ABSTRACT
Non-small cell lung cancer (NSCLC) is among the most difficult malignancies to treat. Type III collagen (COL3A1) can affect the progression and chemoresistance development of NSCLC. We herein explored the mechanism that drives COL3A1 dysregulation in NSCLC. Potential RNA-binding proteins (RBPs) and transcription factors (TFs) that could bind to COL3A1 were searched by bioinformatics. mRNA expression was detected by quantitative PCR. Protein expression was evaluated using immunoblotting and immunohistochemistry. The effects of the variables were assessed by gauging cell growth, invasiveness, migratory capacity, apoptosis, and cisplatin (DDP) sensitivity. The direct YY1/COL3A1 relationship was confirmed by ChIP and luciferase reporter experiments. Xenograft experiments were done to examine COL3A1’s function in DDP efficacy. COL3A1 showed enhanced expression in DDP-resistant NSCLC. In H460/DDP and A549/DDP cells, downregulation of COL3A1 exerted inhibitory functions in cell growth, invasiveness, and migration, as well as promoting effects on cell DDP sensitivity and apoptosis. Mechanistically, ELAV-like RNA binding protein 1 (ELAVL1) enhanced the mRNA stability and expression of COL3A1, and Yin Yang 1 (YY1) promoted the transcription and expression of COL3A1. Furthermore, upregulation of COL3A1 reversed ELAVL1 inhibition- or YY1 deficiency-mediated functions in DDP-resistant NSCLC cells. Additionally, COL3A1 downregulation enhanced the anti-tumor efficacy of DDP in vivo. Our investigation demonstrates that COL3A1 upregulation, induced by both RBP ELAVL1 and TF YY1, exerts important functions in phenotypes of NSCLC cells with DDP resistance, offering an innovative opportunity in the treatment of drug-resistant NSCLC.
Introduction
As the most prevalent subtype of malignant lung tumor, non-small cell lung cancer (NSCLC) is among the most difficult malignancies to treat.Citation1 Due to the absence of clinical presentations, NSCLC patients are confirmed at the advanced stage or metastasis stage and have the poorest prognosis.Citation2 In China, the advance of screening programs has elevated the survival rates of NSCLC due to the increase in early detection.Citation3 Furthermore, substantial improvements in therapeutic regimens, including chemoradiotherapy and neoadjuvant chemoimmunotherapy, have resulted in a remarkable increase in survival times and transformed outcomes for numerous NSCLC cases.Citation3 Nevertheless, chemotherapeutic regimens have limited utility in curbing these NSCLC cases with drug resistance.Citation1 Unveiling disease molecular biology has been generally accepted as an innovative opportunity for the development of targeted therapies against drug-resistant NSCLC.
Type III collagen (COL3A1), a crucial member of extracellular matrix proteins, is the main structural element in various organs and is involved in fibrosis and wound healing process.Citation4 Tumor-associated fibroblasts contribute to cancer microenvironment. Some molecules (including COL3A1) produced by these fibroblasts are related to tumorigenesis and metastasis and have been found to be probable targets for cancer therapy.Citation5 In breast cancer, radiation therapy leads to a striking elevation of COL3A1 expression.Citation6 In gastric cancer, upregulation of COL3A1 has close relevance to poor prognostic significance.Citation7 COL3A1 is also identified as a promising independent predictor for prognosis of head and neck cancer and colorectal cancer.Citation8,Citation9 Abnormal expression of COL3A1 mediated by RNF185 exhibits a potent regulatory activity in the metastatic potential of prostate cancer.Citation10 Moreover, in triple negative breast cancer, knocking down COL3A1 plays a repressive role in cancer cell growth and metastasis depending on PD-L1 reduction.Citation11 In lung cancer, COL3A1 is deregulated and is associated with patient survival outcomes.Citation12,Citation13 COL3A1-positive endothelial cells can affect tumor immune microenvironment, thereby modulating NSCLC development.Citation14 Importantly, high COL3A1 expression is found in NSCLC cells with cisplatin (DDP) resistance, which suggests the implication of COL3A1 in chemoresistance development of NSCLC.Citation15 These lead us to investigate the mechanism that drives COL3A1 upregulation in NSCLC.
RNA-binding proteins (RBPs) are vital for gene expression cascade at the post-transcriptional level by controlling RNA metabolism.Citation16 A previous report noted the post-transcriptional modulation of RBPs hnRNP A1, E1, and K in COL3A1.Citation17 Transcription factors (TFs) guide the transcription and expression of the genome by recognizing gene promoter.Citation18 Moreover, TF STAT1 can activate COL3A1 transcription and induce its expression, thereby sustaining cancer cell dormancy.Citation19 Nonetheless, the RBPs and TFs driving COL3A1 expression have yet to be elucidated in NSCLC development and drug resistance processes.
The current investigation first elucidated the biological action of COL3A1 in cell phenotypes and drug sensitivity of DDP-resistant NSCLC cells. Mechanistically, we demonstrated the molecular determinants modulating the expression level of COL3A1 from two aspects: RBPs (post-transcriptional modulation) and TFs (transcriptional regulation). Our results identify ELAV-like RNA binding protein 1 (ELAVL1), a vital RBP in controlling mRNA stability,Citation20 as a crucial post-transcriptional promoter of COL3A1 and Yin Yang 1 (YY1), a cancer target TF,Citation21 as a transcriptional activator of COL3A1 in NSCLC. Our research provides a strong theoretical basis for the development of COL3A1-basic targeted therapies against DDP-resistant NSCLC.
Materials and methods
Bioinformatics
We utilized the GEO DataSet GSE21656 (https://www.ncbi.nlm.nih.gov/search/all/?term=GSE21656) to find out the aberrantly expressed factors in H460 NSCLC cells with DDP resistance (H460/DDP) versus its sensitive parents. We used the TCGA dataset (https://www.cancer.gov/ccg/research/genome-sequencing/tcga) and ENCORI project (https://rnasysu.com/encori/) to examine the expression patterns of COL3A1, ELAVL1, and YY1 in lung adenocarcinoma (LUAD) specimens versus normal lung specimens. The ENCORI project and GEPIA web (http://gepia2.cancer-pku.cn/#index) were utilized to examine the expression relationship between COL3A1 and ELAVL1 or YY1. We also utilized ENCORI online project to predict the potential RBPs of COL3A1 and Jaspar online project (https://jaspar.elixir.no/) to search TFs that can bind to the COL3A1 promoter.
Patient specimens
We collected clinical NSCLC tumors from two patient cohorts at Affiliated Hospital of Jiangxi University of Chinese Medicine. The first one was from NSCLC patients (n = 20) whose tumors had not further progressed after DDP-based first-line chemotherapy (the tumors were regarded as sensitive specimens). The second was from patients (n = 21) with cancer progression after DDP-based chemotherapy (the tumors were regarded as DDP-resistant NSCLC tumors). Each tumor specimen was paired with a normal lung tissue control from the same case. Their clinicopathologic features are presented in Supplementary Table S1. Human samples were obtained at the time of surgical operation with approval by the Institutional Ethics Committee for Human Research at Affiliated Hospital of Jiangxi University of Chinese Medicine. Each patient provided formal consent.
Cell lines
We procured two NSCLC cell lines A549 (#CL-0016) and H460 (#CL-0299) from Procell (Wuhan, China) and cultivated them under the recommended media (Ham’s F-12K for A549 and RPMI-1640 for H460) containing 10% FBS and 1% Penicillin-Streptomycin, which were obtained from the manufacturer (Procell), at conventional conditions (37°C, 85% humidity, 5% carbon dioxide). As a control, we utilized the normal HBE cell line (#CL-0346, Procell), which was grown in 10% FBS RPMI-1640 medium (Servicebio, Wuhan, China).
To study the effect on DDP resistance, we also procured A549 cells with DDP resistance (A549/DDP, #CL-0519) from Procell. The H460/DDP cell line derived from H460 cell line was generated and maintained as described by Wang and colleaguesCitation15 using DDP reagent (Selleck, Shanghai, China).
Transfection and transduction of cell lines
siRNAs employed were as follows: si-COL3A1#1, si-COL3A1#2, si-COL3A1#3 (si-COL3A1), si-ELAVL1, si-YY1, and si-NC, which were made by GenScript (Nanjing, China). ELAVL1, COL3A1, and YY1 expression constructs, which were produced by Tsingke Biotech (Beijing, China), were based on the pcDNA3.1 vehicle. We also procured sh-COL3A1 lentivirus particles and sh-NC controls from Obiosh (Shanghai, China). For transient transfection of H460/DDP and A549/DDP cell lines, we used RFect siRNA Transfection Reagent as recommended by the producer (BaiDai Biotech, Changzhou, China). For in vivo COL3A1 depletion assays, we established an A549/DDP stable cell line with COL3A1 downregulation by infecting the cell line with lentivirus particles. Selection of transduced cells was carried out in media containing puromycin (2 µg/mL) for 2 weeks.
Analysis of mRNA expression
These precise procedures as indicated below were done in accordance with the specifications of the producers. We utilized RNAeasyTM RNA Preparation Kit from Beyotime (Beijing, China) to extract total RNA from cultured cell lines and harvested patient specimens. After that, we used PrimeScript RT Kit (Takara, Otsu, Japan) to perform reverse transcription (RT) PCR and subsequent SYBR Green mix (Takara) to quantify mRNA expression (quantitative PCR assay) of COL3A1, ELAVL1, and YY1 with pre-designed primers (Supplementary Table S2, Tsingke Biotech). mRNA expression (2−ΔΔCt method) was given relative to that in control group.
Analysis of protein expression
We used the immunoblotting method to examine the protein levels of COL3A1, ELAVL1, and YY1 as described.Citation11 Protein extracts were separated by 10%–12% SDS polyacrylamide gels and eletroblotted to nitrocellulose membranes (Servicebio), which were probed with anti-COL3A1 rabbit pAb (#GB111629, Servicebio), anti-ELAVL1 rabbit pAb (#11910–1-AP, Proteintech, Wuhan, China), anti-YY1 rabbit pAb (#22156-1-Ap, Proteintech), or anti-GAPDH rabbit mAb as a loading control (#GB15004, Servicebio). The secondary antibody was anti-rabbit IgG with HRP label (#GB23303, Servicebio).
Analysis of cell viability and growth inhibition
We assessed the viability of transfected cells and the growth inhibition effect of DDP using CCK-8 assay (MedChemExpress, Shanghai, China). In a 96-well culture plate, H460/DDP and A549/DDP cells were subjected to the 24-h relevant transfection before 24-h treatment without (for viability assessment) or with (for growth inhibition assessment) the range of 0–200 µM of DDP. Afterward, CCK-8 reagent (10%) in growth media was added prior to 37°C incubation for 2–3 h. We gauged the optical density at a wavelength of 450 nm and determined the IC50 value from a plot of the percentage of viable cells versus DDP concentration.
Proliferation evaluation by EdU assay
Each cell line after 48 h transfection was subjected to EdU reagent incubation and Apollo567 staining (red) using the Cell-Light EdU Apollo567 Kit as suggested by the producer (Ribobio, Guangzhou, China). Subsequently, we stained the cell nucleus (blue) with a DAPI reagent (Servicebio). We scored the EdU-positive cells as a percentage of total cells after photographing using a fluorescence microscope.
Apoptosis detection by flow cytometry
For apoptosis detection, we applied the Annexin V-FITC/PI Double Staining Kit from Servicebio as per the concomitant specifications. Within 60 min after staining, we read the results using BD FACSAria (BD Biosciences, North Ryde, Australia). We defined the apoptotic rate of transfected cells by calculating the sum of the percentage of early and late apoptotic cells.
Invasiveness evaluation by transwell assay
Under the use of 24-well Matrigel-coated transwell inserts (8.0 µm pore size, BD Biosciences), we determined cell invasiveness after 48-h transfection. Briefly, after transfected H460/DDP and A549/DDP cells were re-suspended with non-serum media, 200 µL of cell suspension (1 × 105 cells) was seeded to Transwell inserts. The cells were allowed to traverse the inserts into 10% FBS medium. Twenty-four hours later, we washed the inserts with PBS and stained them with crystal violet (0.5%). After elimination of non-invaded cells on the upper surface, the number of the invaded cells was counted under bright-field microscopy.
Migration evaluation by wound-healing assay
After a transfection of 48 h, we seeded transfected H460/DDP and A549/DDP cells into a 6-well culture plate. After achieving 100% confluence, we made vertical wounds using 1 mL of sterile tips. The cells were then maintained in regular conditions for 24 h. Using bright-field microscopy, the wounds were photographed at 0 h, 12 h, and 24 h.
Evaluation of mRNA stability via actinomycin D treatment
To examine the influence of ELAVL1 on COL3A1 mRNA stability, we maintained H460/DDP and A549/DDP cells in media containing actinomycin D (Act D, 10 µg/mL, Selleck). At 0, 3, 6, and 9 h, we utilized quantitative PCR to examine intracellular mRNA level of COL3A1.
ChIP experiments
The ChIP experiments were conducted to verify the direct YY1/COL3A1 relationship with BeyoChIPTM Assay Kit as suggested by the producer (Beyotime). After fixation in 1% formaldehyde and incubation with glycine solution, H460/DDP and A549/DDP cells were cleaned with PBS containing inhibitors of protease and phosphatase (Beyotime), lysed (0°C; 10 min) with SDS lysis buffer, and processed by ultrasonication for DNA fragmentation. Lysates were subsequently incubated with 50 µL Protein A/G beads and anti-YY1 rabbit pAb (#22156–1-AP, Proteintech) or IgG control (#GB111738, Servicebio) overnight at 4°C. Utilizing a magnetic separator, we harvested beads, purified DNA using a purification kit, and performed quantitative PCR to detect the enrichment content of the COL3A1 promoter.
Luciferase reporter assay
We cloned the fragment of the COL3A1 promoter carrying the possible pairing sequence (ACCATC, WT) or mutation sequence (GATCGT, MUT) into the pGL3-basic vector (Miaoling, Wuhan, China), respectively, to produce COL3A1 promoter reporter constructs (WT-COL3A1 and MUT-COL3A1). The pRL-TK Renilla vector (for normalization), WT-COL3A1 or MUT-COL3A1, and si-YY1 or si-NC were co-transfected into H460/DDP and A549/DDP cells grown in 24-well culture plates. After 48 h, harvested cells were lysed with lysis buffer, followed by determination of the quotient of firefly/Renilla luciferase activities using the Dual-luciferase Assay System (Promega, Leiden, The Netherlands).
Xenograft experiments and immunohistochemistry
Mouse handling and procedures complied with International Guidelines and approved protocols from the Animal Ethical Commission of Affiliated Hospital of Jiangxi University of Chinese Medicine. BALB/c female nude mice (18–20 g, Cyagen, Suzhou, China) were randomly grouped into designed groups: sh-NC+PBS (n = 5), sh-NC+DDP (n = 5), sh-COL3A1+PBS (n = 5), and sh-COL3A1+DDP (n = 5). Xenograft tumors were generated by subcutaneous injection of the stable A549/DDP cell line (4 × 106 cells/mouse) transduced with sh-COL3A1 lentivirus particles or sh-NC controls into the right dorsal side of nude mice. DDP administration (10 mg/kg) began on 7th day of cell injection and was done every 3 days. Tumor size was periodically evaluated using the (shortest diameter)2 × (longer diameter)/2 formula. At the endpoint of the experiment, we harvested A549/DDP xenograft tumors for the subsequent protein expression analyses using immunoblotting and immunohistochemistry.
For immunohistochemistry assay as described,Citation22 sections of paraffin-embedded xenografts were probed with anti-Ki67 rabbit pAb (#GB121499), anti-MMP9 rabbit pAb (#GB11132, Servicebio) or anti-COL3A1 mouse mAb (#68320–1-Ig, Proteintech) and stained with DAB Kit (Beyotime).
Statistics
We determined the significance using the Mann-Whitney U-test, unpaired t-test, or ANOVA test with Tukey’s post hoc. Significance was regarded to be significant at p < .05. Error bars presented the standard error of the mean of at least three independent repeats.
Results
Enhanced expression of COL3A1 in DDP-resistant NSCLC
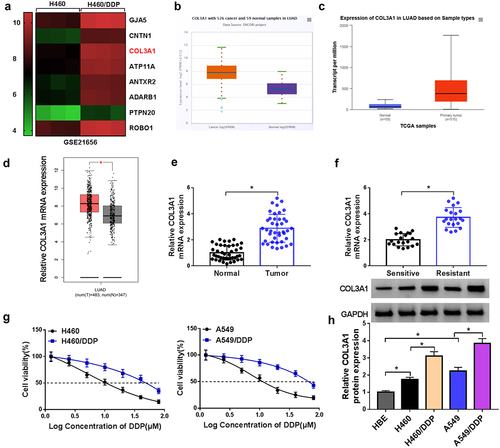
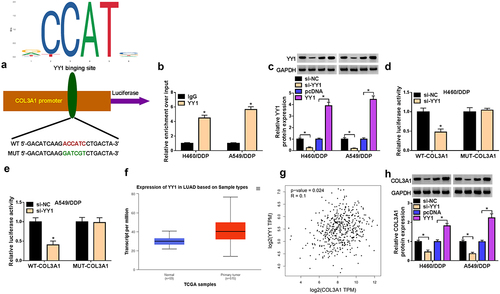
We first examined the expression pattern of COL3A1 in DDP-resistant or nonresistant NSCLC via bioinformatic algorithms. The GSE21656 dataset showed that relative to the sensitive cell line, COL3A1 was highly expressed in DDP-resistant H460 cell line (H460/DDP) (). The TCGA dataset and ENCORI project revealed the upregulation of COL3A1 in lung adenocarcinoma (LUAD) samples versus normal lung specimens (). We further collected the clinical specimens, including NSCLC tumors that had not further progressed after DDP-based first-line chemotherapy (20 cases, we defined them as sensitive samples), samples from patients with cancer progression after DDP-based chemotherapy (21 cases, we defined them as DDP-resistant samples), and adjacent non-cancerous lung specimens (41 cases) from 41 NSCLC patients to verify the overexpression of COL3A1. Through quantitative PCR, we found that the mRNA expression of COL3A1 was enhanced in NSCLC tumors (), and DDP-resistant tumors had higher mRNA levels of COL3A1 than sensitive tumors (). To demonstrate the findings, we also generated two NSCLC cell lines resistant to DDP (H460/DDP and A549/DDP) and validated their successful establishment, revealed by the elevated IC50 value for DDP (). As expected, in agreement with tissue expression, COL3A1 was highly expressed in NSCLC cells at protein level versus normal HEB cells; relative to their respective parent cells, H460/DDP and A549/DDP cell lines exhibited enhanced protein expression of COL3A1 (). In support of these findings, we observed that exposure of DDP resulted in elevated expression of COL3A1 protein in A549 cells (Supplementary Figure S1).
Figure 1. COL3A1 is upregulated in NSCLC as a result of DDP resistance.
Downregulation of COL3A1 modulates the growth, invasiveness, motility, and apoptosis of H460/DDP and A549/DDP cells and sensitizes them to DDP
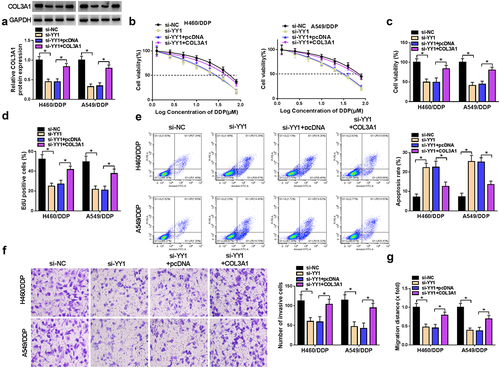
We transfected H460/DDP and A549/DDP cells with three COL3A1-siRNAs (si-COL3A1#1, si-COL3A1#2, and si-COL3A1#3), respectively, or si-NC mock. Transfection of si-COL3A1#3 (also called si-COL3A1) and si-COL3A1#1 caused the more significant downregulation in COL3A1 protein expression and the more significant repression in cell viability (Supplementary Figure S2A and S2B). Therefore, in this study, we used si-COL3A1 and si-COL3A1#1 to deplete COL3A1 in H460/DDP and A549/DDP cells ( and Supplementary Figure S2A). The CCK-8 assay presented that COL3A1 inhibition reduced the IC50 value for DDP of the two cell lines ( and Supplementary Figure S2C). Additionally, in the presence of DDP exposure, inhibition of COL3A1 impaired cell growth, induced cell apoptosis, as well as hindered cell migration and invasion in H460/DDP and A549/DDP cells (Supplementary Figure S3). These data indicate that COL3A1 inhibition sensitizes the two resistant cells to DDP.
Figure 2. COL3A1 inhibition results in a disadvantage of growth and metastasis and an induction of DDP sensitivity and apoptosis.
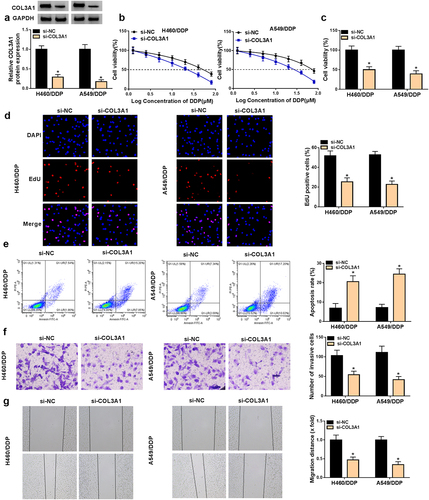
Furthermore, COL3A1 inhibition weakened cell viability and proliferation, but induced apoptosis ( and Supplementary Figure S2D and S2E). We also examined the regulation of COL3A1 in cell invasiveness and migratory ability. Notably, downregulated COL3A1 level caused a repression in cell invasiveness and motility in both H460/DDP and A549/DDP cell lines ( and Supplementary Figure S2F and S2G). Therefore, we conclude that knocking down COL3A1 exerts inhibitory functions in cell malignant phenotypes and promoting effects on cell DDP sensitivity and apoptosis.
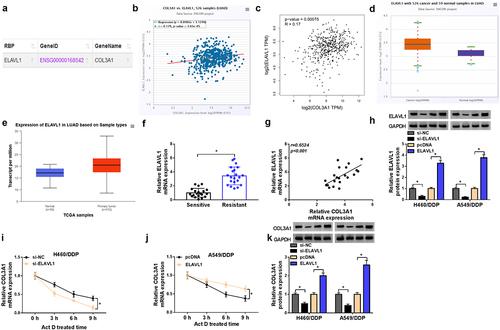
RBP ELAVL1 induces augmentation of COL3A1 expression by enhancing COL3A1 mRNA stability
The post-transcriptional modulation of COL3A1 by RNA-binding proteins (RBPs) has been previously reported.Citation17 Having highlighted the regulation of COL3A1 in malignant phenotypes of DDP-resistant NSCLC cells, we wanted to determine whether COL3A1 upregulation can be caused by certain RBPs. We utilized ENCORI online algorithm to predict the potential RBPs of COL3A1 and found that there were a lot of RBPs that can bind to COL3A1. We then selected several candidate RBPs (ELAVL1, METTL3, FUS, and IGF2BP1) that have been shown to be highly expressed in NSCLC and can function as an oncogenic driver in NSCLC. Through qPCR analysis, we found that only ELAVL1 significantly affected COL3A1 expression in H460/DDP and A549/DDP cells (Supplementary Figure S4). ELAVL1, a vital RBP in controlling mRNA stability,Citation20 was predicted as a putative RBP of COL3A1 using ENCORI algorithm (). Via the data source of ENCORI project, we also observed a positive expression association of COL3A1 and ELAVL1 in LUAD samples (), which was also confirmed by the data of GEPIA database (). The ENCORI project and TCGA database showed the upregulation of ELAVL1 in LUAD samples relative to normal lung tissues (). In DDP-resistant NSCLC tumors, our quantitative PCR data revealed the high expression of ELAVL1 mRNA and a significantly positive association between COL3A1 and ELAVL1 (). Furthermore, we examined the regulation of ELAVL1 in the mRNA stability and protein expression of COL3A1 in H460/DDP and A549/DDP NSCLC cells. The transfection efficiencies of ELAVL1-siRNA (si-ELAVL1) and ELAVL1 expression plasmid were validated by immunoblotting (). Under treatment of actinomycin D (Act D), COL3A1 mRNA stability was weakened by ELAVL1 downregulation and reinforced by the overexpression of ELAVL1 (). Immunoblotting data showed the positive modulation of ELAVL1 in COL3A1 protein expression, as evidenced by COL3A1 increase following ELAVL1 upregulation and COL3A1 downregulation after ELAVL1 inhibition ().
Figure 3. ELAVL1 enhances COL3A1 mRNA stability in H460/DDP and A549/DDP cells.
Upregulation of COL3A1 reverses ELAVL1 inhibition-mediated functions in H460/DDP and A549/DDP cells
Having demonstrated the suppression of ELAVL1 downregulation in COL3A1 expression (), we wanted to determine the role of ELAVL1 in cell phenotypes. ELAVL1 inhibition reduced the IC50 value for DDP of H460/DDP and A549/DDP NSCLC cells (). Inhibition of ELAVL1 also impeded cell viability () and proliferation () and induced apoptosis (), as well as diminished cell invasiveness and migratory capacity ().
Figure 4. ELAVL1-induced COL3A1 upregulation affects DDP sensitivity and NSCLC cell phenotypes.
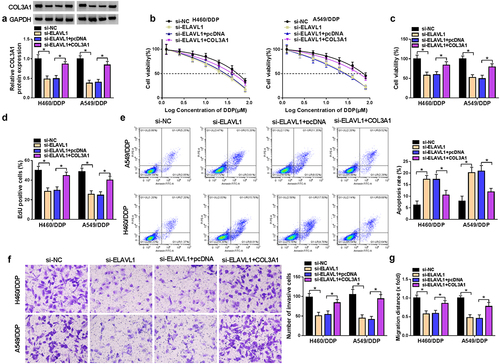
The striking resemblance for the regulatory functions of COL3A1 and ELAVL1 prompted us to examine COL3A1 as a downstream effector of ELAVL1. To address this hypothesis, we elevated COL3A1 expression by COL3A1 expression plasmid introduction in ELAVL1-silenced H460/DDP and A549/DDP cells (). The re-expression of COL3A1 significantly counteracted the influence of ELAVL1 inhibition in sensitizing cells to DDP (). More importantly, the re-expression of COL3A1 offsets ELAVL1 inhibition-caused alterations in cell viability (), proliferation (), apoptosis (), invasiveness, and migratory capacity ( and Supplementary Figure S5). Thus, we conclude that ELAVL1-induced COL3A1 upregulation modulates DDP sensitivity and NSCLC cell phenotypes.
TF YY1 enhances the transcription and expression of COL3A1
We next employed the Jaspar online project to search TFs that can bind to the COL3A1 promoter and found that YY1, a cancer-related target TF,Citation21 might bind to the COL3A1 promoter via a putative pairing site (ACCATC) (). To demonstrate this, we utilized the antibody against YY1 to perform ChIP experiments. Results revealed that in contrast to the IgG control, the incubation of the anti-YY1 antibody drastically elevated the enrichment level of the COL3A1 promoter (), indicating the direct YY1/COL3A1 relationship. The transfection efficiencies of si-YY1 and YY1 expression plasmid were first confirmed by Immunoblotting (). From luciferase reporter assays using pGL3-basic constructs, in which the COL3A1 promoter fragment harbors the pairing sequence, we found that downregulation of YY1 led to a clear suppression of the luciferase activity of cells transfected with wild-type (WT) COL3A1 reporter constructs; downregulation of YY1 did not affect the luciferase activity of mutant (MUT) COL3A1 reporters (). Moreover, TCGA database revealed the overexpression of YY1 in LUAD samples (), and GEPIA online project showed the expression correlation between COL3A1 and YY1 (). Importantly, in H460/DDP and A549/DDP cells, COL3A1 protein expression was diminished by downregulation of YY1, while it was conversely augmented by elevated YY1 level ().
Figure 5. YY1 promotes COL3A1 transcription by binding to COL3A1 promoter and upregulates its expression.
Upregulation of COL3A1 reverses YY1 deficiency-mediated regulation in H460/DDP andA549/DDP cells
We further explored if YY1 can modulate cell phenotypes by the upregulation of COL3A1. The co-introduction of COL3A1 expression construct remarkably abated si-YY1-caused deficiency in COL3A1 protein level in both H460/DDP and A549/DDP cell lines (). The IC50 value for DDP was diminished in YY1-silenced cells; however, COL3A1 re-expression reversed the reduction by YY1 silencing (). YY1-silenced cells exhibited suppressed cell viability () and proliferation (), enhanced apoptosis (), as well as attenuated invasiveness () and migratory ability () in the two cell lines; however, re-expression of COL3A1 abrogated these impacts ().
Figure 6. YY1-induced COL3A1 upregulation modulates DDP sensitivity and NSCLC cell phenotypes.
Downregulation of COL3A1 enhances the anti-tumor efficacy of DDP in vivo
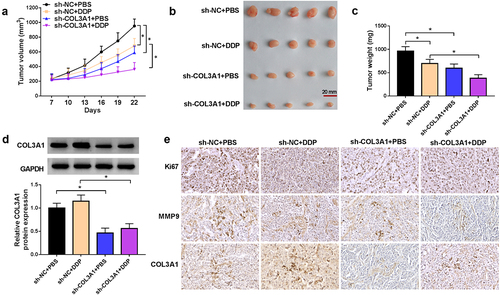
Finally, we carried out mouse xenograft studies to examine the function of COL3A1 in DDP efficacy. Separate administration of DDP or sh-COL3A1 lentivirus dramatically hindered xenograft growth; combination of DDP and sh-COL3A1 lentivirus led to a more striking repression in A549/DDP xenograft growth (). In the presence and absence of sh-COL3A1 lentivirus administration, DDP treatment did not affect COL3A1 protein level in A549/DDP xenografts (). Through immunohistochemistry assay, we also validated that DDP treatment or sh-COL3A1 transduction reduced the cells stained for Ki67 and suppressed MMP9 expression in A549/DDP xenografts; combination of DDP and sh-COL3A1 lentivirus led to a more significant change in the two factors ().
Figure 7. COL3A1 downregulation enhances the anti-tumor efficacy of DDP in A549/DDP xenografts.
Discussion
Development of drug resistance is increasingly being thought to be a great obstacle for successful treatment in most cancers, including NSCLC.Citation23 A series of studies have cataloged the implication of deregulated proteins in drug resistance of NSCLC.Citation24,Citation25 Previous work unveils the important role of COL3A1 in DDP resistance development of NSCLC,Citation15 but its regulatory factors are not fully understood. In this work, we focused on COL3A1 and found that COL3A1 expression is enhanced in DDP-resistant NSCLC. Furthermore, we demonstrated that COL3A1, upregulated by both RBP ELAVL1 and TF YY1, remarkably affects drug sensitivity and malignant phenotypes in DDP-resistant NSCLC cells, pointing to a crucial role of COL3A1 in NSCLC.
Elevated COL3A1 expression has been found in various cancers, such as gastric cancer, esophageal cancer, and colorectal cancer,Citation7,Citation9,Citation26 which highlights the potential role of COL3A1 as a promising predictor for these diseases prognosis. In view of function, knocking down COL3A1 can lead to a potent repression of gastric tumorigenesisCitation27; COL3A1 downregulation is able to hinder breast cancer cell growth and invasiveness through PD-L1Citation11; inhibition of COL3A1 also impedes xenograft tumor growth and metastasis in esophageal squamous cell carcinoma.Citation28 These findings prove the tumor-promoting role of COL3A1 in these cancers. Importantly, in NSCLC, COL3A1 also modulates this disease progression by accelerating cancer cell growth and hindering apoptosis.Citation15 Additionally, Wang et al. illustrated the enhanced COL3A1 expression in NSCLC cells with DDP resistance.Citation15 Therefore, we utilized two NSCLC cells with DDP resistance (H460/DDP and A549/DDP) to examine the action of COL3A1 in NSCLC cell chemoresistance. We found that COL3A1 downregulation provides anti-cancer effects in vitro and elevates the anti-tumor efficacy of DDP in vivo, indicating that inhibition of COL3A1 has a possible value for treatment of DDP-resistant NSCLC. Additionally, our data highlighted the suppressive effect of COL3A1 depletion on the cell invasion of A549 and H460 NSCLC cells (Supplementary Figure S6), suggesting the anti-invasion function of COL3A1 inhibition in NSCLC.
ELAVL1 (also called HuR), an important RBP in controlling mRNA stability,Citation20 exerts a strong tumorigenic function in cancer biology by controlling key oncogenic pathways and causing abnormal gene expression programs.Citation29,Citation30 For example, in hepatocellular carcinoma, ELAVL1 augmentation contributes to cancer cell growth, and it is related to post-operative recurrence in patients with hepatitis B virus-positive cancer.Citation31 The abundant level of ELAVL1 results in enhanced cell proliferation in prostate cancer in an m6A-dependent pattern.Citation30 In lung cancer, upregulated ELAVL1 can induce cancer cell growth, invasiveness enhancement, and 5-fluorouracil resistance development.Citation32 By functioning as a decoy of ELAVL1, lncRNA OSER1-AS1 is a tumor-repressive factor in NSCLC.Citation33 Furthermore, ELAVL1 augments the expression of the oncogene LINC00366 by enhancing its stability via the post-transcriptional regulatory manner in lung cancer.Citation34 Utilizing ENCORI project, we found that ELAVL1 might be a potential RBP in regulating COL3A1, which is demonstrated by the fact that ELAVL1 enhances the mRNA stability and expression of COL3A1 in H460/DDP and A549/DDP cells. More interestingly, our rescue experiments proved that ELAVL1-induced COL3A1 upregulation modulates DDP sensitivity and NSCLC cell phenotypes.
YY1, a ubiquitously expressed TF implicated in diverse biological processes, is a critical factor in human NSCLC.Citation35,Citation36 The dual role of YY1 in NSCLC as an anti-tumor factor or a promoting driver by exerting its transcriptional regulation function has been supported by numerous researches.Citation37,Citation38 As an example, YY1-mediated LINC01089 reduction leads to enhanced progression of lung cancer via promotion of growth and metastasis.Citation39 YY1-induced β-catenin transcriptional activation is crucial for the sustainability of the properties of NSCLC stem cells.Citation40 Conversely, the suppression of YY1 in the oncogenic factor DCBLD1 expression after the variant rs17079281C>T can cause a risk reduction in LUAD.Citation41 Our findings for the first time established the YY1/COL3A1 relationship that YY1 can bind to the COL3A1 promoter to activate the transcription of COL3A1 and augment its expression in NSCLC cells with DDP resistance, thereby affecting these cell phenotypes and DDP sensitivity. In addition, the in vivo assays showed that DDP can enhance ELAVL1 protein expression and does not affect YY1 expression, and COL3A1 does not affect the protein expression of ELAVL1 and YY1 (Supplementary Figure S7). These findings together suggest that COL3A1 can be regulated by RBP ELAVL1 and TF YY1, and COL3A1 cannot in turn affect the protein expression of ELAVL1 and YY1. The in vivo assays also showed the regulation of COL3A1 in metastasis-related marker MMP9 expression, suggesting that COL3A1 may influence tumor metastasis status. Related research is warranted in future work via various animal experimental models. In clinical, the combination chemotherapy of DDP and pemetrexed is the most commonly used first-line chemotherapy regimen for NSCLC. We collected these clinical tumor specimens from patients who received DDP-based first-line chemotherapy combined with pemetrexed. The possibility that pemetrexed application could affect the expression of COL3A1 and ELAVL1 in NSCLC samples is a major limitation of this study.
In summary, our current investigation demonstrates that COL3A1 upregulation, induced by both RBP ELAVL1 at the post-transcriptional level and TF YY1 in transcriptional regulation, exerts important functions in cell growth, invasiveness, migratory ability, and DDP sensitivity of NSCLC cells with DDP resistance. Elucidating this mechanism will offer an innovative opportunity in the treatment of drug-resistant NSCLC.
Highlights
ELAVL1 enhances the mRNA stability and expression of COL3A1.
YY1 promotes the transcription and expression of COL3A1.
COL3A1 upregulation induced by ELAVL1 and YY1 exerts an important function in DDP-resistant NSCLC.
Authors’ contributions
Songwei Zhao designed and performed the research; Junyu Lai analyzed the data; Jiankun Ren wrote the manuscript. All authors read and approved the final manuscript.
Animal studies
Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Consent for publication
Written informed consents were obtained from all participants.
Data availability material
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Written informed consents were obtained from all participants, and this study was permitted by the Ethics Committee of Affiliated Hospital of Jiangxi University of Chinese Medicine.
Supplemental Material
Download Zip (8.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2024.2328382
Additional information
Funding
References
- Slatore C, Lareau SC, Fahy B. Staging of Lung Cancer. Am J Respir Crit Care Med. 2022;205(9):17–12. doi:10.1164/rccm.2059P17.
- Mithoowani H, Febbraro M. Non-small-cell lung cancer in 2022: a review for general practitioners in oncology. Curr Oncol. 2022;29(3):1828–1839. doi:10.3390/curroncol29030150.
- Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun (Lond). 2022;42(10):937–970. doi:10.1002/cac2.12359.
- Kuivaniemi H, Tromp G. Type III collagen (COL3A1): gene and protein structure, tissue distribution, and associated diseases. Gene. 2019;707:151–171. doi:10.1016/j.gene.2019.05.003.
- Kamali Zonouzi S, Pezeshki PS, Razi S, Rezaei N. Cancer-associated fibroblasts in colorectal cancer. Clin Transl Oncol. 2022;24(5):757–769. doi:10.1007/s12094-021-02734-2.
- Yao G, Zhao K, Bao K, Li J. Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer. Open Med (Wars). 2022;17(1):329–340. doi:10.1515/med-2022-0436.
- Ucaryilmaz Metin C, Ozcan G. Comprehensive bioinformatic analysis reveals a cancer-associated fibroblast gene signature as a poor prognostic factor and potential therapeutic target in gastric cancer. BMC Cancer. 2022;22(1):692. doi:10.1186/s12885-022-09736-5.
- Shen Y, Li X, Wang D, Zhang L, Li X, Su L, Fan X, Yang X. COL3A1: potential prognostic predictor for head and neck cancer based on immune-microenvironment alternative splicing. Cancer Med. 2023;12(4):4882–4894. doi:10.1002/cam4.5170.
- Hosseini ST, Nemati F. Identification of GUCA2A and COL3A1 as prognostic biomarkers in colorectal cancer by integrating analysis of RNA-Seq data and qRT-PCR validation. Sci Rep. 2023;13(1):17086. doi:10.1038/s41598-023-44459-y.
- Van Espen B, Oo HZ, Collins C, Fazil L, Molinolo A, Yip K, Murad R, Gleave M, Ronai ZE. RNF185 control of COL3A1 expression limits prostate cancer migration and metastatic potential. Mol Cancer Res. 2023;22(1):41–54. doi:10.1158/1541-7786.mcr-23-0512.
- Yang F, Lin L, Li X, Wen R, Zhang X. Silencing of COL3A1 represses proliferation, migration, invasion, and immune escape of triple negative breast cancer cells via down-regulating PD-L1 expression. Cell Biol Int. 2022;46(11):1959–1969. doi:10.1002/cbin.11875.
- Chen Y, Pan Y, Ji Y, Sheng L, Du X. Network analysis of differentially expressed smoking-associated mRnas, lncRnas and miRnas reveals key regulators in smoking-associated lung cancer. Exp Ther Med. 2018;16(6):4991–5002. doi:10.3892/etm.2018.6891.
- Yu DH, Ruan XL, Huang JY, Liu XP, Ma HL, Chen C, Hu W-D, Li S. Analysis of the interaction network of hub miRnas-Hub genes, being involved in idiopathic pulmonary fibers and its emerging role in non-small cell lung cancer. Front Genet. 2020;11:11302. doi:10.3389/fgene.2020.00302.
- Zhang M, Liang Y, Song P. COL3A1-positive endothelial cells influence LUAD prognosis and regulate LUAD carcinogenesis by NCL–PI3K–AKT axis. J Gene Med. 2023;26(1):e3573. doi:10.1002/jgm.3573.
- Wang L, Sun Y, Guo Z, Liu H. COL3A1 overexpression associates with poor prognosis and cisplatin resistance in lung cancer. Balkan Med J. 2022;39(6):393–400. doi:10.4274/balkanmedj.galenos.2022.2022-6-16.
- Singh A. RNA-binding protein kinetics. Nat Methods. 2021;18(4):335. doi:10.1038/s41592-021-01122-6.
- Thiele BJ, Doller A, Kähne T, Pregla R, Hetzer R, Regitz-Zagrosek V. RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circ Res. 2004;95(11):1058–1066. doi:10.1161/01.res.0000149166.33833.08.
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT, et al. The human transcription factors. Cell. 2018;172(4):650–665. doi:10.1016/j.cell.2018.01.029.
- Di Martino JS, Nobre AR, Mondal C, Taha I, Farias EF, Fertig EJ, Naba A, Aguirre-Ghiso JA, Bravo-Cordero JJ. A tumor-derived type III collagen-rich ECM niche regulates tumor cell dormancy. Nat Cancer. 2022;3(1):90–107. doi:10.1038/s43018-021-00291-9.
- Rothamel K, Arcos S, Kim B, Reasoner C, Lisy S, Mukherjee N, Ascano M. ELAVL1 primarily couples mRNA stability with the 3′ UTRs of interferon-stimulated genes. Cell Rep. 2021;35(8):109178. doi:10.1016/j.celrep.2021.109178.
- Hosea R, Hillary S, Wu S, Kasim V. Targeting transcription factor YY1 for cancer treatment: Current strategies and future directions. Cancers Basel. 2023;15(13):3506. doi:10.3390/cancers15133506.
- Zhang X, Xu Y, Ma L, Yu K, Niu Y, Xu X, Shi Y, Guo S, Xue X, Wang Y, et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun (Lond). 2022;42(4):287–313. doi:10.1002/cac2.12275.
- Meador CB, Hata AN. Acquired resistance to targeted therapies in NSCLC: updates and evolving insights. Pharmacol Ther. 2020;210:210107522. doi:10.1016/j.pharmthera.2020.107522.
- Wang D, Zhao C, Xu F, Zhang A, Jin M, Zhang K, Liu L, Hua Q, Zhao J, Liu J, et al. Cisplatin-resistant NSCLC cells induced by hypoxia transmit resistance to sensitive cells through exosomal PKM2. Theranostics. 2021;11(6):2860–2875. doi:10.7150/thno.51797.
- Lei T, Xu T, Zhang N, Zou X, Kong Z, Wei C, Wang Z. Anlotinib combined with osimertinib reverses acquired osimertinib resistance in NSCLC by targeting the c-MET/MYC/AXL axis. Pharmacol Res. 2023;188:188106668. doi:10.1016/j.phrs.2023.106668.
- Zhang SW, Zhang N, Wang N. Role of COL3A1 and POSTN on pathologic stages of esophageal cancer. Technol Cancer Res Treat. 2020;19:191533033820977489. doi:10.1177/1533033820977489.
- Han S, Wang Z, Liu J, Wang HD, Yuan Q. miR-29a-3p-dependent COL3A1 and COL5A1 expression reduction assists sulforaphane to inhibit gastric cancer progression. Biochem Pharmacol. 2021;188:188114539. doi:10.1016/j.bcp.2021.114539.
- Zhou J, Yang Y, Zhang H, Luan S, Xiao X, Li X, Fang P, Shang Q, Chen L, Zeng X, et al. Overexpressed COL3A1 has prognostic value in human esophageal squamous cell carcinoma and promotes the aggressiveness of esophageal squamous cell carcinoma by activating the NF-κB pathway. Biochem Biophys Res Commun. 2022;613:193–200. doi:10.1016/j.bbrc.2022.05.029.
- Palomo-Irigoyen M, Pérez-Andrés E, Iruarrizaga-Lejarreta M, Barreira-Manrique A, Tamayo-Caro M, Vila-Vecilla L, Moreno-Cugnon L, Beitia N, Medrano D, Fernández-Ramos D, et al. HuR/ELAVL1 drives malignant peripheral nerve sheath tumor growth and metastasis. J Clin Invest. 2020;130(7):3848–3864. doi:10.1172/jci130379.
- Cai Z, Xu H, Bai G, Hu H, Wang D, Li H, Wang Z. ELAVL1 promotes prostate cancer progression by interacting with other m6A regulators. Front Oncol. 2022;12:12939784. doi:10.3389/fonc.2022.939784.
- Kanzaki H, Chiba T, Kaneko T, Ao J, Kan M, Muroyama R, Nakamoto S, Kanda T, Maruyama H, Kato J, et al. The RNA-Binding protein ELAVL1 regulates hepatitis B virus replication and growth of hepatocellular carcinoma cells. Int J Mol Sci. 2022;23(14):7878. doi:10.3390/ijms23147878.
- Mao G, Mu Z, Wu DA. Exosomal lncRNA FOXD3-AS1 upregulates ELAVL1 expression and activates PI3K/Akt pathway to enhance lung cancer cell proliferation, invasion, and 5-fluorouracil resistance. Acta Biochim Biophys Sin (Shanghai). 2021;53(11):1484–1494. doi:10.1093/abbs/gmab129.
- Xie W, Wang Y, Zhang Y, Xiang Y, Wu N, Wu L, Li C, Cai T, Ma X, Yu Z, et al. Single-nucleotide polymorphism rs4142441 and MYC co-modulated long non-coding RNA OSER1-AS1 suppresses non-small cell lung cancer by sequestering ELAVL1. Cancer Sci. 2021;112(6):2272–2286. doi:10.1111/cas.14713.
- Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, Shi Y, Chen L, Xiao D, Yu F, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26(11):2329–2343. doi:10.1038/s41418-019-0304-y.
- Ali A, Ali A, Devi HS, Daddam JR, Sarwar R, Badrealam FK. The HDAC2/YY1/c-myc signaling axis regulates lung cancer cell migration and proliferation. Environ Toxicol. 2023;38(8):1989–2001. doi:10.1002/tox.23825.
- Zhu Y, Chen B, Pan H, Sun L, Yu T. PLIC11 drives lung cancer progression through regulating the YY1/PIWIL4 axis. Mol Carcinog. 2023;62(4):427–437. doi:10.1002/mc.23496.
- Huang X, Zhou W, Zhang Y. Transcription factor YY1 enhances the stemness of lung cancer cells by stabilizing hypoxia factor HIF-1α under a hypoxic microenvironment. Environ Toxicol. 2021;36(1):114–122. doi:10.1002/tox.23017.
- Huang T, Wang G, Yang L, Peng B, Wen Y, Ding G, Wang Z. Transcription factor YY1 modulates lung cancer progression by activating lncRNA-PVT1. DNA Cell Biol. 2017;36(11):947–958. doi:10.1089/dna.2017.3857.
- Yang R, Liu Z, Cao H, Shi Y. LINC01089, suppressed by YY1, inhibits lung cancer progression by targeting miR-301b-3p/HPDG axis. Cell Biol Toxicol. 2022;38(6):1063–1077. doi:10.1007/s10565-021-09643-8.
- Zheng JY, Zhu T, Zhuo W, Mao XY, Yin JY, Li X, He Y-J, Zhang W, Liu C, Liu Z-Q, et al. eIf3a sustains non-small cell lung cancer stem cell-like properties by promoting YY1-mediated transcriptional activation of β-catenin. Biochem Pharmacol. 2023;213:115616. doi:10.1016/j.bcp.2023.115616.
- Wang Y, Ma R, Liu B, Kong J, Lin H, Yu X, Wang R, Li L, Gao M, Zhou B, et al. SNP rs17079281 decreases lung cancer risk through creating an YY1-binding site to suppress DCBLD1 expression. Oncogene. 2020;39(20):4092–4102. doi:10.1038/s41388-020-1278-4.
