ABSTRACT
Pancreatic ductal adenocarcinoma (PDAC) is the most malignant tumor. Zinc finger and SCAN domain-containing protein 23 (ZSCAN23) is a new member of the SCAN domain family. The expression regulation and biological function remain to be elucidated. In this study, we explored the epigenetic regulation and the function of ZSCAN23 in PDAC. ZSCAN23 was methylated in 60.21% (171/284) of PDAC and its expression was regulated by promoter region methylation. The expression of ZSCAN23 inhibited cell proliferation, colony formation, migration, invasion, and induced apoptosis and G1/S phase arrest. ZSCAN23 suppressed Panc10.05 cell xenograft growth in mice. Mechanistically, ZSCAN23 inhibited Wnt signaling by interacting with myosin heavy chain 9 (MYH9) in pancreatic cancer cells. ZSCAN23 is frequently methylated in PDAC and may serve as a detective marker. ZSCAN23 suppresses PDAC cell growth both in vitro and in vivo.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is currently the third leading cause of cancer-related deaths in the United States and is predicted to be the second leading cause of cancer-related deaths by 2030. Only 15–20% patients are eligible for surgical resection, with approximately 75% of these cases experiencing recurrence within 2 years. Regrettably, systemic therapies remain largely ineffective.Citation1–3 Although oncogenic KRAS mutations are found in more than 90% of PDAC cases, the development of inhibitors remains a challenge. Familial pancreatic cancer accounts for 5–10% of cases with mutations in DNA damage repair genes, including breast cancer type 1 (BRCA1), BRCA2, partner and localizer of BRCA2 (PALB2), serine/threonine kinase 11 (STK11), and MutL homolog 1 (MLH1).Citation4–6 However, the utilization of precision-based therapy is limited to only 7% of patients.Citation7,Citation8 Therefore, the identification of novel biomarkers remains a critical topic in PDAC diagnosis and therapy.
Epigenetic regulation is involved in carcinogenesis, tumor development, and metastasis.Citation9,Citation10 Epigenetic heterogeneity increases the complexity of cancer diagnosis and therapy.Citation11,Citation12 Epigenetic regulators consist of three distinct categories: writers, readers and erasers. Writer is responsible for the addition of chemical groups to DNA or histone (such as DNA methyltransferases, DNMTs). Eraser, such as histone deacetylases (HDACs), is involved in the removal of these modifications. Reader, such as methyl-binding domain protein, is to recognize specific epigenetic marks and acts as an effector.Citation13,Citation14 Epidrugs are small molecular inhibitors to target the epigenome or epigenetic regulatory enzymes. The role of epidrugs has been extensively investigated in various cancers. However, their application remains restricted to hematological malignancies and is infrequently implemented in clinical practice.Citation10,Citation13 Oncologists have been pursuing the precise targeting of cancer cells without damaging normal cells. Many new anticancer drugs have been registered for clinical trials by combination of epidrugs with other therapeutics in solid tumors, yet a significant portion of them have ultimately been excluded (such as CRUKD/07/065, NCT00481078, NCT01100944).Citation15–17
This limitation is mainly attributed to the complexity of diverse cellular compositions and imprecise targeting. “Synthetic lethality” based on epigenetic loss-of-function is a novel strategy for cancer therapy.Citation15,Citation18,Citation19 Therefore, understanding the abnormal epigenetic changes involved in the signaling pathways and regulatory networks becomes imperative.
Zinc finger and SCAN domain-containing protein 23 (ZSCAN23) is a member of the SCAN domain zinc-finger family, which is located on chromatin 6p22.1, a tumor-susceptible locus.Citation20 The ZSCAN23 protein contains two domains: a zinc- finger domain at the C-terminus and a SCAN domain at the N-terminus.Citation21 The zinc-finger domain binds to specific DNA segments and interacts with other cellular factors to regulate the transcription of target genes.Citation22 The SCAN domain, a highly conserved vertebrate-specific protein domain, is involved in protein-protein interactions via self-binding and mediates oligomerization, leading to formation of homodimers or heterodimers that regulate various transcriptional activities.Citation23–26 Other members of the ZSCAN family are involved in cancer development, and their functions differ in various cancers.Citation27–29 The role of ZSCAN23 in cancer remains unclear. Therefore, the aim of this study is to elucidate the role of ZSCAN23 in PDAC development.
Results
ZSCAN23 was frequently methylated in PDAC and its expression was regulated by promoter region methylation
The TCGA database was used to analyze the association between ZSCAN23 mRNA expression and CpG site methylation using the extracted data of 194 PDAC cases (http://xena.ucsc.edu/). The mRNA levels were inversely associated with methylation at CpG sites around the transcription start site (cg21486944, cg06818710, cg00651523, and cg07836142; all p < .0001; ), suggesting that ZSCAN23 expression is regulated by promoter region methylation.
Figure 1. Expression and methylation status of ZSCAN23 in PDAC. a, association of ZSCAN23 expression and methylation status of CpG sites around the TSS site. b, scatter plots show that ZSCAN23 expression is negatively associated with representative CpG site methylation status (cg21486944, cg06818710, cg00651523, and cg07836142). c, detection of ZSCAN23 expression in PDAC cells via semi-quantitative RT-PCR. 5-AZA-DC: 5-aza-2’-deoxycytidine; GAPDH: internal control of RT-PCR; H2O: double distilled water; (-): absence of 5-AZA-DC; (+): presence of 5-AZA-DC. d, MSP results of ZSCAN23 in PDAC cells. U: unmethylated alleles; M: methylated alleles; IVD: in vitro methylated DNA, serves as methylation control; NL: normal lymphocytes DNA, serves as unmethylation control. e, Bisulfite sequencing results of ZSCAN23 in SW1990, Panc10.05, and T3M4 cells. The size of unmethylated MSP products was 136 bp. Bisulfite sequencing was performed in a 264 bp region around the ZSCAN23 transcription start site (−198 bp to 65 bp). Filled circles: methylated CpG sites; open circles: unmethylated CpG sites; TSS: transcription start site. f, representative MSP results of ZSCAN23 in primary PDAC samples. PN: adjacent normal tissue of pancreas solid pseudopapillary neoplasm; PC: primary PDAC samples.
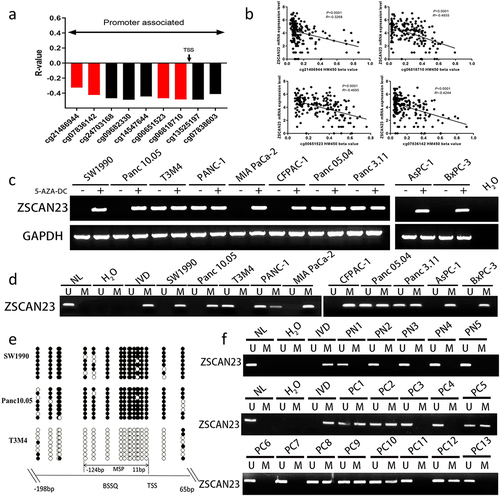
ZSCAN23 was found to be highly expressed in T3M4, PANC-1, Panc 05.04, and Panc3.11 cells and no expression was detected in SW1990, Panc10.05, MIAPaCa-2, CFPAC-1, AsPC-1, and BxPC-3 cells, determined by the semi-quantitative reverse transcription-polymerase chain reaction (RT‐PCR) detection (). ZSCAN23 was unmethylated in T3M4 cells, completely methylated in SW1990, Panc 10.05, MIAPaCa-2, CFPAC-1, AsPC-1, and BxPC-3 cells, and partially methylated in PANC-1, Panc 05.04, and Panc3.11 cells (). The loss of ZSCAN23 expression correlated with complete methylation. ZSCAN23 expression was restored after 5-AZA-2′-deoxycytidine (5-AZA-DC) treatment in SW1990, Panc10.05, MIAPaCa-2, CFPAC-1, AsPC-1, and BxPC-3 cells, whereas no changes were observed in T3M4, PANC-1, Panc 05.04, and Panc3.11 cells (), demonstrating the epigenetic regulation of ZSCAN23 expression. Validation of the methylation-specific PCR (MSP) primer efficiency and methylation density was performed using bisulfite sequencing in SW1990, Panc 10.05, and T3M4 cells ().
No methylation was found in the 17 cases of adjacent tissue samples of serous cystadenoma (benign cyst in pancreas), excluding tissue/cell type-specific methylation. ZSCAN23 was methylated in 60.21% (171/284) of the PDAC samples (). No association was found between ZSCAN23 methylation and clinical factors, including sex, age, tumor size, differentiation, TNM stage, tumor location, smoking or alcohol consumption (all p > .05, ).
Table 1. Clinical factors and ZSCAN23 methylation status in PDAC.
ZSCAN23 expression inhibited PDAC cell proliferation
MTT assays were used for evaluating the effect of ZSCAN23 on cell proliferation. The optical density (OD) values before and after restoration of ZSCAN23 expression were 1.245 ± 0.052 vs 1.034 ± 0.050 in SW1990 cells (p < .001) and 0.799 ± 0.014 vs 0.719 ± 0.024 in Panc10.05 cells (p < .0001) (). The OD value was reduced in ZSCAN23-re-expressed SW1990 (16.9%) and Panc10.05 cells (10.0%), indicating an inhibitory role of ZSCAN23 expression in cell proliferation.
Figure 2. Effect of ZSCAN23 expression on cell proliferation, invasion, and migration. a, growth curves represent the effects of ZSCAN23 expression on PDAC cells. b, the average number of cell clones is presented in the bar diagram. c, cell migration and invasion experiments on SW1990 and Panc10.05 cells before and after re-expression of ZSCAN23. The average number of migrating cells is presented in the bar diagram. Each experiment was repeated in triplicate. *p < .05, **p < .01, ***p < .001, and ****p < .0001. D, the expression levels of flag, MMP2, MMP7, and MMP9 were detected via western blotting.
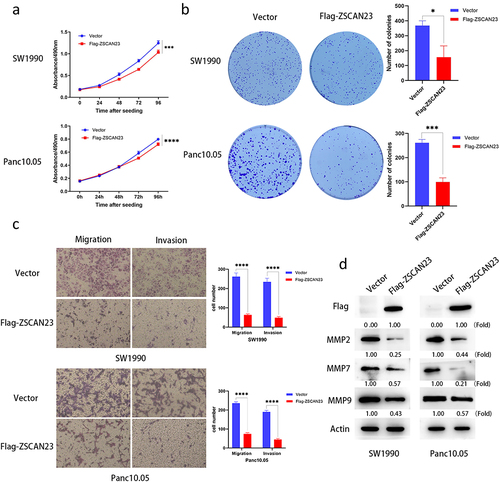
In colony formation analysis, the colony number before and after restoration of ZSCAN23 expression was 384.000 ± 22.627 vs 199.500 ± 14.849 in SW1990 cells (p < .05) and 261.333 ± 14.012 vs 99.333 ± 17.388 in Panc10.05 cells (p < .001) (). The number of colonies reduced after restoring ZSCAN23 expression in SW1990 (48.0%) and Panc10.05 cells (62.0%). These data indicated that ZSCAN23 expression inhibited PDAC cell proliferation.
ZSCAN23 expression inhibited PDAC cell migration and invasion
Cell migration and invasion were evaluated using a Transwell assay. The number of migrating cells with and without ZSCAN23 expression was 261.667 ± 18.930 vs 63.667 ± 4.933 cells (SW1990 cells; p < .0001) and 236.000 ± 7.350 vs 74.000 ± 6.928 cells (Panc10.05 cells; p < .0001). Cell migration was decreased in SW1990 (75.7%) and Panc10.05 cells (68.6%) after restoring ZSCAN23 expression (). The number of invading cells with and without ZSCAN23 expression was 234.333 ± 19.140 vs 49.000 ± 6.245 cells (SW1990 cells; p < .0001) and 190.333 ± 8.327 vs 44.333 ± 8.859 cells (Panc10.05 cells; p < .0001). The number of invading cells was reduced in SW1990 (79.1%) and Panc10.05 cells (76.6%) after restoring ZSCAN23 expression (), demonstrating that ZSCAN23 expression inhibited cell invasion.
To further validate the inhibitory role of ZSCAN23, the migration and invasion related proteins, matrix metallopeptidase-2 (MMP2), MMP7 and MMP9 were detected using western blotting. The levels of MMP2, MMP7 and MMP9 were reduced after restoration of ZSCAN23 expression ().
ZSCAN23 expression induced G1/S arrest in PDAC cells
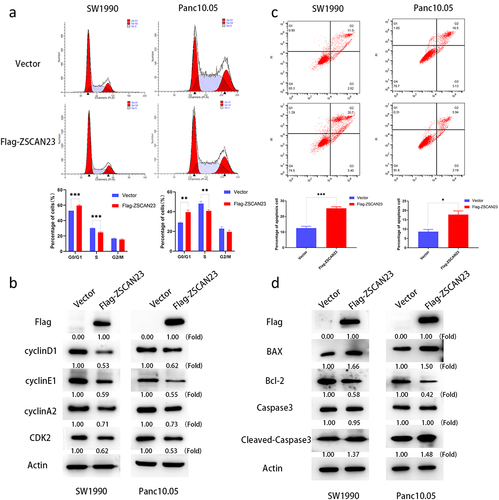
The cell cycle phase distributions of G0/G1, S, and G2/M in ZSCAN23 unexpressed and overexpressed SW1990 cells were 53.0 ± 0.0% vs 59.7 ± 1.2% (p < .001), 30.2 ± 0.6% vs 24.8 ± 0.8% (p < .001), and 16.8 ± 0.7% vs 15.5 ± 0.9%, respectively. In ZSCAN23 unexpressed and re-expressed Panc10.05 cells, the distributions of G0/G1, S, and G2/M cell phases were 29.0 ± 0.1% vs 39.6 ± 2.2% (p < .01), 48.2 ± 2.5% vs 40.8 ± 1.2% (p < .01), and 22.8 ± 1.8% vs 19.6 ± 1.7%, respectively (). The proportion of G0/G1 phase cells increased in SW1990 (6.7%) and Panc10.05 cells (10.6%), and that of S phase cells decreased in SW1990 (5.4%) and in Panc10.05 cells (7.4%) upon restoration of ZSCAN23 expression. These results indicated that ZSCAN23 induced G1/S arrest. Cyclin D1, cyclin E1, cyclin A2, and cyclin-dependent kinase 2 (CDK2) are G1/S checkpoint regulators and their expression was detected by western blotting. The expression of cyclin D1, cyclin E1, cyclin A2, and CDK2 was reduced following re-expression of ZSCAN23 in SW1990 and Panc10.05 cells (), validating that ZSCAN23 expression induced G1/S arrest in PDAC cells.
Figure 3. Effect of ZSCAN23 expression on cell cycle and apoptosis. a, the effect of ZSCAN23 expression on cell cycle phase in PDAC cells. The bar diagram presents the proportions of cells at different cell cycle phases. b, Western blots present the levels of flag, cyclin D1, cyclin E1, cyclin A2, and CDK2. c, effect of ZSCAN23 expression on apoptosis in PDAC cells. Each experiment was repeated in triplicate. *p < .05, **p < .01, and ***p < .001. d, Western blotting shows the levels of flag, BAX, Bcl-2, Caspase 3 and Cleaved-caspase 3.
ZSCAN23 expression induced PDAC cell apoptosis
Flow cytometry analysis revealed that the proportions of apoptotic cells among ZSCAN23 silenced and overexpressed cells were 12.5 ± 1.2% vs 25.2 ± 1.0% (SW1990 cells; p < .001) and 8.6 ± 1.2% vs 17.8 ± 1.9% (Panc10.05 cells; p < .05) (). These results indicated that ZSCAN23 expression induces apoptosis. Subsequently, the levels of apoptosis-related proteins were analyzed. Increased levels of BAX and Cleaved caspase-3 along with reduced levels of Bcl-2 were detected after re-expression of ZSCAN23 in SW1990 and Panc10.05 cells (), further demonstrating that ZSCAN23 induced apoptosis.
ZSCAN23 expression suppressed Panc10.05 cell xenograft growth
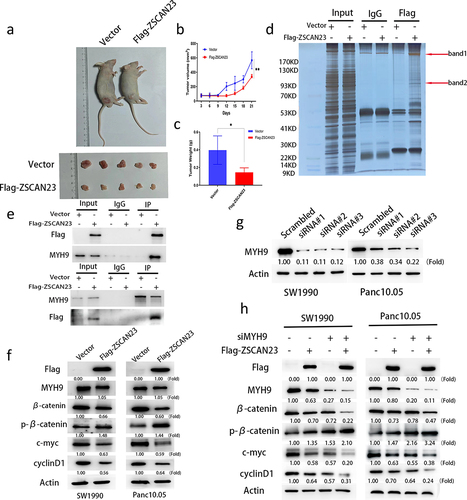
A mouse xenograft model was used to evaluate the role of ZSCAN23 in PDAC development. Before and after re-expression of ZSCAN23, the tumor volume was 570.874 ± 118.293 mm3 and 340.482 ± 32.191 mm3 in Panc10.05 cell xenografts (p < .01; ), respectively. The tumor volume decreased after restoring ZSCAN23 expression (). The tumor weight in ZSCAN23 unexpressed and re-expressed xenografts was 0.396 ± 0.160 g and 0.146 ± 0.050 g, respectively (p < .05). Tumor weight was reduced after ZSCAN23 re-expression (), suggesting that ZSCAN23 expression suppressed PDAC growth in vivo.
Figure 4. ZSCAN23 suppresses human PDAC cell xenograft growth in mice and ZSCAN23 suppresses the wnt signaling pathway through MYH9. a, representative tumors from ZSCAN23 unexpressed and re-expressed Panc10.05 cell xenografts. b, tumor weights of nude mice at the 21st day after inoculation of ZSCAN23 unexpressed and re-expressed Panc10.05 cells. Bars present the mean values for five mice. **p < .01. c, tumor growth curves of ZSCAN23 unexpressed and ZSCAN23 re-expressed Panc10.05 cells. *p < .05. d, Co-IP assay and silver staining. e, Co-IP assay shows the binding between ZSCAN23 and MYH9 in SW1990 cells. IgG: negative control. f, Western blotting shows the levels of flag, MYH9, β-catenin, p-β-catenin, c-myc and cyclinD1 in PDAC cells. g, Western blotting shows the effects of MYH9 knockdown by different siRNAs. Scrambled: siRNA negative control; siRNA#1 and siRNA#2: siRNA for MYH9. h, Western blotting shows the levels of flag, MYH9, β-catenin, p-β-catenin, c-myc and cyclinD1 before and after knockdown of MYH9 in PDAC cells.
ZSCAN23 expression inhibited Wnt signaling by interacting with MYH9
Co-immunoprecipitation assays were performed to further evaluate the mechanism of action of ZSCAN23 in PDAC. An extra band, which was pulled down using a Flag antibody, was excised and analyzed using mass spectrometry (). The protein with the highest score was myosin heavy chain 9 (MYH9), and its interaction with ZSCAN23 was validated using co-IP and reciprocal co-IP assay (). MYH9 has been recently reported to promote the progression of PDAC and other cancers via Wnt signaling.Citation30–32 By analyzing the potential molecules in the mass spectrometry results, Wnt signaling proteins were found to represent an important part of the complex (Supplementary Table S1 and Supplementary Figure S1). Subsequently, we analyzed the role of ZSCAN23 in Wnt signaling. The expression levels of β-catenin, c-myc, and cyclin D1 were reduced, while p-β-catenin expression level was increased after re-expression of ZSCAN23 in SW1990 and Panc10.05 cells (). The effect of ZSCAN23 on Wnt signaling via MYH9 was validated by knocking down MYH9 in these cells (). These results demonstrate that ZSCAN23 inhibited Wnt signaling by interacting with MYH9 in PDAC cells.
Discussion
Epigenetic based “synthetic lethality” represents a novel strategy for cancer therapy. Applying this theory to cell fate or DNA damage repair genes with abnormal epigenetic changes may provide additional opportunities for targeted therapy without damaging normal cells.Citation18,Citation19 Therefore, the discovery of novel epigenetic markers involved in cell fate determination or DNA damage repair is becoming increasingly important. The SCAN domain proteins play multifaceted roles in embryonic development, DNA damage repair, and cancers.Citation33–35 However, most of these members have not been studied extensively. ZKSCAN3 is involved in Wnt/β-catenin signaling and causes chromosomal instability.Citation36 ZSCAN18 is a tumor suppressor in breast cancer and has been found to inhibit Wnt/β-catenin and glycolysis signaling pathways.Citation34 ZSCAN23 is a new member of this family and has recently been reported to contain a C2H2 zinc-finger structure.Citation21 However, its regulation of expression and role in cancer remain unclear. In this study, we found that ZSCAN23 was frequently methylated in PDAC and its expression was regulated by promoter region methylation, suggesting that ZSCAN23 methylation is a potential PDAC diagnostic marker and epigenetic regulation involves in PDAC development. Then, we studied the function of ZSCAN23 in PDAC. It was observed that ZSCAN23 exerted inhibitory effects on cell proliferation and colony formation in PDAC cells. Furthermore, the use of a transwell assay demonstrated that ZSCAN23 suppressed cell migration and invasion, which was further confirmed at the molecular level through the assessment of MMP2, MMP7 and MMP9 expression in PDAC cells. G1/S arrest and apoptosis were induced by ZSCAN23. The G1/S checkpoint regulatory proteins, cyclin D1, cyclin E1, cyclin A2 and CDK2, were reduced. Pro-apoptotic proteins, BAX and cleaved caspase-3, were increased. While Bcl-2, an anti-apoptotic protein, was decreased by restoration of ZSCAN23 expression. ZSCAN23 suppressed Panc10.05 cell xenografts growth in mice. To further understand the mechanism of ZSCAN23 in PDAC, immunoprecipitation and mass spectrometry assay were applied. The interaction of ZSCAN23 and MYH9 was discovered, and further validation was performed using reciprocal co-IP assay and western blotting. MYH9 was reported to be involved in Wnt, PI3K-AKT and NOTCH signaling pathways in cancers.Citation31,Citation37–40 The analysis of the binding proteins in the complex of ZSCAN23 antibody revealed the presence of significant components related to Wnt signaling. Wnt signaling is a well-known signaling pathway for development and disease.Citation41 It is divided into canonical and non-canonical pathways. The canonical pathway is involved in cell fate, proliferation, differentiation and migration. While the non-canonical pathway regulates cell polarity and migration.Citation42 Recent findings have indicated that inhibiting Wnt signaling can induce a BRCA-like state in cancer.Citation43,Citation44 The dysregulation of Wnt signaling varies across different types of cancer, with hyperactivation of the canonical Wnt pathway observed in colorectal, pancreatic, ovarian and gastric cancers.Citation44–46 Then, we studied the role of ZSCAN23 on Wnt signaling. Our findings revealed that ZSCAN23 acted as an inhibitor of Wnt signaling by interacting with MYH9, and epigenetic silencing of ZSCAN23 activated Wnt signaling in PDAC cells. The impact of ZSCAN23 on Wnt signaling through MYH9 was validated by small interfering RNAs (siRNAs) knockdown technique. The complexity of epigenetics in cancer requires a more extensive study. Epigenetic-based synthetic lethality is becoming a new therapeutic strategy in human cancer.Citation18,Citation47–49 Our finding paves the way for further investigation of synthetic lethality in PDAC. Better understanding the role of ZSCAN23, especially in the regulatory network, may improve the therapeutic strategies in PDAC in the future.
In conclusion, ZSCAN23 is frequently methylated in PDAC, and its expression is regulated by promoter region methylation. ZSCAN23 serves as a novel tumor suppressor that inhibits Wnt signaling in PDAC.
Materials and methods
Human tissue samples and cell lines
A total of 284 cases of primary PDAC and 17 cases of adjacent tissue of serous cystadenoma were obtained from surgically resected tissue samples at the Chinese PLA General Hospital without chemo-radiotherapy before surgery. Tumors were classified according to the standard TNM staging system (AJCC on Cancer 8th Edition Cancer Staging Manual). All human tissue samples were collected according to the guidelines approved by the Institutional Review Board of the Chinese PLA General Hospital.
Ten PDAC cell lines were established from primary pancreatic cancer cells and cultured in RPMI-1640 medium (Invitrogen, Waltham, MA, USA) supplemented with 10% fetal bovine serum (GeminiBio, West Sacramento, CA, USA).
5-AZA-2′-deoxycytidine treatment, RNA preparation, and semiquantitative RT-PCR
5-AZA-DC (Sigma Aldrich, St. Louis, MO, USA) treatment for cell lines, total RNA preparation, and first-strand cDNA synthesis were performed as described previously.Citation50 The semi-quantitative RT‐PCR primer sequences used for ZSCAN23 were 5′-CATCTCCTCTCACGGATGCC-3′ (F) and 5′- TCTGAAGGGTCAAGGTTATGGC-3′ (R). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers were used as described previously.Citation48
Sodium bisulfite treatment and methylation detection
DNA extraction, sodium bisulfite, MSP, and bisulfite sequencing were performed as previously described.Citation51 The following primers were used: 5′-AAGATTATAGACGGGATTTAGAGTGAC-3′ (methylation forward) and 5′-ACTACAAAAACCGAAAATACGTCACCG-3′ (methylation reverse); 5′-TTAAGATTATAGATGGGATTTAGAGTGAT-3′ (unmethylation forward) and 5′-TAAACTACAAAAACCAAAAATACATCACCA-3′ (unmethylation reverse). The following bisulfite sequencing primers were used: 5′-AATAYGTGTTAAAATATTATGTTGG-3′ (forward) and 5′-CTAACRCAAATCACATCTACTC-3′ (reverse).
Construction of ZSCAN23 expression plasmid and screening of ZSCAN23 stably expressing cells
The full-length CDS sequence (NM_001012455.2) and 3 × Flag sequences of ZSCAN23 were synthesized by Azenta Life Sciences (Suzhou, China), and cells that stably expressed ZSCAN23 were generated by constructing pCDH-CMV-MCS-puro vectorand screening cells via puromycin-based selection for 3 d.Citation52 The puromycin concentration used was 6.0 μg/ml for SW1990 cells and 2.0 μg/ml for Panc10.05 cells; monoclonal cells were selected via dilution in 96-well plates.
Cell proliferation analysis
A total of 1 × 103 SW1990 and 2 × 103 Panc10.05 cells (ZSCAN23-silenced and overexpressed) were seeded in 96-well plates and grown for 4 d. The OD values were measured using a microplate reader. Six-well plates were used for colony formation analysis, as previously described.Citation53
Transwell, cell cycle, and apoptosis analysis
For cell migration analysis, 4 × 104 SW1990 and 2 × 104 Panc10.05 cells (ZSCAN23-silenced and overexpressed) were used for Transwell analysis. Each experiment was repeated three times. After coating the upper chamber with matrigel, the same cell numbers and procedures were used for studying cell invasion. Cell cycle and apoptosis were analyzed as described previously.Citation50
Western blotting
Cell lysis was prepared as previously described.Citation54 Antibodies were used as follows: MMP2 (Huaxingbio, China), caspase 3/cleaved-caspase 3, MMP9, c-myc, MYH9, β-catenin, actin (Proteintech, Rosemont, IL, USA), Flag Tag Rabbit mAb, MMP7, and p-β-catenin (ZENBIO, China).
PDAC cell xenograft mice
Four-week-old BABL/c nude mice were used to establish ZSCAN23-silenced and overexpressing Panc10.05 cells (4.5 × 106 cells) in a xenograft tumor model by following previously described procedures, according to the Animal Ethics Committee of the Chinese PLA General Hospital.Citation55
Co-immunoprecipitation and mass spectrometry
ZSCAN23 silenced and re-expressing SW1990 cells were lysed using cold IP buffer (1% Triton X-100, 50 mM Tris-HCl [pH 7.4], 1 mM EDTA, 150 mM NaCl, 0.1% DTT, 1% PIC, 1% PI, and 1% PMSF). Specific antibodies or IgG (Beyotime Biotech, China) were added to the cell lysates and incubated overnight. Protein A/G beads (Yeasen Biotechnology, Shanghai, China) were incubated with the antibody – protein complex for at least 4 h. The bead-antibody-protein complex was washed with IP buffer, and then the protein loading buffer was added to the complex. The IP products were analyzed via silver staining (Thermo Fisher Scientific, Waltham, MA, USA) and mass spectrometry, and then validated by western blotting.
RNA interference assay
SiRNAs used to target MYH9 and scrambled control duplex sequences were as follows: siRNA#1: 5′-GCAAGCUGCCGAUAAGUAUTT-3′ (sense, S) and 5′-AUACUUAUCGGCAGCUUGCTG-3′ (antisense, AS); siRNA#2: 5′-CCAGAAGGCGCAGACUAAATT-3′ (S) and 5′-UUUAGUCUGCGCCUUCUGGAC-3′ (AS); siRNA#3: 5′-CCAGUCCUCUGACAAGUUUTT-3′ (S) and 5′-AAACUUGUCAGAGGACUGGTG-3′ (AS); siRNA scrambled control duplex: 5′-UUCUCCGAACGUGUCACGUTT-3′ (S) and 5′-ACGUGACACGUUCGGAGAATT-3′ (AS).
Statistical analysis
SPSS software (version 22.0; IBM, Armonk, NY, USA) was used for the statistical analysis. The association between ZSCAN23 methylation and clinicopathological factors was analyzed using the chi-squared test. Quantitative data were evaluated using the Student’s t-test. Values of p < .05 were considered significant.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the Chinese PLA General Hospital.
Availability of data and materials
The data in the present study are available from the corresponding author.
Author contributions
QD and AG performed experiments. QD and MZ helped analyze the data. QD and MG wrote the manuscript. MG designed the study and edited the manuscript. TH and MG interpreted the data. All the authors have read and approved the final version of the manuscript.
supplementary Table 1.docx
Download MS Word (97.1 KB)supplementary Figure1.tif
Download TIFF Image (277.6 KB)supplementary.xlsx
Download MS Excel (350.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2024.2302924
Additional information
Funding
Notes on contributors
Qian Du
Qian Du, Master Degree of Biology and Medicine, The Third Affiliated Hospital of Xinxiang Medical University.
Meiying Zhang
Meiying Zhang, PhD, Assistant Researcher, Department of Gastroenterology and Hepatology, First Medical Center of Chinese PLA General Hospital.
Aiai Gao
Aiai Gao, MD, PhD, Department of Gastroenterology and Hepatology, First Medical Center of Chinese PLA General Hospital.
Tao He
Tao He, MD, PhD, Department of Pathology, Characteristic Medical Center of the Chinese People’s Armed Police Force.
Mingzhou Guo
Mingzhou Guo, MD, PhD, Professor, Supervisor, Department of Gastroenterology and Hepatology, National Key Laboratory of Kidney Diseases, the First Medical Center of Chinese PLA General Hospital.
References
- Kisling SG, Natarajan G, Pothuraju R, Shah A, Batra SK, Kaur S. Implications of prognosis-associated genes in pancreatic tumor metastasis: lessons from global studies in bioinformatics. Cancer Metastasis Rev. 2021;40(3):721–10. doi:10.1007/s10555-021-09991-1.
- Halbrook CJ, Lyssiotis CA, Pasca di Magliano M, Maitra A. Pancreatic cancer: advances and challenges. Cell. 2023;186(8):1729–54. doi:10.1016/j.cell.2023.02.014.
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–21. doi:10.1158/0008-5472.CAN-14-0155.
- Goldstein JB, Zhao L, Wang X, Ghelman Y, Overman MJ, Javle MM, Shroff RT, Varadhachary GR, Wolff RA, McAllister F, et al. Germline DNA sequencing reveals novel mutations predictive of overall survival in a cohort of patients with pancreatic cancer. Clin Cancer Res. 2020;26(6):1385–94. doi:10.1158/1078-0432.CCR-19-0224.
- Overbeek KA, Goggins MG, Dbouk M, Levink IJM, Koopmann BDM, Chuidian M, Konings ICAW, Paiella S, Earl J, Fockens P, et al. Timeline of development of pancreatic cancer and implications for successful early detection in high-risk individuals. Gastroenterology. 2022;162(3):772–85.e4. doi:10.1053/j.gastro.2021.10.014.
- Goggins M, Overbeek KA, Brand R, Syngal S, Del Chiaro M, Bartsch DK, Bassi C, Carrato A, Farrell J, Fishman EK, et al. Management of patients with increased risk for familial pancreatic cancer: updated recommendations from the international cancer of the pancreas screening (CAPS) consortium. Gut. 2020;69(1):7–17. doi:10.1136/gutjnl-2019-319352.
- Hosein AN, Dougan SK, Aguirre AJ, Maitra A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat Cancer. 2022;3(3):272–286. doi:10.1038/s43018-022-00349-2.
- Lowery MA, Jordan EJ, Basturk O, Ptashkin RN, Zehir A, Berger MF, Leach T, Herbst B, Askan G, Maynard H, et al. Real-time genomic profiling of pancreatic ductal adenocarcinoma: potential actionability and correlation with clinical phenotype. Clin Cancer Res. 2017;23(20):6094–100. doi:10.1158/1078-0432.CCR-17-0899.
- Ma K, Cao B, Guo M. The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin Epigenetics. 2016;8(1):43. doi:10.1186/s13148-016-0210-9.
- Davalos V, Esteller M. Cancer epigenetics in clinical practice. CA Cancer J Clin. 2023;73(4):376–424. doi:10.3322/caac.21765.
- Guo M, Peng Y, Gao A, Du C, Herman JG. Epigenetic heterogeneity in cancer. Biomarker Res. 2019;7:23. doi:10.1186/s40364-019-0174-y.
- Zhu C, Zhang M, Wang Q, Jen J, Liu B, Guo M. Intratumor epigenetic heterogeneity-a panel gene methylation study in thyroid cancer. Front Genet. 2021;12:714071. doi:10.3389/fgene.2021.714071.
- Yan W, Herman JG, Guo M. Epigenome-based personalized medicine in human cancer. Epigenomics. 2016;8(1):119–33. doi:10.2217/epi.15.84.
- Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics. 2019;14(12):1164–1176. doi:10.1080/15592294.2019.1640546.
- Cossío FP, Esteller M, Berdasco M. Towards a more precise therapy in cancer: exploring epigenetic complexity. Curr Opin Chem Biol. 2020;57:41–9. doi:10.1016/j.cbpa.2020.04.008.
- Morel D, Jeffery D, Aspeslagh S, Almouzni G, Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol. 2020;17(2):91–107. doi:10.1038/s41571-019-0267-4.
- Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet. 2019;20(2):109–27. doi:10.1038/s41576-018-0074-2.
- Gao A, Guo M. Epigenetic based synthetic lethal strategies in human cancers. Biomarker Res. 2020;8:44. doi:10.1186/s40364-020-00224-1.
- Hu Y, Guo M. Synthetic lethality strategies: beyond BRCA1/2 mutations in pancreatic cancer. Cancer Sci. 2020;111(9):3111–21. doi:10.1111/cas.14565.
- Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015;47(2):164–71. doi:10.1038/ng.3185.
- Edelstein LC, Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi:10.1016/j.gene.2005.06.022.
- Sander TL, Stringer KF, Maki JL, Szauter P, Stone JR, Collins T. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003;310:29–38. doi:10.1016/S0378-1119(03)00509-2.
- Schumacher C, Wang H, Honer C, Ding W, Koehn J, Lawrence Q, Coulis CM, Wang LL, Ballinger D, Bowen BR, et al. The SCAN domain mediates selective oligomerization. J Biol Chem. 2000;275(22):17173–9. doi:10.1074/jbc.M000119200.
- Williams AJ, Blacklow SC, Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol Cell Biol. 1999;19(12):8526–35. doi:10.1128/MCB.19.12.8526.
- Stone JR, Maki JL, Blacklow SC, Collins T. The SCAN domain of ZNF174 is a dimer. J Biol Chem. 2002;277(7):5448–52. doi:10.1074/jbc.M109815200.
- Li X, Han M, Zhang H, Liu F, Pan Y, Zhu J, Liao Z, Chen X, Zhang B. Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma. Biomarker Res. 2022;10(1):2. doi:10.1186/s40364-021-00345-1.
- Lee K, Gollahon LS. ZSCAN4 and TRF1: a functionally indirect interaction in cancer cells independent of telomerase activity. Biochem Bioph Res Co. 2015;466(4):644–9. doi:10.1016/j.bbrc.2015.09.107.
- Zhang X, Bai Y, Huang L, Liu S, Mo Y, Cheng W, Wang G, Cao Z, Chen X, Cui H, et al. CHD1L augments autophagy-mediated migration of hepatocellular carcinoma through targeting ZKSCAN3. Cell Death Disease. 2021;12(10):950. doi:10.1038/s41419-021-04254-x.
- Kim CW, Roh SA, Tak KH, Koh BM, Ha YJ, Cho DH, Kim SY, Kim YS, Kim JC. ZKSCAN3 facilitates liver metastasis of colorectal cancer associated with CEA-expressing tumor. Anticancer Res. 2016;36(5):2397–2406.
- Li Q, Luo H, Dai FQ, Wang RT, Fan XQ, Luo YY, Deng M-S, Wang Y, Long T, Guo W, et al. SAMD9 promotes postoperative recurrence of esophageal squamous cell carcinoma by stimulating MYH9-mediated GSK3β/β-catenin signaling. Adv Sci (Weinheim, Baden-Wurttemberg, Germany). 2023;10(11):e2203573. doi:10.1002/advs.202203573.
- Hu S, Ren S, Cai Y, Liu J, Han Y, Zhao Y, Yang J, Zhou X, Wang X. Glycoprotein PTGDS promotes tumorigenesis of diffuse large B-cell lymphoma by MYH9-mediated regulation of wnt–β-catenin–STAT3 signaling. Cell Death Differ. 2022;29(3):642–656. doi:10.1038/s41418-021-00880-2.
- Zhou P, Li Y, Li B, Zhang M, Liu Y, Yao Y, Li D. NMIIA promotes tumor growth and metastasis by activating the Wnt/β-catenin signaling pathway and EMT in pancreatic cancer. Oncogene. 2019;38(27):5500–5515. doi:10.1038/s41388-019-0806-6.
- Thool M, Sundaravadivelu PK, Sudhagar S, Thummer RP. A comprehensive review on the role of ZSCAN4 in embryonic development, stem cells, and cancer. Stem Cell Rev Rep. 2022;18(8):2740–56. doi:10.1007/s12015-022-10412-1.
- Wang Y, Luo Y, Fu S, He L, Pan G, Fan D, Wen Q, Fan Y. Zinc finger and SCAN domain-containing protein 18 is a potential DNA methylation-modified tumor suppressor and biomarker in breast cancer. Front Endocrinol (Lausanne). 2023;14:1095604. doi:10.3389/fendo.2023.1095604.
- Srinivasan R, Nady N, Arora N, Hsieh LJ, Swigut T, Narlikar GJ, Wossidlo M, Wysocka J. Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Sci Adv. 2020;6(12):eaaz9115. doi:10.1126/sciadv.aaz9115.
- Cho YE, Kim JH, Che YH, Kim YJ, Sung JY, Kim YW, Choe B-G, Lee S, Park J-H. Role of the WNT/β-catenin/ZKSCAN3 pathway in regulating chromosomal instability in colon cancer Cell lines and tissues. Int J Mol Sci. 2022;23(16):9302. doi:10.3390/ijms23169302.
- Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang H, Song X, Han D, Wang X, Liu Y, et al. Circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta-catenin pathway. Mol Ther. 2022;30(1):415–30. doi:10.1016/j.ymthe.2021.08.026.
- Kai JD, Cheng LH, Li BF, Kang K, Xiong F, Fu JC, Wang S. MYH9 is a novel cancer stem cell marker and prognostic indicator in esophageal cancer that promotes oncogenesis through the PI3K/AKT/mTOR axis. Cell Biol Int. 2022;46(12):2085–2094. doi:10.1002/cbin.11894.
- Zhao R, Ge Y, Gong Y, Li B, Xiao B, Zuo S. NAP1L5 targeting combined with MYH9 inhibit HCC progression through PI3K/AKT/mTOR signaling pathway. Aging. 2022;14(22):9000–19. doi:10.18632/aging.204377.
- Yang SB, Zhou ZH, Lei J, Li XW, Chen Q, Li B, Zhang Y-W, Ge Y-Z, Zuo S. TM4SF1 upregulates MYH9 to activate the NOTCH pathway to promote cancer stemness and lenvatinib resistance in HCC. Biol Direct. 2023;18(1):18. doi:10.1186/s13062-023-00376-8.
- Eubelen M, Bostaille N, Cabochette P, Gauquier A, Tebabi P, Dumitru AC, Koehler M, Gut P, Alsteens D, Stainier DYR, et al. A molecular mechanism for Wnt ligand-specific signaling. Sci (New York, NY). 2018;361(6403). doi:10.1126/science.aat1178.
- Zhao H, Ming T, Tang S, Ren S, Yang H, Liu M, Tao Q, Xu H. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer. 2022;21(1):144. doi:10.1186/s12943-022-01616-7.
- Kaur A, Lim JYS, Sepramaniam S, Patnaik S, Harmston N, Lee MA, Petretto E, Virshup DM, Madan B. WNT inhibition creates a BRCA-like state in Wnt-addicted cancer. EMBO Mol Med. 2021;13(4):e13349. doi:10.15252/emmm.202013349.
- Angers S. Wnt signaling inhibition confers induced synthetic lethality to PARP inhibitors. EMBO Mol Med. 2021;13(4):e14002. doi:10.15252/emmm.202114002.
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–4. doi:10.1038/nature11282.
- Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, Saksena G, Lawrence MS, Qian ZR, Nishihara R, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46(12):1264–6. doi:10.1038/ng.3127.
- Gao A, Bai P, Zhang M, Yao Y, Herman JG, Guo M. RASSF1A promotes ATM signaling and RASSF1A methylation is a synthetic lethal marker for ATR inhibitors. Epigenomics. 2023;15(22):1205–20. doi:10.2217/epi-2023-0306.
- Li H, Yang W, Zhang M, He T, Zhou F, GH J, Hu L, Guo M. Methylation of TMEM176A, a key ERK signaling regulator, is a novel synthetic lethality marker of ATM inhibitors in human lung cancer. Epigenomics. 2021;13(17):1403–1419. doi:10.2217/epi-2021-0217.
- Du W, Gao A, Herman JG, Wang L, Zhang L, Jiao S, Guo M. Methylation of NRN1 is a novel synthetic lethal marker of PI3K-Akt-mTOR and ATR inhibitors in esophageal cancer. Cancer Sci. 2021;112(7):2870–2883. doi:10.1111/cas.14917.
- Li H, Zhang M, Linghu E, Zhou F, Herman JG, Hu L, Guo M. Epigenetic silencing of TMEM176A activates ERK signaling in human hepatocellular carcinoma. Clin Epigenetics. 2018;10(1):137. doi:10.1186/s13148-018-0570-4.
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93(18):9821–9826. doi:10.1073/pnas.93.18.9821.
- Yang W, Guo C, Herman JG, Zhu C, Lv H, Su X, Zhang L, Zhang M, Guo M. Epigenetic silencing of JAM3 promotes esophageal cancer development by activating Wnt signaling. Clin Epigenetics. 2022;14(1):164. doi:10.1186/s13148-022-01388-3.
- Liu Y, Zhang M, He T, Yang W, Wang L, Zhang L, Guo M. Epigenetic silencing of IGFBPL1 promotes esophageal cancer growth by activating PI3K-AKT signaling. Clin Epigenetics. 2020;12(1):22. doi:10.1186/s13148-020-0815-x.
- Yan W, Wu K, Herman JG, Brock MV, Fuks F, Yang L, Zhu H, Li Y, Yang Y, Guo M, et al. Epigenetic regulation of DACH1, a novel Wnt signaling component in colorectal cancer. Epigenetics. 2013;8(12):1373–83. doi:10.4161/epi.26781.
- Wang Y, He T, Herman JG, Linghu E, Yang Y, Fuks F, Zhou F, Song L, Guo M. Correction to: methylation of ZNF331 is an independent prognostic marker of colorectal cancer and promotes colorectal cancer growth. Clin Epigenetics. 2018;10:36. doi:10.1186/s13148-018-0467-2.
