ABSTRACT
LncRNAs can be transported to tumor cells where they exert regulatory effects by bone marrow mesenchymal stem cells (BMSC)-derived exosomes. Here, we aimed to investigate the functional mechanism of BMSC-derived exosomal lncRNA PTENP1 in the progression of bladder cancer (BC). Methods of BMSC were identified by detecting surface markers through flow cytometry. Exosomes from BMSC were identified by transmission electron microscopy, nanoparticle tracking analysis (NTA), and western blot analysis of exosome markers. Cellular internalization of BMSC-derived exosomes (BMSC-Exo) into BC cells was detected by confocal microscopy. CCK-8, colony formation, flow cytometry, wound healing, and transwell assays were adopted to estimate cell proliferation, apoptosis, migration, and invasion abilities, respectively. Interplay between miR-17 and lncRNA PTENP1 or SCARA5 was verified by dual-luciferase reporter, RNA pull down, and/or RNA immunoprecipitation (RIP) assays. Tumor xenograft assay was conducted in nude mice to study the role of exosomal lncRNA PTENP1 in BC progression in vivo. We showed exosomal lncRNA PTENP1 can be delivered into and suppress the malignant phenotypes of BC cells. LncRNA PTENP1 was identified as a sponge of miR-17, and SCARA5 was identified as a target gene of miR-17. The exosomes derived from PTENP1-overexpressing BMSC (BMSCOE-PTENP1-Exo) abolished the promotive effects of miR-17 overexpression or SCARA5 knockdown on the malignant phenotypes of BC cells. Moreover, exosomal lncRNA PTENP1 was demonstrated to inhibit BC tumor growth in nude mice by miR-17/SCARA5 axis. In conclusion, BMSC-derived exosomal PTENP1 suppressed the BC progression by upregulating the expression of SCARA5 via sponging miR-17, offering a potential novel therapeutic target for BC therapy.
Introduction
Bone mesenchymal stem cells (BMSC) are a group of multipotent cells with the abilities of self-renewal and differentiation into osteoblasts, chondrocytes, and adipocytes.Citation1 Evidence has shown that BMSC can secrete various bioactive agents that significantly affect the critical cellular functions of neighboring cells, such as apoptosis, proliferation, and maturation.Citation2 This secretome-based property of BMSC is widely believed to be primarily mediated by the smallest subset of extracellular vesicles, exosomes.Citation2,Citation3 Exosomes are bilayer lipid vesicles containing various cellular agents, such as nucleic acids, proteins, and RNAs, that are actively released by mammalian cells with a diameter ranging from 50 to 150 nm.Citation4 Accumulating evidence has shown that BMSC-derived exosomes can regulate tumor cell proliferation, angiogenesis, apoptosis, and metastasis by transferring signaling molecules and controlling cellular pathways.Citation2,Citation5,Citation6 Bladder cancer (BC) is a common tumor in the urinary system with 80470 new cases diagnosed annually.Citation7 The regulatory effects of BMSC-derived exosomes on the initiation, development, and metastasis of BC have been recently investigated by several studies; however, the underlying mechanisms remain largely unclear.Citation6,Citation8
Multiple species of non-coding RNAs (ncRNAs), such as long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs), were found to be enriched in BMSC-derived exosomes.Citation9 These functional ncRNAs transported by exosomes to tumor cells can modulate the expression of target genes to participate in the growth, apoptosis, and metastasis of tumor cells.Citation2,Citation10 Among these species of ncRNAs, the lncRNAs in exosomes have been extensively investigated. LncRNAs have been revealed to be dysregulated in various human cancers and play as oncogenes or suppressors through a diversity of mechanisms.Citation11 LncRNA PTENP1, the pseudogene of PTEN, was revealed to be an inhibitor in numerous malignancies.Citation12,Citation13 Nevertheless, only a few studies have focused on the role of lncRNA PTENP1 in the progression of BC,Citation14 the underlying molecular mechanisms of lncRNA PTENP1 in BC and whether BMSC-derived exosomes can carry and transport lncRNA PTENP1 to BC cells remain unclear.
Notably, the critical function of lncRNAs rested on regulating biological events of tumor cells by controlling gene expression or competitively sponging miRNAs, rather than encoding proteins.Citation15 As an important subset of ncRNAs, miRNAs are a group of transcripts with approximately 22 nucleotides.Citation16 It is now widely recognized that miRNAs participate in multiple biological events of cancer cells, thus becoming one of the most promising diagnostic and therapeutic targets for tumors.Citation17 MiR-17 has been revealed to be overexpressed in BC.Citation18 In hepatocellular carcinoma, increased PTENP1 was proved to inhibit the oncogenic PI3K/AKT pathway and promote pro-death autophagy by sequestering miR-17.Citation19 Nevertheless, whether PTENP1/miR-17 axis has a role in BC progression remains unknown. Moreover, based on the prediction results of bioinformatics, there are binding sites between miR-17 and lncRNA PTENP1 or scavenger receptor class A member 5 (SCARA5), which was proved to be a key tumor suppressor in various cancers.Citation20,Citation21 It was also regarded as a promising biomarker for various human cancers, including colorectal cancer, oral squamous cell carcinoma, and breast cancer.Citation22–24 However, whether SCARA5 plays a role in BC tumorigenesis remains undetermined.
Here, we aimed to investigate whether lncRNA PTENP1 could be transported by BMSC-derived exosomes to BC cells, and whether the impacts of lncRNA PTENP1 on BC cell malignant phenotypes are mediated by miR-17/SCARA5 axis. Our results will certainly enrich the understanding of the role of BMSC-derived exosomes in BC pathogenesis and provide new ideas for BC treatment.
Results
Identification of BMSC and BMSC-derived exosomes
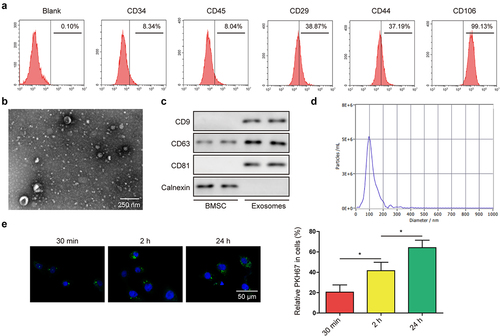
Previous studies suggested that CD29, CD44, and CD106 could be used as positive biomarkers, and CD34 and CD45 could be used as negative biomarkers for BMSC.Citation25 As expected, we found positive expression of CD29, CD44, and CD106, as well as negative expression of CD34 and CD45 in collected BMSC (). Next, exosomes were isolated from the medium of BMSC and observed under a transmission electron microscope (), results indicated that most exosomes had a diameter of around 100 nm. For further verification of BMSC-Exo, the expression of CD9, CD63, and CD81 (exosomal markers) were tested, which indicated a higher expression of these markers in exosomes from BMSC; moreover, calnexin was not expressed in BMSC-Exo (). In addition, nanoparticle tracking analysis (NTA) was performed to detect the size of the particles. Results indicated that the main peak of the particle size was among the typical size arrangement of exosomes (). To examine whether BMSC-derived exosomes can be transported into bladder cancer cell line 5637, PKH67-labeled BMSC-Exo were co-cultured with 5637 cells for 30 min, 2 h, and 24 h, respectively. Using confocal microscopy, we can observe a gradual increase in green fluorescence of PKH67-labeled exosomes in 5637 cells (), suggesting that BMSC-derived exosomes can be taken up by 5637 cells.
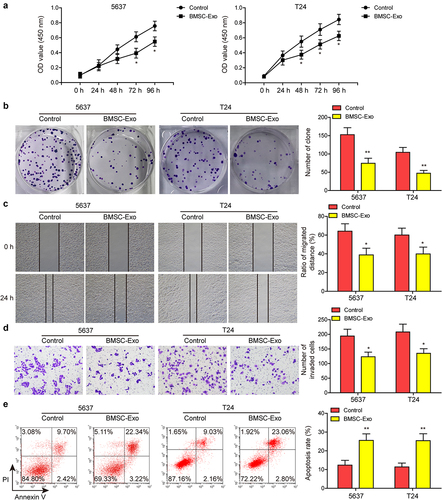
Figure 1. Identification of BMSC and BMSC-derived exosomes.(a and b) proliferative ability of BC cells after incubating with BMSC-derived exosomes (BMSC-Exo) or not was evaluated by CCK-8 and colony formation assays. (c and d) impacts of BMSC-Exo incubation on BC cell migration and invasion were estimated by wound healing and transwell assays, respectively. (e) flow cytometry analysis was carried out to assess the apoptosis of BC cells treated with BMSC-Exo. *P < .05, **P < .01.
BMSC-derived exosomes suppressed the malignant phenotypes of bladder cancer cells
To test the impacts of BMSC-Exo on the malignant phenotypes of BC cells in vitro, 5637 and another bladder cancer cell-line T24 were cultured with BMSC-Exo. As shown in ) BMSC-Exo significantly repressed the proliferative ability of bladder cancer cells. Impacts of BMSC-Exo administration on BC cell migration and invasion were also evaluated. Compared to the control group, administering BMSC-Exo dramatically suppressed the capacities of migration and invasion of these two bladder cancer cells ). Moreover, BC cell apoptosis was measured after the administration of BMSC-Exo, which indicated that BMSC-Exo treatment markedly increased the BC cell apoptosis rate (). These findings implied that BMSC-Exo inhibited BC cell proliferation, migration, and invasion, and promoted cell apoptosis.
Figure 2. BMSC-derived exosomes suppressed the malignant phenotypes of BC cells.(a) surface markers of BMSC including CD34, CD45, CD29, CD44, and CD106, were detected through flow cytometry. (b) morphology of BMSC-derived exosomes was detected by transmission electron microscopy. (c) positive markers (CD9, CD63, and CD81) and negative marker (calnexin) of exosomes were tested by western blot. (d) NTA was performed to detect the size of the particles. (e) cellular internalization of BMSC-derived exosomes into 5637 cells was detected by confocal microscopy. Scale bar: 50 μm.
BMSC-derived exosomal PTENP1 regulated the malignant phenotypes of 5637 cells
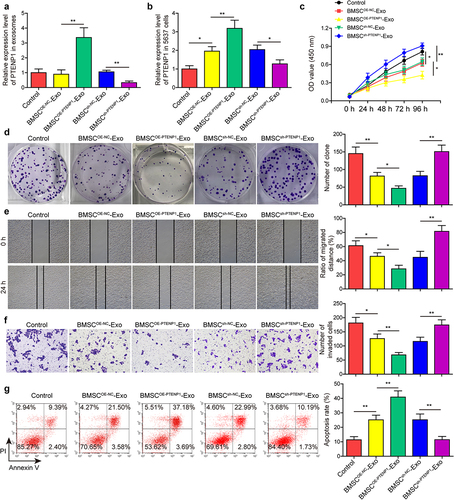
Based on the previous conclusion that lncRNAs are enriched in exosomes, and lncRNA PTENP1 was reported to be a tumor suppressor of BC, we speculate that the repressive effects of BMSC-Exo on BC are achieved by transporting lncRNA PTENP1 into BC cells. To address this, BMSC were transfected with PTENP1 overexpression or knockdown vectors, and the expression of lncRNA PTENP1 in BMSC-Exo was measured after 48 h. Results indicated that lncRNA PTENP1 was sharply increased in the exosomes from PTENP1-overexpressing BMSC, while it was decreased in the exosomes from PTENP1-silencing BMSC (). Next, the exosomes derived from PTENP1-overexpressing or PTENP1-silencing BMSC (BMSCOE-PTENPCitation1-Exo or BMSCsh-PTENPCitation1 -Exo) were co-cultured with 5637 cells, lncRNA PTENP1 level in 5637 cells was measured, which indicated an upregulation of lncRNA PTENP1 in BMSCOE-NC-Exo or BMSCsh-NC-Exo treatment groups, and a further significant upregulation of lncRNA PTENP1 in BMSCOE-PTENPCitation1-Exo-treated 5637 cells, while a significant downregulation of lncRNA PTENP1 in BMSCsh-PTENPCitation1-Exo-treated 5637 cells (). Functional experiments showed that BMSCOE-NC-Exo or BMSCsh-NC-Exo treatment significantly repressed 5637 cell proliferation ), migration (), and invasion (), while promoted cell apoptosis (), these effects were found to be further enhanced in BMSCOE-PTENPCitation1-Exo-treated 5637 cells, but these effects were abolished by BMSCsh-PTENPCitation1-Exo treatment ). These findings indicated that exosomal lncRNA PTENP1 from BMSC transported into and repressed the malignant behaviors of BC cells.
Figure 3. BMSC-derived exosomal PTENP1 regulated the malignant phenotypes of 5637 cells.(a) BMSC-derived exosomes were isolated and the expression of lncRNA PTENP1 in exosomes was examined by qRT-PCR. (b) after 48 h of co-culture with the exosomes derived from NC- or PTENP1-overexpressing BMSC (BMSCOE-NC-Exo, BMSCOE-PTENPCitation1-Exo), 5637 cells were collected and subjected for qRT-PCR analysis of lncRNA PTENP1. (c and d) proliferative ability of 5637 cells treated with BMSCOE-NC-Exo or BMSCOE-PTENPCitation1-Exo was evaluated by CCK-8 and colony formation assays. Impacts of treatment with BMSCOE-NC-Exo or BMSCOE-PTENPCitation1-Exo on 5637 (e) cell migration, (f) invasion, and (g) apoptosis were measured by wound healing, transwell, and flow cytometry assays. *P < .05, **P < .01.
LncRNA PTENP1 promoted the expression of SCARA5 by sponging miR-17
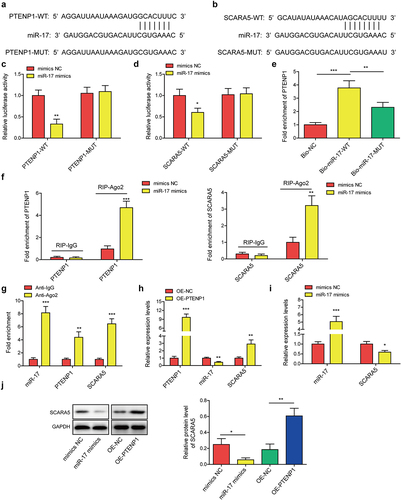
Since lncRNAs have been reported to serve as competing endogenous RNAs (ceRNAs) by sponging miRNAs, we then predicted potential binding sites of PTENP1 on miR-17 using RNA Interactome Database (). Moreover, we found miR-17 binding sites in SCARA5 by employing Starbase Database (). These conclusions implied that PTENP1/miR-17/SCARA5 axis may work as a functional pathway for BC progression. Interplay between miR-17 and PTENP1 or SCARA5 was determined using dual-luciferase reporter experiment. Luciferase activities driven by PTENP1-WT or SCARA5-WT were significantly suppressed by miR-17 mimics, while the activities driven by PTENP1-MUT or SCARA5-MUT were not affected by miR-17 mimics or mimics NC ). Moreover, PTENP1 was found to be pulled down by Bio-miR-17-WT probe rather than Bio-NC and Bio-miR-17-MUT probes (). RIP assay was carried out to examine whether PTENP1, miR-17, and SCARA5 could form RNA-induced silencing complex. Results indicated that treatment with miR-17 mimic increased the enrichment of PTENP1 and SCARA5 in Ago2 immunoprecipitation complex compared to mimics NC group (). Moreover, miR-17, PTENP1 and SCARA5 were all found to be enriched in Ago2 group compared to IgG group (), indicating that PTENP1, miR-17, and SCARA5 could form RNA-induced silencing complex. Additionally, we found that transfection of PTENP1 overexpression vector elevated the expression levels of PTENP1 and SCARA5 mRNA, while reduced the level of miR-17 (). MiR-17 mimics transfection elevated miR-17 level, while it reduced SCARA5 mRNA level (). Western blot analysis indicated that SCARA5 protein was decreased or increased in miR-17 mimics or OE-PTENP1 group (). These findings suggested that lncRNA PTENP1 could indirectly regulate the expression of SCARA5 by sponging miR-17.
Figure 4. LncRNA PTENP1 promoted the expression of SCARA5 by sponging miR-17.(a and b) the binding sites of miR-17 with PTENP1 or SCARA5. (c and d) interplay between miR-17 and PTENP1 or SCARA5 was validated by dual-luciferase reporter assay. (e-g) RNA pull down and RIP assays were performed to examine the interaction between miR-17 and PTENP1 or SCARA5 in 5637 cells. (h and i) after transfecting with OE-PTENP1 or miR-17 mimics, as well as their negative controls, the expression levels of lncRNA PTENP1, miR-17, and SCARA5 in 5637 cells were tested by qRT-PCR. (j) western blot analysis of SCARA5 in 5637 cells treated with OE-PTENP1 or miR-17 mimics, as well as their negative controls. *P < .05, **P < .01, and ***P < .001.
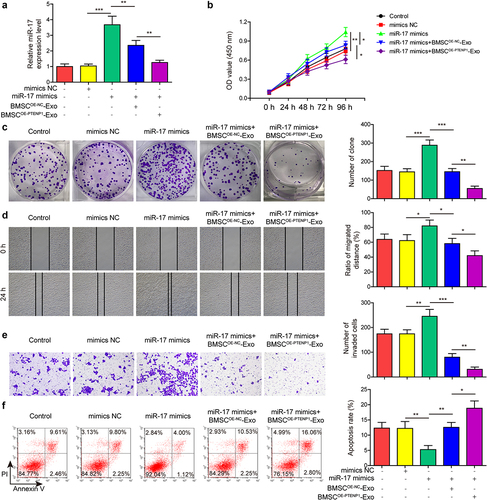
Exosomes derived from PTENP1-overexpressing BMSC abolished the promotive effects of miR-17 overexpression on the malignant phenotypes of 5637 cells
Next, rescue experiments were conducted to determine the influence of BMSCOE-PTENPCitation1-Exo on miR-17-mediated promotive impacts on 5637 cells. Compared with mimics NC group, miR-17 level was significantly elevated in miR-17 mimics-transfected 5637 cells (). The upregulation of miR-17 was partly reversed by the treatment of BMSCOE-NC-Exo, and further abolished by the treatment of BMSCOE-PTENPCitation1-Exo (). Results from functional experiments indicated that miR-17 overexpression promoted 5637 cell proliferation ) migration (), and invasion (), and reduced cell apoptosis (). Moreover, the promotive effects of miR-17 overexpression on 5637 cell malignant phenotypes were partly reversed by the treatment of BMSCOE-NC-Exo, and totally abolished by the treatment of BMSCOE-PTENPCitation1-Exo ). These results indicated that exosomal lncRNA PTENP1/miR-17 axis plays a role in regulating the malignant phenotypes of BC cells.
Figure 5. Exosomes derived from PTENP1-overexpressing BMSC abolished the promotive effects of miR-17 overexpression on the malignant phenotypes of 5637 cells.(a) relative expression level of miR-17 in 5637 cells treated with miR-17 mimics and BMSCOE-PTENPCitation1-Exo was measured by qRT-PCR. (b and c) proliferative ability of 5637 cells treated with miR-17 mimics and BMSCOE-PTENPCitation1-Exo was evaluated by CCK-8 and colony formation assays. impacts of treatment with miR-17 mimics and BMSCOE-PTENPCitation1-Exo on 5637 (d) cell migration, (e) invasion, and (f) apoptosis were measured by wound healing, transwell, and flow cytometry assays. *P < .05, **P < .01, and ***P < .001.
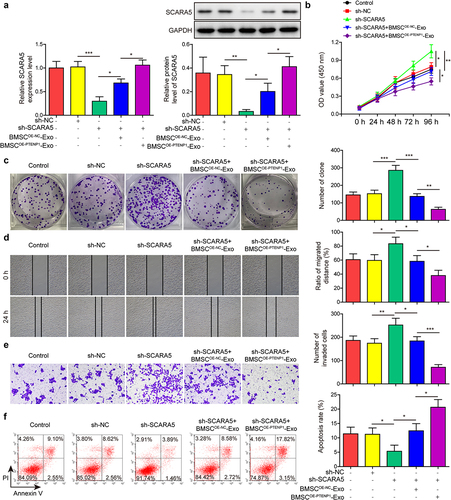
Exosomes derived from PTENP1-overexpressing BMSC abolished the promotive effects of SCARA5 knockdown on the malignant phenotypes of 5637 cells
Similarly, rescue assays were performed on 5637 cells to test the roles of SCARA5 in exosomal PTENP1-mediated repressive impacts on BC cells. SCARA5 mRNA and protein levels were significantly decreased in sh-SCARA5-transfected 5637 cells compared with control group (). The sh-SCARA5-induced downregulation of SCARA5 was partly reversed by the treatment of BMSCOE-PTENPCitation1-Exo and totally abolished by the treatment of BMSCOE-PTENPCitation1-Exo (). Results from functional experiments indicated that SCARA5 knockdown promoted 5637 cell proliferation ) migration (), and invasion (), and reduced cell apoptosis (). Moreover, the promotive effects of SCARA5 knockdown on 5637 cell malignant phenotypes were partly reversed by the treatment of BMSCOE-NC-Exo and totally abolished by the treatment of BMSCOE-PTENPCitation1-Exo ). These results indicated that SCARA5 played as a downstream regulator of exosomal lncRNA PTENP1 in BC cells.
Figure 6. Exosomes derived from PTENP1-overexpressing BMSC abolished the promotive effects of SCARA5 knockdown on the malignant phenotypes of 5637 cells.(a) relative mRNA and protein expression of SCARA5 in 5637 cells treated with sh-SCARA5 and BMSCOE-PTENPCitation1-Exo were measured by qRT-PCR and western blot, respectively. After treatment with sh-SCARA5 and BMSCOE-PTENPCitation1-Exo, 5637 cells were subjected for the analysis of (b and c) proliferation, (d) migration, (e) invasion, and (f) apoptosis. *P < .05, **P < .01, and ***P < .001.
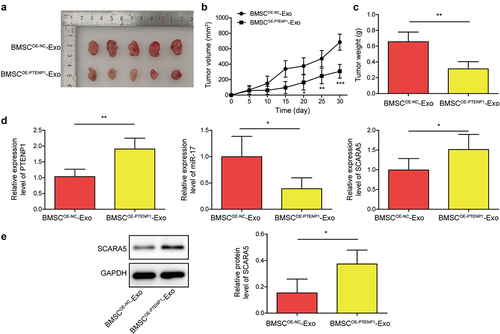
Exosomes derived from PTENP1-overexpressing BMSC repressed BC tumor growth in vivo
Xenograft tumor growth assay was carried out to evaluate the impacts of exosomal lncRNA PTENP1 on tumor growth in nude mice. Treatment with BMSCOE-PTENPCitation1-Exo significantly repressed the volume and weight of tumors compared to BMSCOE-NC-Exo group ). In addition, we found that PTENP1 and SCARA5 mRNA were sharply increased, while miR-17 was decreased in tumors treated with BMSCOE-PTENPCitation1-Exo compared to those treated with BMSCOE-NC-Exo (). Western blot analysis confirmed the upregulation of SCARA5 protein in BMSCOE-PTENPCitation1-Exo group (). These results showed that exosomes derived from PTENP1-overexpressing BMSC repressed BC tumor growth in nude mice.
Figure 7. Exosomes derived from PTENP1-overexpressing BMSC repressed BC tumor growth in vivo.(a) representative images of tumors formed by the 5637 cells treated with BMSCOE-NC-Exo or BMSCOE-PTENPCitation1-Exo. (b and c) tumor volume and weight in each group. (d) relative expression levels of lncRNA PTENP1, miR-17, and SCARA5 mRNA in the tumors formed by the 5637 cells treated with BMSCOE-NC-Exo or BMSCOE-PTENPCitation1-Exo were examined by qRT-PCR. (e) western blot analysis of SCARA5 protein in the tumors formed by the 5637 cells treated with BMSCOE-NC-Exo or BMSCOE-PTENPCitation1-Exo. *P < .05, **P < .01, and ***P < .001.
Discussion
Accumulating studies have suggested that exosomes containing lncRNAs can be captured by tumor cells and trigger a series of cellular response, which may also affect the dysregulation of lncRNA expression profile in tumor cells.Citation26 With the tremendous advancement of sequencing technologies, lncRNA dysregulation has been evaluated more accurately during each step of the malignant process.Citation15 The abundance and specific stable expression of exosomal lncRNAs in blood and tissues make them ideal biomarkers for early diagnosis and metastasis prediction in cancer.Citation27 For instance, plasma exosomal lncRNA PTENP1 and serum exosomal lncRNA H19 were demonstrated to be promising noninvasive markers for BC diagnosis.Citation28,Citation29 Moreover, exosomal lncRNA PTENP1 was revealed to be transported to BC cells and repressed its malignant behaviors by promoting cell apoptosis.Citation28 Here, we demonstrated that BMSC-derived exosomes can transport lncRNA PTENP1 into BC cells. Exosomal lncRNA PTENP1 indirectly increased the expression of SCARA5 by sponging miR-17, and thus alleviating the malignant phenotypes of BC cells.
Tumor growth and metastasis require the formation of a favorable microenvironment in which mesenchymal stem cells (MSCs) act as important participants. MSCs interact with the surrounding cells and exchange the information that is orchestrated by the cancer cells,Citation30 thus regulating the progression of tumors. The regulatory impacts of MSCs on tumor cells are believed to be dependent on paracrine mechanisms and exosomes have been implicated as one of the most common mediators of MSC actions.Citation31–33 For instance, lncRNA PVT1 encapsulated in MSC-derived exosomes was found to facilitate osteosarcoma growth and metastasis by miR-183-5p/ERG pathway.Citation10 Other research groups indicated that MSC-derived exosomes regulated survival of tumor cells by involving lncRNA RUNX2-AS1 and lncRNA Sox2ot.Citation34,Citation35 For BC, exosomal lncRNA LNMAT2 was demonstrated to facilitate lymphatic metastasis by transcriptionally increasing the expression of PROX1.Citation36 Moreover, lncRNA PTENP1 was found to be mainly wrapped by exosomes, and normal cells secreted exosomal PTENP1 and transported it to BC cells, thus repressing the malignant phenotypes of BC cells.Citation28 Here, we examined the shape and size of BMSC-derived exosomes using transmission electron microscopy and NTA, and the exosomal markers were also detected by Western blot. Consistent with other studies, fluorescence microscopy suggested that BMSC-derived exosomes labeled with PKH67 could be transported into BC cells. The malignant phenotypes, including proliferation, migration, and invasion of BC cells exposed to BMSC-derived exosomes, were significantly repressed. Moreover, we found that exosomes derived from PTENP1-overexpressing BMSC further inhibited the malignant phenotypes of BC cells in vitro. These findings suggested that BMSC-derived exosomes transport PTENP1 to BC cells, thus inhibiting malignant phenotypes of BC cells. The repressive effects of exosomal lncRNA PTENP1 on BC were also verified in in vivo assays. To the best of our knowledge, we are the first group to report this finding.
Evidence has shown that lncRNA PTENP1 could competitively bind miRNAs to regulate the expression of PTEN in various tumor cells.Citation37–39 Therefore, we speculate that lncRNA PTENP1 may serve as a competing endogenous RNA (ceRNA), participating in the regulation of BC cell malignant phenotypes. Consistently, we found that BMSC-derived lncRNA PTENP1 can regulate BC tumorigenesis by sponging miR-17. SCARA5 was found to be modulated by MEG3/miR-141 axis to suppress BC cell growth.Citation40 Here, we firstly reported that SCARA5 can be targeted by miR-17 in BC cells. As a mechanism, we revealed that lncRNA PTENP1 indirectly increased the expression of SCARA5 by sponging miR-17. Additionally, we showed that miR-17 overexpression or SCARA5 knockdown abolished the repressive effects of exosomal lncRNA PTENP1 on BC cell malignant phenotypes. These findings suggested exosomal lncRNA PTENP1 inhibited BC cell malignant phenotypes by sponging miR-17 to increase the expression of SCARA5.
Taken together, our results indicated that exosomal lncRNA PTENP1 secreted by BMSC suppresses BC cell malignant phenotypes by regulating miR-17/SCARA5 axis. Our findings not only enriched the understanding of exosomal lncRNA PTENP1 in the tumorigenesis of BC but also provided a novel therapeutic target for BC treatment.
Materials and methods
Cell lines and BMSC identification
Human BMSC and BC cell lines 5637 and T24 were all offered by the American Type Culture Collection (ATCC, USA). Both BC cells and BMSC were cultured in DMEM (Hyclone, USA), supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and penicillin/streptomycin (100 U/mL, Gibco, USA). To identify BMSC, the BMSC were collected and re-suspended on PBS, adjusting the cell density to 2 × 105 cells/mL. Then, 10 μL CD29-PE, CD44-PE, CD106-FITC, CD34-FITC, or CD45-FITC antibodies (Invitrogen, USA) were added into 200 μL cell suspension and allowed for 30 min incubation in dark. Cell suspension was then centrifuged for 5 min at 1000 g. The sediment was re-suspended in PBS, and the expression levels of these biomarkers were determined by flow cytometry (FACSCalibur, BD Biosciences, USA).
BMSC-derived exosomes (BMSC-Exo) isolation and identification
An exosome isolation reagent (Thermo Fisher Scientific, USA) was employed to separate exosomes from the culture medium of BMSC. In brief, the medium of BMSC cultured in DMEM containing exosome depleted FBS was collected and centrifuged for half an hour at 2000 g to remove cells and debris, then transferred the supernatant (1 mL) into new Eppendorf tubes. We added 0.5 mL of the Total Exosome Isolation reagent into the supernatant and mixed them well by vortexing. After incubation at 4°C overnight, the samples were centrifuged at 10,000 g for 1 h. Aspirated and discarded the supernatant, then re-suspend the pellet in 100 μL PBS. The isolated BMSC-Exo were observed under transmission electron microscopy (1200EX, Jeol, Japan), and identified by nanoparticle tracking analysis (NTA) and detecting the exosomal markers (CD9, CD63, CD81, and calnexin) using western blot.
Nanoparticle tracking analysis (NTA)
The size of isolated exosomes was measured by a ZETASIZER Nano series-Nano ZS (Malvern Panalytical, Malvern, United Kingdom). In brief, 1 mL PBS was used to resuspend the exosomes, and then exosomes were injected into a clean sample pool. Next, the sample tool was placed into the instrument to detect the size of the exosomes.
Fluorescent labeling and transfer of exosomes
Extracted BMSC-Exo were incubated with the lipophilic dye PKH67 (10 μM, Sigma-Aldrich, USA) for 30 min at 37°C. After washed with PBS and removed the residual PKH67 by gel filtration chromatography, the 20 μg/mL PKH67-labeled exosomes were incubated with 5637 cells for 24 h. After washed with PBS, the 5637 cells were stained with DAPI (Sigma-Aldrich, USA) to label the cell nucleus. Finally, the internalization of BMSC-Exo into 5637 cells was observed under a confocal microscope (Olympus, Japan).
Cell transfection
LncRNA PTENP1 overexpression (OE-PTENP1) and knockdown (sh-PTENP1) plasmids, miR-17 mimics, sh-SCARA5, as well as their negative controls, were all purchased from GenePharma (Shanghai, China). Transfection was conducted using Lipofectamine 3000 (Invitrogen, USA).
Evaluation of cell proliferation
For CCK-8 experiment, treated cells were collected and cultured in 96-well plates. After cultured for the indicated time, CCK-8 solution (10 μL) was added and allowed for 2-h incubation. The OD values of each well were measured in a microplate reader (450 nm, Bio-Rad, USA). For colony formation, 1 × 103 cells were seeded into a petri dish (diameter: 35 mm) and incubated at 37°C for 2 weeks. The cultural medium changed every 2 days. Next, colonies were fixed in 4% paraformaldehyde and stained by 0.5% crystal violet. Colony number was counted under a microscope (Olympus, Japan).
Evaluation of cell migration and invasion
For cell migration analysis, treated cells at exponential growth phase were collected and seeded into 6-well plates. After cells reached to 100% confluence, a straight line was made in the middle of the cell layer by a pipette tip, cells were cultured for another 24 h. Images were made 0 and 24 h of making the scratch, and the width of the scratch was measured under a microscope (Olympus, Japan). For cell invasion evaluation, 8 μm pore size transwell chambers coated with Matrigel were used. Culture medium (500 μL) containing 10% FBS was added into the lower chamber, 1 × 106 cells cultured in 200 μL serum-free medium were placed into the upper chamber. Cells that remained in the apical chamber were removed after 24 h, and those invaded into the undersurface were fixed with paraformaldehyde and stained with crystal violet.
Evaluation of cell apoptosis
Treated cells were collected and re-suspended in PBS, adjusting cell density to 1 × 106 cells/mL. Cells were fixed by cold 70% ethanol solution for 12 h at 4°C, and then 100 μL cell suspension was re-suspended in 200 μL binding buffer containing 10 μL Annexin V-FITC and 5 μL propidium iodide (PI). After 15 min of incubation in the dark, cell apoptosis was analyzed by flow cytometry (FACSCalibur, BD Biosciences, USA).
Dual-luciferase reporter assay
The recombinant luciferase reporter plasmids: wild type of lncRNA PTENP1 (PTENP1-WT), mutant lncRNA PTENP1 (PTENP1-MUT), wild type of SCARA5 (SCARA5-WT), and mutant SCARA5 (SCARA5-MUT) were designed and provided by Genepharma (Shanghai, China). For the validation of the interplay between lncRNA PTENP1 and miR-17, 5637 cells were co-transfected with miR-17 mimics or mimics NC (Genepharma, China) and PTENP1-WT or PTENP1-MUT, and allowed for incubation at 37°C for 48 h. Next, luciferase activity of cells was determined by Dual-Luciferase Reporter Assay System (Promega, USA). Interplay between miR-17 and SCARA5 was also verified in the same way as above.
RNA pull down assay
Wild-type and mutant biotinylated miR-17 probes (Bio-miR-17-WT and Bio-miR-17-MUT, Genepharma, China) were used to pull down lncRNA PTENP1 according to the MagCapture RNA Pull Down Kit (Lumiprobe, USA). In total, 5637 cells were lysed in a cell lysis buffer from this kit, and cell lysates were incubated with Bio-miR-17-WT or Bio-miR-17-MUT probes for 2 h. Next, cell lysates were incubated with pre-coated M-280 streptavidin beads for another 4 h. After washed with washing buffer for three times, the enrichment of lncRNA PTENP1 was examined using qRT-PCR.
RNA immunoprecipitation (RIP) assay
The binding between miR-17 and lncRNA PTENP1 or SCARA5 was further examined using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA). Cells were lysed in RIPA buffer (Beyotime, China) and then centrifuged at 12,000 g for 10 min. Collected the supernatant and incubated with a RIP buffer containing magnetic beads conjugated to human anti-argonaute2 (Ago2) antibody (Abcam, USA). IgG antibody (Abcam, USA) was used as negative control. RNA was isolated from the sample and detected by qRT-PCR.
Tumor xenograft assay
BALB/c nude mice (male, 4–6 weeks old, n = 5 mice in each group) provided by SLAC Laboratory Animal Center (Shanghai, China) were housed at SPF grade animal room at a constant temperature of around 26°C. In total, 5637 cells at logarithmic growth phase were collected and re-suspended in PBS, adjusting cell concentration to 1 × 107 cells/mL. Next, 100 μL cell suspension was subcutaneously delivered into the right armpit of nude mice. After 7 days of inoculation, the exosomes derived from lncRNA PTENP1 overexpressing BMSC or its negative control exosomes (BMSCOE-PTENPCitation1-Exo or BMSCOE-NC-Exo) were injected into nude mice through tail vein injection. Tumor size was measured every 5 days after inoculation according to the formula: (Long × WidthCitation2)/2. Tumors were collected and weighed after 30 days of inoculation. All the animal experiment procedures were approved by the Animal Ethics Committee of the First Affiliated Hospital, Hengyang Medical School, University of South China.
Western blot
Proteins from exosomes, cells, and xenograft tumors were extracted by RIPA buffer (Beyotime, China). After determining the concentration of proteins using BCA Kit (Beyotime), 50 μg (proteins from cells and xenograft tumors), or 20 μg (proteins from exosomes) samples were subjected for the isolation through SDS-PAGE. Next, proteins were transferred into nitrocellulose membranes and blocked in 5% BSA for 2 h. Primary antibodies against CD9 (1:1000, ab263019, Abcam), CD63 (1:1000, ab271286, Abcam), CD81 (1:5000, ab109201, Abcam), calnexin (1:2000, ab22595, Abcam), SCARA5 (1:2000, ab118894, Abcam), and GAPDH (1:5000, ab9485, Abcam) were applied to probe indicated antigens, and HRP-conjugated secondary antibodies were employed to probe primary antibodies. Bands were visualized by enhanced chemiluminescence reagent (Millipore, USA) and analyzed in Image J (NIH, USA).
Quantitative real-time PCR (qRT-PCR) analysis
Total RNAs of exosomes, cells, and xenograft tumors were isolated by TRIzol reagent (Invitrogen, USA), and the concentration was determined by NanoDrop 2000 (Thermo Fisher Scientific, USA) following the manufacturer’s protocols. Primers were designed and synthesized by Sangon Biotech (Shanghai, China). The RNA sample (2 μg) was reversely transcribed into cDNA by PrimeScript RT reagent Kit (Takara, Japan), and the PCR process was conducted in StepOnePlus Real-Time PCR system (Applied Biosystems, Thermo Fisher Scientific, USA) with the SYBR Premix Dimer Eraser Kit (Takara, Japan). GAPDH or U6 was employed as internal control to detect gene expression in cells and xenograft tumors. The synthetic miRNA Caenorhabditis elegans miR-39 (cel-miR-39) was used as internal control to detect gene expression in exosomes. Relative expression levels were calculated using the 2−ΔΔCt method.
Statistical analysis
All the experiments were repeated at least three times. GraphPad Prism (version 7.0, USA) was adopted for data analysis and data were expressed as mean ± standard deviation (SD). Statistical analysis was conducted by two tailed Student’s t-test between two groups or one-way analysis of variance (ANOVA) with Tukey’s post hoc test for multiple groups. P value less than 0.05 was considered statistically significant.
Ethical Approval
All the animal experiment procedures were approved by the Animal Ethics Committee of the First Affiliated Hospital, Hengyang Medical School, University of South China.
Consent for Publication
Not Applicable. This article does not contain any studies with human participants performed by any of the authors.
Availability of Data and Material
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable requests.
Authors’ Contribution
SCL: Conceptualization; Writing-original draft; Methodology; Formal analysis; YHC: Supervision; Validation; LBC: Data curation; Resources; RK: Investigation; Software; ZXH: Visualization; XSL: Funding acquisition; Project administration; Writing-review & editing. All authors have read and approved the final version of this manuscript to be published.
Acknowledgements
We would like to express our sincere gratitude to the reviewers for their constructive comments.
Disclosure statement
The authors declare that there is no conflict of interest.
Additional information
Funding
References
- Ruiz M, Cosenza S, Maumus M, Jorgensen C, Noel D. Therapeutic application of mesenchymal stem cells in osteoarthritis. Expert Opin Biol Ther. 2016;16:33–42. doi:10.1517/14712598.2016.1093108.
- Fu D, Liu B, Jiang H, Li Z, Fan C, Zang L. Bone marrow mesenchymal stem cells-derived exosomal microRNA-19b-1-5p reduces proliferation and raises apoptosis of bladder cancer cells via targeting abl2. Genomics. 2021;113:1338–1348. doi:10.1016/j.ygeno.2021.03.011.
- Liu W, Wang Y, Gong F, Rong Y, Luo Y, Tang P, Zhou Z, Zhou Z, Xu T, Jiang T, et al. Exosomes derived from bone mesenchymal stem cells repair traumatic spinal cord injury by suppressing the activation of a1 neurotoxic reactive astrocytes. J Neurotrauma. 2019;36(3):469–484. doi:10.1089/neu.2018.5835.
- He C, Zheng S, Luo Y, Wang B. Exosome theranostics: biology and translational medicine. Theranostics. 2018;8(1):237–255. doi:10.7150/thno.21945.
- Jiang S, Mo C, Guo S, Zhuang J, Huang B, Mao X. Human bone marrow mesenchymal stem cells-derived microRNA-205-containing exosomes impede the progression of prostate cancer through suppression of rhpn2. J Exp Clin Cancer Res. 2019;38(1):495. doi:10.1186/s13046-019-1488-1.
- Cai H, Yang X, Gao Y, Xu Z, Yu B, Xu T, Li X, Xu W, Wang X,Hua L, et al. Exosomal microRNA-9-3p secreted from bmscs downregulates esm1 to suppress the development of bladder cancer. Mol Ther Nucleic Acids. 2019;18:787–800. doi:10.1016/j.omtn.2019.09.023.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551.
- Jia Y, Ding X, Zhou L, Zhang L, Yang X. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting prc1. Oncogene. 2021;40(2):246–261. doi:10.1038/s41388-020-01486-7.
- Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Perez Lanzon M, Zini N, Naaijkens B, Perut F, Niessen HWM, Baldini N, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6(1):127. doi:10.1186/s13287-015-0116-z.
- Zhao W, Qin P, Zhang D, Cui X, Gao J, Yu Z, Chia Y, Wang J, Li J. Long non-coding RNA pvt1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing erg and sponging mir-183-5p. Aging (Albany NY). 2019;11:9581–9596. doi:10.18632/aging.102406.
- Leveille N, Baglio SR. Exosome-transferred lncrnas at the core of cancer bone lesions. Crit Rev Oncol Hematol. 2019;139:125–127. doi:10.1016/j.critrevonc.2019.03.002.
- Fan Y, Sheng W, Meng Y, Cao Y, Li R. Lncrna ptenp1 inhibits cervical cancer progression by suppressing mir-106b. Artif Cells Nanomed Biotechnol. 2020;48:393–407. doi:10.1080/21691401.2019.1709852.
- Hu S, Xu L, Li L, Luo D, Zhao H, Li D, Peng B. Overexpression of lncrna ptenp1 suppresses glioma cell proliferation and metastasis in vitro. Onco Targets Ther. 2019;12:147–156. doi:10.2147/OTT.S182537.
- Zhong XL, Wang L, Yan X, Yang XK, Xiu H, Zhao M, Wang X, Liu J. Mir-20a acted as a cerna of lncrna PTEN and promoted bladder cancer cell proliferation and migration by regulating pdcd4. Eur Rev Med Pharmacol Sci. 2020;24:2955–2964. doi:10.26355/eurrev_202003_20660.
- Chi Y, Wang D, Wang J, Yu W, Yang J. 2019. Long non-coding RNA in the pathogenesis of cancers. Cells. 8. DOI:10.3390/cells8091015
- Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. microRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2019;70:3–20. doi:10.1016/j.mam.2018.07.003.
- Ganju A, Khan S, Hafeez BB, Behrman SW, Yallapu MM, Chauhan SC, Jaggi M. miRNA nanotherapeutics for cancer. Drug Discov Today. 2017;22:424–432. doi:10.1016/j.drudis.2016.10.014.
- Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q, et al. Circular RNA circ-itch inhibits bladder cancer progression by sponging mir-17/mir-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19. doi:10.1186/s12943-018-0771-7.
- Chen CL, Tseng YW, Wu JC, Chen GY, Lin KC, Hwang SM, Hu YC. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA ptenp1 and microRNA regulation. Biomaterials. 2015;44:71–81. doi:10.1016/j.biomaterials.2014.12.023.
- You K, Su F, Liu L, Lv X, Zhang J, Zhang Y, Liu B. Scara5 plays a critical role in the progression and metastasis of breast cancer by inactivating the erk1/2, stat3, and akt signaling pathways. Mol Cell Biochem. 2017;435:47–58. doi:10.1007/s11010-017-3055-4.
- Wen X, Wang N, Zhang F, Dong C. Overexpression of scara5 inhibits tumor proliferation and invasion in osteosarcoma via suppression of the fak signaling pathway. Mol Med Rep. 2016;13:2885–2891. doi:10.3892/mmr.2016.4857.
- Liu J, Zeng ML, Shi PC, Cao YP, Zhang JL, Xie YP. Scara5 is a novel biomarker in colorectal cancer by comprehensive analysis. Clin Lab. 2020;66. doi:10.7754/Clin.Lab.2019.191015.
- Liu Y, Tan Y-R, Sun -W-W, Ju W-T, Fu Y, Wang L-Z, Li J, Zhang C-P, Zhang Z-Y, Zhong L-P, et al. Identification of scara5 as a potential biomarker for oral squamous cell carcinoma using maldi-tof-ms analysis. Proteomics Clin Appl. 2018;12(5):e1700180. doi:10.1002/prca.201700180.
- Wang F, Cao X, Yin L, Wang Q, and He Z. Identification of scara5 gene as a potential immune-related biomarker for triple-negative breast cancer by integrated analysis. DNA Cell Biol. 2020;39(10):1813–1824. doi:10.1089/dna.2020.5449.
- Baker N, Boyette LB, Tuan RS. Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone. 2015;70:37–47. doi:10.1016/j.bone.2014.10.014.
- Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Xu J, Xia K, Chang Y, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17(1):82. doi:10.1186/s12943-018-0831-z.
- Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Science. 2018;109(7):2093–2100. doi:10.1111/cas.13642.
- Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, Lv Q, Qin C, Chu H, Wang M, et al. Exosome–transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17(1):143. doi:10.1186/s12943-018-0880-3.
- Wang J, Yang K, Yuan W, Gao Z. Determination of serum exosomal h19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–9316. doi:10.12659/MSM.912018.
- Ridge SM, Sullivan FJ, Glynn SA. Mesenchymal stem cells: key players in cancer progression. Mol Cancer. 2017;16(1):31. doi:10.1186/s12943-017-0597-8.
- Klopp AH, Gupta A, Spaeth E, Andreeff M, Marini F. Concise review: dissecting a discrepancy in the literature: do mesenchymal stem cells support or suppress tumor growth? Stem Cells. 2011;29(1):11–19. doi:10.1002/stem.559. 3rd.
- Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et al. Vegfr1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438(7069):820–827. doi:10.1038/nature04186.
- Biancone L, Bruno S, Deregibus MC, Tetta C, Camussi G. Therapeutic potential of mesenchymal stem cell-derived microvesicles. Nephrol Dial Transplant. 2012;27(8):3037–3042. doi:10.1093/ndt/gfs168.
- Li B, Xu H, Han H, Song S, Zhang X, Ouyang L, Qian C, Hong Y, Qiu Y, Zhou W, et al. Exosome-mediated transfer of lncrunx2-as1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene. 2018;37(41):5508–5519. doi:10.1038/s41388-018-0359-0.
- Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, Chen K, Zhang Y, Liu H, Gan L, et al. Tumor-derived exosomal lnc-sox2ot promotes emt and stemness by acting as a cerna in pancreatic ductal adenocarcinoma. Oncogene. 2018;37(28):3822–3838. doi:10.1038/s41388-018-0237-9.
- Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, Zhong G, Li Y, Li J, Huang J, et al. Exosomal long noncoding RNA lnmat2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404–421. doi:10.1172/JCI130892.
- Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W, Chen K, Xiao H, Li H, Tang K, et al. Pseudogene ptenp1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13(12):3086–3097. doi:10.1158/1535-7163.MCT-14-0245.
- Zhang R, Guo Y, Ma Z, Ma G, Xue Q, Li F, Liu L. Long non-coding RNA ptenp1 functions as a cerna to modulate PTEN level by decoying mir-106b and mir-93 in gastric cancer. Oncotarget. 2017;8(16):26079–26089. doi:10.18632/oncotarget.15317.
- Qian -Y-Y, Li K, Liu Q-Y, Liu Z-S. Long non-coding RNA ptenp1 interacts with mir-193a-3p to suppress cell migration and invasion through the PTEN pathway in hepatocellular carcinoma. Oncotarget. 2017;8(64):107859–107869. doi:10.18632/oncotarget.22305.
- Lu X-S, Huang M-L, Chen L-B, Liu S-C, Huang Z-X, Liu S-M. WITHDRAWN: SCARA5 regulated by MEG3/miR-141 axis attenuates proliferation, migration and invasion of bladder cancer. Life Sci. 2021:119619. doi:10.1016/j.lfs.2021.119619
