more. In this context, molecules that mediate cell glycolysis and thereby stimulate cancer cell proliferation also drew major attention.Citation39–42 Adding to these findings, we here identified another signaling pathway of cancer cell glycolysis that involves two key molecules: NEDD4L and ENO1. In OSCC, the downregulation of NEDD4L induces the overexpression of ENO1 and leads to enhanced glycolysis and proliferation, whereas artificially induced overexpression of NEDD4L manifests anti-cancer effects by downregulating ENO1 expression and compromising cell glycolysis and proliferation, as well as increasing cell cycle arrest. Overall, our work unraveled a new molecular mechanism of OSCC pathogenesis, and also identified the NEDD4L/ENO1 signaling axis as a potential target for anti-cancer therapeutics.
NEDD4L is an ubiquitin-protein ligaseCitation3 that plays many different biological roles due to the large ensemble of its substrates. Up to now, multiple-signaling pathways in cancer have already been identified to be mediated by molecules in the NEDD4 family via their E3 ligase activity. As two examples, NEDD4 induces the ubiquitination and degradation of LATS1 and inhibits the Hippo signaling pathway,Citation43 and also facilitates the ubiquitination and degradation of both Notch and Deltex to control the Notch signaling pathway.Citation44 This suggests a large “signaling network” centered on NEDD4 that influences many aspects of cancer biology. To better understand the precise functions of NEDD4L on cancer requires us to identify all its substrates and dissect their functions. In this context, our identification of ENO1 as another substrate of NEDD4L for cancer regulation adds to our current understanding of the wide repertoire of functions of NEDD4L. Furthermore, miRNAsCitation45,Citation46 have been reported to regulate NEDD4L expression in cancers. Whether NEDD4L expression in OSCC is reduced by miRNAs will be explore in the future.
NEDD4L and ENO1 are both well-known key players in many types of cancers and in mediating glycolysis.Citation12,Citation20,Citation21 However, whether they have any functional relationship was never clear. Here, we provided direct evidence showing that NEDD4L binds to and regulates ubiquitination of ENO1, inducing ENO1 degradation and impairing OSCC cell glycolysis. Downregulation of NEDD4L in OSCC and the resulting upregulation of ENO1 should serve as a mechanism that facilitates OSCC progression, which is supported by the prognostic correlation of NEDD4L in human patients. More importantly, NEDD4L/ENO1 signaling pathway is unlikely specific for OSCC, but may contribute to other cancer types that involve these two molecules, including glioma, hepatocellular carcinoma, and gastric cancer.Citation6,Citation9,Citation11,Citation15,Citation21 Thus, our findings have important clinical implications and raise intriguing questions for future investigations.
Glycolysis is important to cancer cell development. Inhibition of glycolysis not only impairs cancer cell proliferation, but also transforms tumor cells to become more susceptible to immunotherapy, and therefore shows great promise in fighting against cancer.Citation18 Carbohydrate-restricted diets were shown to have certain benefits in cancer patients.Citation47 However, this approach strictly relies on the self-discipline of the patients themselves, and may not show satisfactory effects in relatively short time. Circumventing these issues, an alternative approach that was explored in numerous previous works is to target the glycolytic enzymes and their relevant signaling pathways so as to look for next-generation anti-cancer drugs.Citation18 For instance, compounds like Phloretin and WZB117 inhibit GLUTs and suppress glucose entry into cancer cells, and demonstrated anti-cancer effects in animal models.Citation48 Inhibitors of pyruvate kinase M2, which converts phosphoenolpyruvate to pyruvate to generate ATP, also showed specific inhibitory effects on tumor cells.Citation49 However, up to now, all studies on the inhibition of glycolytic enzymes for anti-cancer purposes either are still in progress, or have already failed due to side effects or off-target effects.Citation50 In our work, the discovery of the NEDD4L/ENO1 signaling pathway provides a new target for the development of anti-cancer agents. For example, an agent that specifically increases the association affinity of NEDD4L with ENO1, but not other substrates, should serve as a precision drug to inhibit cancer cell glycolysis and proliferation with minimized off-target effects.
In summary, we uncovered that an E3 ubiquitin ligase, NEDD4L, is suppressed in OSCC and serves as a prognostic marker of OSCC. Furthermore, we found that NEDD4L inhibits glycolysis and proliferation of OSCC cells by inducing ubiquitination and degradation of its target substrate, ENO1. Overexpression of ENO1 can counteract the functional effects of NEDD4L and promote OSCC cell glycolysis and proliferation. Our study underscores an anti-cancer function of NEDD4L and a cancer-facilitating role of ENO1, and emphasizes the NEDD4L/ENO1 signaling pathway for clinical and therapeutic inspirations.
Methods
Tissue samples and patient information
This study was approved by the Ethics Committee of Shengjing Hospital, China Medical University. Written consent was obtained from all the participant. For quantitative RT-PCR and Western blotting, 40 OSCC tissues (20 each in Stage I/II and III/IV) and 15 non-tumorous adjacent tissues were collected from patients who received surgical treatment at Shengjing Hospital, and stored at −80°C. Paraffin sections of 110 human patient OSCC tissues (81 in Stage I/II and 29 in Stage III/IV) and 10 adjacent tissues were purchased from Outdo Biotechnology (Shanghai, China).
Immunohistochemistry (IHC) analysis
Paraffin sections were deparaffinized with xylene and rehydrated with ethanol,Citation51 heated for 10 min in 0.1 M citric acid buffer (pH 6.0), cooled, and blocked for 15 min (0.3% hydrogen peroxide). The sections were incubated overnight with anti-NEDD4L (Abcam, ab240753) or anti-ENO1 (Abcam, ab227978) and then with horseradish peroxidase-conjugated secondary antibodies. Finally, results were read with a 3, 3’-diaminobenzidine (DAB) kit (Long Island, Shanghai, China), with intensity graded as: IRS = staining intensity (SI: 0, negative; 1, weak; 2, moderate; 3, strong; >3, overexpression) × percentage of positive cells (PP) by two trained pathologists. IRS ≥ 3 was considered as indicative of high expression, whereas IRS < 3 was deemed low expression.
Cell culture
Human oral gingival epithelial cell line (HOEC cells) and OSCC cell lines (CAL27, HSC4, SCC4, SCC9, and SCC15) were acquired from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). All the cell lines were cultured at 37°C and 5% CO2, in high glucose Dulbecco’s modified Eagle’s medium (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and penicillin/streptomycin (Solarbio, Beijing, China).
Lentivirus mediated overexpression of NEDD4L and ENO1
NEDD4L coding sequence was amplified using the primers below: NEDD4L (NM_001144966.2), forward 5’ -CGGAATTCATGGAGCGACCCTATACATTTAAG-3’ and reverse 5’-CGGGATCCTTAATCCACCCCTTCAAATCC-3’. After digestion, the DNA fragments were cloned into a linearized pLVX-puro expression vector (Clontech, Palo Alto, CA, USA). For virus production, 293 T cells were transfected with the above constructs in combination with psPAX2 and pMD2G (Addgene, Cambridge, MA, USA). After 48–72 h, lentiviruses were collected from the culture medium and transfected into indicated cells.
The full-length human ENO1 was amplified using the primers below: ENO1 (NM_001201483.4), forward 5’-CCCAAGCTTATGATCGAGATGGATGGAACAG-3’ and reverse 5’-CCCAAGCTTATGATCGAGATGGATGGAACAG-3’, and cloned into pcDNA3.1 vector (Life Technology) by Genewiz Company.
Lentivirus mediated RNA interference (RNAi)
RNAi oligos targeting NEDD4L were cloned into linearized pLKO.1 plasmids (Addgene). The interference sites are as follows: sh-1, 5’- GAGCGACCCTATACATTTA −3’; sh-2, 5’- GGGAAGTTGTTGACTCAAA −3’; and sh-3, 5’- GCTCTTTGATTCAAAGAGA −3’. The lentivirus was packaged as described above.
RNA isolation and quantitative RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen). The mRNA levels of indicated genes were determined by quantitative RT-PCR using SYBR®Green (Thermo Fisher Scientific) under the following cycling: 95°C for 10 min, followed by 40 cycles of 95°C for 15s, and finally 60°C for 45s. Verification of specific product amplification was determined by dissociation curve analysis. Comparative Ct method was used for quantifying the transcripts. Fold-change for target genes was determined by the formula 2−ΔΔCT.
Primers are as follows: NEDD4L, forward 5’- TCTGGAAGGCTGTGCTAC −3’ and reverse 5’- TCTGGGCAGTTTCTCAGG −3’; ENO1, forward 5’- CTCCGGGACAATGATAAGAC −3’ and reverse 5’- TGCACTGCTTCCATCAAC −3’; GAPDH, forward 5’- AATCCCATCACCATCTTC −3’ and reverse 5’- AGGCTGTTGTCATACTTC −3’.
Protein isolation and Western blot
Proteins were isolated with RIPA buffer (JRDUN Biotech., Shanghai, China) that contained protease and phosphatase inhibitors, which then went through 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose membranes (Millipore, Billerica, MA, USA), and blocked by 5% nonfat milk. The membranes were probed overnight at 4°C with antibodies against: NEDD4L (Ab240753, Abcam, Cambridge, MA, USA), p21 (Ab107099, Abcam), p27 (Ab32034, Abcam), ENO1 (Ab227978, Abcam) and GAPDH (#5174, Cell Signaling Technology). After wash, membranes were treated with horseradish peroxidase conjugated secondary antibody for 1 h, then with substrate (ECL; Bio-Rad, Richmond, CA, USA) followed by signal reading.
Cell proliferation assay
Cell proliferation was assessed using Cell Counting Kit-8 assay (CCK-8, CP002, SAB biotech., College Park, MD, USA). Briefly, cell suspension with a density of 3 × 104 per ml was cultured overnight in 96-well plates (100 μl/well) and transducted with NEDD4LOE or a control Vector. CCK-8 solution was then added to each well and incubated at 37°C for 1 h. Optical density (OD) was finally read at 450 nm.
Cell cycle analysis
Ethanol-fixed cells were labeled with propidium iodine (PI, Sigma-Aldrich), and cell cycle was assessed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA). Different phases of the cell cycle were identified based on the different DNA content levels.
Measurement of cellular respiration and glycolytic activity
ECAR and OCR were measured with Seahorse XF96 extracellular flux analyzer (Seahorse Bioscience, North Billerica, MA, USA). Briefly, cells were plated in microplates at a density of 4 × 104 /well one day before measurement. Then, glyco-stress test kit (Seahorse Bioscience) was used for examining ECAR, whereas a mito-stress kit was used for examining OCR.
Lactate production
After 48 h of cell culture, lactate level in the medium was measured with a lactic acid (LD) detection kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), and normalized to values of control group.
Measurement of adenosine triphosphate (ATP) level
After 48 h of culture, the ATP level in OSCC cells was measured with a commercial kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Tumor growth
Animal experiments were performed using female athymic nude mice (Shanghai Laboratory Animal Center, China), which were in accordance with the guidelines for the Care and Use of Laboratory Animals and approved by Ethical Committee of Shengjing Hospital, China Medical University. The mice were randomly divided into two groups (n = 6 per group) and injected subcutaneously with 100 μl of SCC15 cell suspension (density: 5 × 107 per ml) expressing NEDD4L or a control Vector. The xenograft size was monitored every three days after formation, and tumor volume was calculated using the following equation: volume = 1/2 × widthCitation2 × length. Mice were sacrificed on day 33 for xenograft extraction.
Coimmunoprecipitation (co-IP) assays
Cell lysates were incubated with anti-NEDD4L (Ab245522, Abcam), anti-ENO1 (Ab155102, Abcam) or control IgG (Santa Cruz Biotech., Santa Cruz, CA, USA) for 1 h, and then with protein A/G-agarose (Santa Cruz Biotech.) for 3 h at 4°C. Precipitates were washed three times and assessed by Western blot.
Statistical analysis
All statistical analysis was performed using GraphPad Prism (GraphPad Software, San Diego, CA, USA). Fisher’s exact test was used to analyze the correlation of NEDD4L expression with the severity of OSCC in patients. Student’s t and ANOVA test were used to calculate the statistical significance between two groups and among more than two groups, respectively. p <.05 was considered statistically significant.
Supplemental Material
Download MS Word (1.1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Markopoulos AK. Current aspects on oral squamous cell carcinoma. The Open Dentistry Journal. 2012;6:126–130. doi:10.2174/1874210601206010126.
- Campbell CD, Mohajeri K, Malig M, Hormozdiari F, Nelson B, Du G, Patterson KM, Eng C, Torgerson DG, Hu D, et al. Whole-genome sequencing of individuals from a founder population identifies candidate genes for asthma. PloS one. 2014;9:e104396. doi:10.1371/journal.pone.0104396.
- Lee DE, Yoo JE, Kim J, Kim S, Kim S, Lee H, Cheong H. NEDD4L downregulates autophagy and cell growth by modulating ULK1 and a glutamine transporter. Cell Death & Disease. 2020;11:38. doi:10.1038/s41419-020-2242-5.
- Ye X, Wang L, Shang B, Wang Z, Wei W. NEDD4: a promising target for cancer therapy. Current Cancer Drug Targets. 2014;14:549–556. doi:10.2174/1568009614666140725092430.
- Yang Q, Zhao J, Cui M, Gi S, Wang W, Han X. Nedd4L expression is decreased in ovarian epithelial cancer tissues compared to ovarian non-cancer tissue. The Journal of Obstetrics and Gynaecology Research. 2015;41:1959–1964. doi:10.1111/jog.12808.
- He S, Deng J, Li G, Wang B, Cao Y, Tu Y. Down-regulation of Nedd4L is associated with the aggressive progression and worse prognosis of malignant glioma. Japanese Journal of Clinical Oncology. 2012;42:196–201. doi:10.1093/jjco/hyr195.
- Jiang X, Zhang S, Yin Z, Sheng Y, Yan Q, Sun R, Lu M, Zhang Z, Li Y. The correlation between NEDD4L and HIF-1alpha levels as a gastric cancer prognostic marker. International Journal of Medical Sciences. 2019;16:1517–1524. doi:10.7150/ijms.34646.
- Hu XY, Xu YM, Fu Q, Yu JJ, Huang J. Nedd4L expression is downregulated in prostate cancer compared to benign prostatic hyperplasia. European Journal of Surgical Oncology: The Journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2009;35:527–531. doi:10.1016/j.ejso.2008.09.015.
- Gao C, Pang L, Ren C, Ma T. Decreased expression of Nedd4L correlates with poor prognosis in gastric cancer patient. Medical Oncology. 2012;29:1733–1738. doi:10.1007/s12032-011-0061-3.
- Yilmaz E, Gul M, Melekoglu R, Inci Coskun E, Sahin N, Gul S, Bastemur AG, Ciplak B. Neural precursor cell-expressed developmentally down-regulated 4-like: a new biomarker in the pathophysiology of endometrial cancer. The Journal of International Medical Research. 2018;46:3709–3716. doi:10.1177/0300060518777944.
- Zhao F, Gong X, Liu A, Lv X, Hu B, Zhang H. Downregulation of Nedd4L predicts poor prognosis, promotes tumor growth and inhibits MAPK/ERK signal pathway in hepatocellular carcinoma. Biochemical and Biophysical Research Communications. 2018;495:1136–1143. doi:10.1016/j.bbrc.2017.11.139.
- Yang H, Zhu J, Wang G, Liu H, Zhou Y, Qian J. STK35 Is ubiquitinated by NEDD4L and promotes glycolysis and inhibits apoptosis through regulating the AKT signaling pathway, influencing chemoresistance of colorectal cancer. Frontiers in Cell and Developmental Biology. 2020;8:582695. doi:10.3389/fcell.2020.582695.
- Tanksley JP, Chen X, Coffey RJ, Liu C. NEDD4L is downregulated in colorectal cancer and inhibits canonical WNT signaling. PloS one. 2013;8:e81514. doi:10.1371/journal.pone.0081514.
- Novellasdemunt L, Kucharska A, Jamieson C, Prange-Barczynska M, Baulies A, Antas P, van der Vaart J, Gehart H, Maurice MM, Li VS, et al. NEDD4 and NEDD4L regulate Wnt signalling and intestinal stem cell priming by degrading LGR5 receptor. The EMBO Journal. 2020;39:e102771. doi:10.15252/embj.2019102771.
- Chen JM, Chiu SC, Chen KC, Huang YJ, Liao YA, Yu CR. Enolase 1 differentially contributes to cell transformation in lung cancer but not in esophageal cancer. Oncology Letters. 2020;19:3189–3196. doi:10.3892/ol.2020.11427.
- Zuo J, Wang B, Long M, Gao Z, Zhang Z, Wang H, Wang X, Li R, Dong K, Zhang H, et al. The type 1 transmembrane glycoprotein B7-H3 interacts with the glycolytic enzyme ENO 1 to promote malignancy and glycolysis in HeLa cells. FEBS Letters. 2018;592:2476–2488. doi:10.1002/1873-3468.13164.
- Yang T, Shu X, Zhang HW, Sun LX, Yu L, Liu J, Sun L-C, Yang Z-H, Ran Y-L. Enolase 1 regulates stem cell-like properties in gastric cancer cells by stimulating glycolysis. Cell Death & Disease. 2020;11:870. doi:10.1038/s41419-020-03087-4.
- Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Molecular Cancer. 2013;12:152. doi:10.1186/1476-4598-12-152.
- Nowak N, Kulma A, Gutowicz J. Up-regulation of key glycolysis proteins in cancer development. Open Life Sciences. 2018;13:569–581. doi:10.1515/biol-2018-0068.
- Yin H, Wang L, Liu HL. ENO1 overexpression in pancreatic cancer patients and its clinical and diagnostic significance. Gastroenterology Research and Practice. 2018;2018:3842198. doi:10.1155/2018/3842198.
- Song Y, Luo Q, Long H, Hu Z, Que T, Zhang X, Li Z, Wang G, Yi L, Liu Z, et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Molecular Cancer. 2014;13:65. doi:10.1186/1476-4598-13-65.
- Li HJ, Ke FY, Lin CC, Lu MY, Kuo YH, Wang YP, Liang K-H, Lin S-C, Chang Y-H, Chen H-Y, et al. ENO1 promotes lung cancer metastasis via HGFR and WNT signaling-driven epithelial-mesenchymal transition. Cancer Research. 2021;81:4094–4109. doi:10.1158/0008-5472.CAN-20-3543.
- Hsiao KC, Shih NY, Fang HL, Huang TS, Kuo CC, Chu PY, Hung Y-M, Chou S-W, Yang -Y-Y, Chang G-C, et al. Surface alpha-enolase promotes extracellular matrix degradation and tumor metastasis and represents a new therapeutic target. PloS one. 2013;8:e69354. doi:10.1371/journal.pone.0069354.
- Principe M, Borgoni S, Cascione M, Chattaragada MS, Ferri-Borgogno S, Capello M, Bulfamante S, Chapelle J, Di Modugno F, Defilippi P, et al. Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. Journal of Hematology & Oncology. 2017;10:16. doi:10.1186/s13045-016-0385-8.
- Liu J, Yang Q, Sun H, Wang X, Saiyin H, Zhang H. The circ-AMOTL1/ENO1 axis implicated in the tumorigenesis of OLP-associated oral squamous cell carcinoma. Cancer Management and Research. 2020;12:7219–7230. doi:10.2147/CMAR.S251348.
- Wu F, Shi X, Zhang R, Tian Y, Wang X, Wei C, Li D, Li X, Kong X, Liu Y, et al. Regulation of proliferation and cell cycle by protein regulator of cytokinesis 1 in oral squamous cell carcinoma. Cell Death & Disease. 2018;9:564. doi:10.1038/s41419-018-0618-6.
- Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Reviews in Molecular Medicine. 2008;10:e19. doi:10.1017/S1462399408000744.
- Shamloo B, Usluer S. p21 in cancer research. Cancers. 2019;11:1178.
- Yang Q, Al-Hendy A. The emerging role of p27 in development of diseases. Cancer Studies and Molecular Medicine: Open Journal. 2018;4:e1–e3. doi:10.17140/CSMMOJ-4-e006.
- Yu L, Chen X, Sun X, Wang L, Chen S. The glycolytic switch in tumors: how many players are involved? Journal of Cancer. 2017;8:3430–3440. doi:10.7150/jca.21125.
- Wang Z, Dang T, Liu T, Chen S, Li L, Huang S, Fang M. NEDD4L protein catalyzes ubiquitination of PIK3CA protein and regulates PI3K-AKT signaling. The Journal of Biological Chemistry. 2016;291:17467–17477. doi:10.1074/jbc.M116.726083.
- Mookerjee SA, Brand MD. Measurement and analysis of extracellular acid production to determine glycolytic rate. Journal of Visualized Experiments: JoVE. 2015:e53464. 10.3791/53464.
- Pelletier M, Billingham LK, Ramaswamy M, Siegel RM. Extracellular flux analysis to monitor glycolytic rates and mitochondrial oxygen consumption. Methods in Enzymology. 2014;542:125–149.
- Sun X, Kaufman PD. Ki-67: more than a proliferation marker. Chromosoma. 2018;127:175–186. doi:10.1007/s00412-018-0659-8.
- Didiasova M, Schaefer L, Wygrecka M. When place matters: shuttling of enolase-1 across cellular compartments. Frontiers in Cell and Developmental Biology. 2019;7:61. doi:10.3389/fcell.2019.00061.
- Nandi D, Tahiliani P, Kumar A, Chandu D. The ubiquitin-proteasome system. Journal of Biosciences. 2006;31:137–155. doi:10.1007/BF02705243.
- Goel P, Manning JA, Kumar S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene. 2015;557:1–10. doi:10.1016/j.gene.2014.11.051.
- Liu L, Chen J, Cai X, Yao Z, Huang J. Progress in targeted therapeutic drugs for oral squamous cell carcinoma. Surgical Oncology. 2019;31:90–97. doi:10.1016/j.suronc.2019.09.001.
- Li M, Gao F, Zhao Q, Zuo H, Liu W, Li W. Tanshinone IIA inhibits oral squamous cell carcinoma via reducing Akt-c-Myc signaling-mediated aerobic glycolysis. Cell Death & Disease. 2020;11:381. doi:10.1038/s41419-020-2579-9.
- Wei J, Wu J, Xu W, Nie H, Zhou R, Wang R, Liu Y, Tang G, Wu J. Salvianolic acid B inhibits glycolysis in oral squamous cell carcinoma via targeting PI3K/AKT/HIF-1alpha signaling pathway. Cell Death & Disease. 2018;9:599. doi:10.1038/s41419-018-0623-9.
- Han L, Cheng J, Li A. hsa_circ_0072387 suppresses proliferation, metastasis, and glycolysis of oral squamous cell carcinoma cells by downregulating miR-503-5p. Cancer Biotherapy & Radiopharmaceuticals. 2021;36:84–94. doi:10.1089/cbr.2019.3371.
- Zheng M, Cao MX, Yu XH, Li L, Wang K, Wang SS, Wang H-F, Tang Y-J, Tang Y-L, Liang X-H, et al. STAT3 promotes invasion and aerobic glycolysis of human oral squamous cell carcinoma via inhibiting FoxO1. Frontiers in Oncology. 2019;9:1175. doi:10.3389/fonc.2019.01175.
- Salah Z, Cohen S, Itzhaki E, Aqeilan RI. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle. 2013;12:3817–3823. doi:10.4161/cc.26672.
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Current Biology: CB. 2004;14:2228–2236. doi:10.1016/j.cub.2004.12.028.
- Guo X-Y, Liu -T-T, Zhu W-J, Liu H-T, Zhang G-H, Song L. CircKDM4B suppresses breast cancer progression via the miR-675/NEDD4L axis. Oncogene 2022:1–12.
- Guarnieri A, Towers C, Drasin D, Oliphant M, Andrysik Z, Hotz T, Vartuli RL, Linklater ES, Pandey A, Khanal S, et al. The miR-106b-25 cluster mediates breast tumor initiation through activation of NOTCH1 via direct repression of NEDD4L. Oncogene. 2018;37:3879–3893. doi:10.1038/s41388-018-0239-7.
- Klement RJ, Kammerer U. Is there a role for carbohydrate restriction in the treatment and prevention of cancer? Nutrition & Metabolism. 2011;8:75. doi:10.1186/1743-7075-8-75.
- Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H, Colvin R, Ding J, Tong L, Wu S, et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Molecular Cancer Therapeutics. 2012;11:1672–1682. doi:10.1158/1535-7163.MCT-12-0131.
- Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, Xian J, Cantley LC. Identification of small molecule inhibitors of pyruvate kinase M2. Biochemical Pharmacology. 2010;79:1118–1124. doi:10.1016/j.bcp.2009.12.003.
- Price GS, Page RL, Riviere JE, Cline JM, Thrall DE. Pharmacokinetics and toxicity of oral and intravenous lonidamine in dogs. Cancer Chemotherapy and Pharmacology. 1996;38:129–135. doi:10.1007/s002800050460.
- Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, Wang M, Chen D, Sun Y, Hung M-C, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15:1486–1494. doi:10.1038/ncb2874.
