Abstract
The induction and maintenance of pluripotency requires the expression of several core factors at appropriate levels (Oct4, Sox2, Klf4, Prdm14). A subset of these proteins (Oct4, Sox2, Prdm14) also plays crucial roles for the establishment of primordial germ cells (PGCs). Here we demonstrate that the Mad2l2 (MAD2B, Rev7) gene product is not only required by PGCs, but also by pluripotent embryonic stem cells (ESCs), depending on the growth conditions. Mad2l2−/− ESCs were unstable in LIF/serum medium, and differentiated into primitive endoderm. However, they could be stably propagated using small molecule inhibitors of MAPK signaling. Several components of the MAPK cascade were up- or downregulated even in undifferentiated Mad2l2−/− ESCs. Global levels of repressive histone H3 variants were increased in mutant ESCs, and the epigenetic signatures on pluripotency-, primitive endoderm-, and MAPK-related loci differed. Thus, H3K9me2 repressed the Nanog promoter, while the promoter of Gata4 lost H3K27me3 and became de-repressed in LIF/serum condition. Promoters associated with genes involved in MAPK signaling also showed misregulation of these histone marks. Such epigenetic modifications could be indirect consequences of mutating Mad2l2. However, our previous observations suggested the histone methyltransferases as direct (G9a) or indirect (Ezh2) targets of Mad2l2. In effect, the intricate balance necessary for pluripotency becomes perturbed in the absence of Mad2l2.
Introduction
Pluripotency is a transient state between totipotency and lineage differentiation in the early embryo. It defines the potential of cells for multi-lineage differentiation, however excluding the extraembryonic ectoderm. Pluripotent cells derived from the inner cell mass (ICM) of the blastocyst can be cultured in vitro as embryonic stem cells (ESCs), if growth conditions support unlimited self-renewal and inhibit differentiation. Oct4, Nanog and Sox2, represent the core circuit of transcription factors in pluripotent ESCs. They function by regulating their own expression in a positive, auto-regulatory loop, by upregulation of stem cell genes necessary for maintenance of the undifferentiated state, and by suppression of differentiation associated genes.Citation1 Many additional factors were found to be required or at least to support the maintenance of ESCs by affecting transcription, cell signaling or epigenetics.Citation2-Citation5 Chromatin organization is typically open in pluripotent stem cells, and closed in differentiated cells. Thus, the stably repressive histone modification H3K9me3 is less abundant in ESCs compared to the differentiated cells.Citation6,7 Frequent features of ESCs are so-called bivalent loci, which are occupied by both activating (e.g. H3K4me3 or H3K9Ac) and repressive (e.g., H3K27me3) histone marks.Citation8-Citation10 This configuration allows the suppression of multi-lineage differentiation genes and at the same time poises them for activation.Citation11 A bivalent epigenetic signature prevents premature differentiation of ESCs, and at the same time places lineage-specific genes in a “stand-by” status, ready to stop self-renewal and to start differentiation. Upon appropriate stimulation, repressive H3K27me3 is removed from the lineage-specification loci, and activating H3K4me3 facilitates accessibility for RNA polymerase (Pol II).
Murine ESCs can be grown in 2 principally different conditions, which render pluripotent cells distinct transcriptional and epigenetic signatures. Traditionally, a growth medium containing serum as well as the cytokine leukemia inhibitory factor (LIF) is used for ESC culture (LIF/serum condition). LIF induces pluripotency-related genes via at least 2 routes: it activates its transducer STAT3 that in turn induces Klf4, and secondly, LIF maintains Nanog expression via the PI3K pathway.Citation12 A major, required component of the serum is the growth factor BMP4, which activates the Smad pathway to induce inhibitor of differentiation (Id) genes.Citation13 In addition, BMP4 inhibits mitogen activated protein kinase (MAPK) signaling, a major pathway leading to ESC differentiation.Citation14 In LIF/serum grown ESCs, the DNA is efficiently methylated, there is less pausing of Pol II, and the epigenetic signature of developmental control genes is characterized by bivalent modifications. In these cells an extensive transcriptional heterogeneity is observed, since the expression of pluripotency-associated genes varies among individual cells. This variation, together with the bivalency of differentiation genes, causes an altogether metastable status of LIF/serum grown ESCs. More recently, a new regime for ESC culture has been developed using a chemically defined, serum-free medium supplemented with LIF and 2 selective, small molecule inhibitors, which block the activation of the MAPK pathway and repress glycogen synthase kinase-3 (GSK3β), respectively. This so-called “LIF/2i” medium supports the maintenance of ESCs in the absence of feeder cells and serum.Citation15 ESCs adapted in LIF/2i manifest some distinctive features as compared to LIF/serum cells. In general, the total number of transcribed genes is lower, pluripotency markers like Nanog are expressed more uniformly, lower levels of c-Myc and lineage priming transcripts are found, whereas the number of transcripts from metabolic genes is higher. The genome is widely demethylated,Citation16 and Pol II is often stalled close to the promoter (promoter-proximal pausing). LIF/2i ESCs contain only low amounts of the polycomb repressive complex 2 (PRC2). Consequently, the repressive histone H3K27me3, and therefore bivalent modifications are rare.Citation17 Altogether LIF/2i ESCs represent a much more stable population as compared to LIF/serum cells.
The earliest developmental decision of ICM cells regards the acquisition of either an epiblast or a primitive endoderm (PrE) fate. In the 8- to 16-cell morula, Nanog and Gata6 are initially co-expressed, before their expression segregates in the blastula.Citation18 Then, only epiblast cells express the pluripotency markers Nanog and Oct4, and go on to contribute to the formation of all somatic lineages, i.e. ectoderm, mesoderm, and endoderm, as well as the germ cell lineage. Epiblast cells secrete FGF4 that acts on PrE precursors to trigger MAPK signaling via the FGF receptor 2 (Ffgr2), the SH2/SH3 adaptor Grb2, and the transcription factors Erk1/2 and Elk1, which finally activate the PrE markers Gata6 and Gata4.Citation19 Such FGF signaling would cause murine ESCs in culture to differentiate.
Many proteins of importance for pluripotent ESCs are also relevant for the development of PGCs, including Oct4, Sox2 and PRDM14.Citation20,21 This may correspond with the remarkable potential of PGCs from early post-implantation embryos to become pluripotent in vitro under LIF/2i conditions.Citation22 The resulting embryonic germ cells are indistinguishable from ESCs.Citation23 We have recently demonstrated the absolute requirement of PGCs for the multi-functional protein Mad2l2, which influences their cell cycle and epigenetic reprogramming.Citation24 The Mad2l2 protein is distantly related to the spindle checkpoint regulator Mad2 (Mad2l1) and consists mostly of a single domain, the HORMA domain that is known to interact with many proteins.Citation25 One of several reported Mad2l2 functions is to act as an accessory subunit of translesion DNA polymerases. In addition, it affects both FGF and Wnt signaling via functional binding to the transcription factors Elk-1 and TCF4, respectively.Citation26,27 Here, we demonstrate the importance of Mad2l2 for the stable maintenance of pluripotent mouse ESCs, preventing their FGF-dependent deviation into primitive endoderm.
Results
Spontaneous differentiation of Mad2l2−/− ESCs in LIF/serum
Both Mad2l2 mRNA and protein are highly expressed in pluripotent mouse ESCs, and levels diminish during embryoid body (EB) differentiation (). Mad2l2 is expressed at a basal level in most differentiated murine cells, including murine embryonic fibroblasts (MEFs).Citation24 In order to establish Mad2l2 deficient ESCs, blastocysts were harvested from intercrossing of heterozygous Mad2l2 mutants, plated on inactivated MEF feeders, and fed with N2B27 medium supplemented with LIF and 2 inhibitors (LIF/2i; see material and methods). To allow the monitoring of pluripotency, Oct4-GFP transgene was introduced to Mad2l2 heterozygous mice by breeding.Citation28 Wild type and heterozygous ESCs appeared prior to Mad2l2−/− cells, and grew faster. Altogether, 89 lines were established, out of which 12 were Mad2l2-deficient (Table S1). Thus, we obtained significantly fewer Mad2l2-deficient ESC lines than expected, probably correlating to the embryonic lethality described previously. No obvious aberrations with regard to gender were observed.Citation24 After the fourth passage, cultures were shifted to conventional LIF/serum medium. Here, knockout lines started to differentiate spontaneously to epithelial-like, Oct4-GFP negative cells. Differentiation commenced at the periphery of the colonies and then expanded upon further passaging (, S1A,C). This phenotype was not restricted to an individual ESC line, but was common in various knockout lines to different extents (, S1A, Table S1). Quantitative PCR with reverse transcription (RT-qPCR) analysis revealed a decreased expression of pluripotency markers like Nanog, Oct4, Sox2, and Prdm14, and an elevated expression of Zfp42 (Rex1) in 3 tested knockout lines, while FGF4 expression was not altered (). In addition, knockout cells expressed lower levels of Sox2, Oct4, E-Ras, and Nanog proteins than the control ESCs, as determined by western blotting (Fig. S1B).
Figure 1. Instability and differentiation of Mad2l2−/− ESCs in LIF/serum. (A) RT-qPCR analysis of Mad2l2 and Nanog mRNA in ESCs and differentiating embryoid bodies (EBs). Representative data of 3 replicates are shown, (B) Western blot analysis of Mad2l2 and Nanog protein expression in ESCs and differentiating EBs. Representative data of 3 replicates are shown. (C) Phase contrast images and their corresponding Oct4-GFP expressions in heterozygous control (CTRL) versus Mad2l2−/− ESCs. Red arrows point to Oct4-GFP negative epithelial differentiating cells in Mad2l2−/− ESC cultures grown in LIF /serum medium; see also the Figure S1A for other knockout line. (D) Immunofluorescent staining of CTRL and Mad2l2−/− ESCs grown in LIF /serum with antibodies against Oct4 and Nanog. Note that differentiating cells in the periphery of a Mad2l2−/− colony lack expression of Nanog and Oct4 and undifferentiated cells at the center of Mad2l2−/− colony expressed Nanog only poorly. Scale bar 25 μm. See also Figure S1C. (E) RT-qPCR analysis of pluripotency-associated gene expression in 3 independent knockout ESC lines in comparison to CTRL ESCs grown in LIF /serum. Expression levels were normalized to Gapdh and then to a heterozygous ESC line as control. The error bar represents the mean ± SEM. (F) Mad2l2−/− ESCs grown in LIF/serum fail to generate chimeric pups. A representative picture of 5 different litters obtained from embryo injection of each cell line is shown. The same result was observed in the injection of 2 other knockout lines (Table S4 and data not shown).
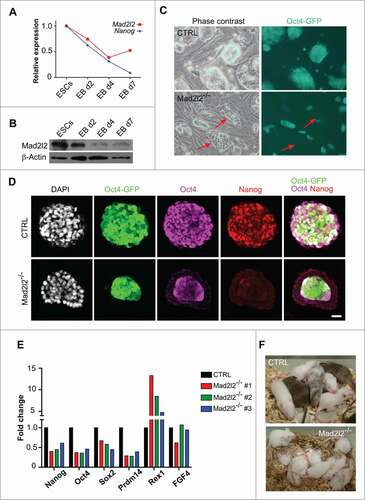
LIF/serum grown, Oct4-GFP positive ESCs were injected into blastocyst stage embryos, which were cultured overnight, and then transferred to the uterus of pseudo-pregnant foster mothers. Chimera formation of ESCs was judged by the fur color, since the host blastocysts were of FVB background (white coat) and the ESCs were of black or agouti background. Both of the 2 control ESC lines contributed successfully to embryogenesis as indicated by incorporation into the inner cell mass (ICM) or post-implantation embryos (data not shown), and by chimeric coat colors (). However, 3 different knockout ESC lines (Mad2l2−/− #1, 2, and 3) cultured in LIF/serum failed to incorporate into the ICM, stayed in the periphery of host blastocysts, lost the Oct4-GFP signal, and failed to hatch with the rest of the blastocyst (data not shown). Finally, transferring them back to the foster mothers did not lead to formation of any chimera (; Table S4). These results indicate that LIF/serum grown Mad2l2−/− ESCs are unstable and do not fulfill the criteria of authentic pluripotent ESCs.
Flowcytometry analysis (Fig. S1D) of the cell cycle status showed that control cells manifest a typical ESC profile (∼30.7%, 30.5%, 35.3% for G1, S and G2/M fractions, respectively). However, Mad2l2−/− ESCs showed a differentiated profile,Citation29 with the G1 fraction increased at the expense of S phase cells (∼41.4%, 22.0%, 34.3% for G1, S and G2/M fractions, respectively). Western blot analysis of the cell cycle-related proteins Cyclin B1, Cdh1 and Geminin showed no difference between knockout and control cells. Although there was a slight increase in phosphorylation of histone H2A (γH2AX), no elevated apoptosis was observed in knockout ESCs. Moreover, no increased activation of checkpoint protein Chk2, of cleaved Caspase 3, or of disrupted DNA (TUNEL assay) were evident (Fig. S1E,F). These observations make it unlikely that DNA damage or cell cycle perturbations cause the differentiation of Mad2l2−/− ESCs.
Mad2l2−/− ESCs deviate to primitive endoderm in LIF/serum
To address the identity of differentiated cells in LIF/serum Mad2l2−/− ESCs, first RT-qPCR was applied to analyze the expression of specific markers of different embryonic as well as extra-embryonic lineages. No striking differences were observed in the expression of examined markers for mesendoderm/trophectoderm (Gata3, Cdx2, Tead4), (neuro-) ectoderm (Sox1, Nestin, Pax6), and mesoderm (T, Eomes, Mixl1) lineages between control and 3 different knockout ESC lines (). Instead, a prominent increase in the expression of primitive endoderm-related markers was detected. Gata6, Gata4, and PDGFRα were upregulated up to 40-fold in Mad2l2−/− ESCs (). Since the expression of Gata6 transcription factor, as a marker for primitive endoderm precursors, precedes the expression of Sox17 and Gata4,Citation30-Citation33 we further focused on these 2 last markers to monitor commitment to the primitive endoderm lineage. Western blot analysis showed prominent levels of Sox17 and Gata4 proteins in Mad2l2−/− ESC cultures (, lanes 1 and 2). Furthermore, these observations were supported by immunocytochemistry, which revealed that Sox17 () and Gata4 () were preferentially expressed in differentiating Mad2l2−/− cells located in the periphery of colonies, with a concomitant lack of Oct4-GFP expression. No appreciable levels of other lineage-specific markers e.g. for mesoderm (T), ectoderm (Sox1), or extraembryonic ectoderm (Cdx2), were detected in knockout ESCs by protein gel blotting (data not shown). In order to rescue the Mad2l2-deficient ESC phenotype in LIF/serum, mutant cells were infected with a lentivirus vector allowing the inducible expression of Mad2l2. In the absence of doxycycline, infected ESCs differentiated spontaneously into primitive endoderm, as demonstrated by substantial expression of Gata4 and Sox17 (). However, upon doxycycline treatment, expression of primitive endoderm markers was attenuated. Thus, differentiating cells in the Mad2l2−/− ESC cultures in LIF/serum possess the characteristics of primitive endoderm cells, and their appearance is directly correlated with the presence or absence of Mad2l2.
Figure 2. Mad2l2−/− ESCs deviate into primitive endoderm in LIF/serum. (A) RT-qPCR analysis of differentiation markers in 3 independent knockout ESC lines grown in LIF/serum in comparison to CTRL ESCs. Expression levels were normalized to Gapdh and then to a heterozygous ESC line as control. The error bar represents the mean ± SEM. Immunocytochemistry demonstrates expression of Sox17 (B) and Gata4 (C) in the differentiating cells at the periphery of Mad2l2−/− ESC colonies (LIF/serum). Scale bar, 20 μM. (D) Mad2l2−/− ESCs were infected with lentiviral particles allowing for Mad2l2 expression under the control of doxycycline (Dox) as described in the material section. Note the reduced level of Gata4 and Sox17 in ESCs grown in LIF/serum/ doxycycline, while levels of Rex 1 remained unaffected.
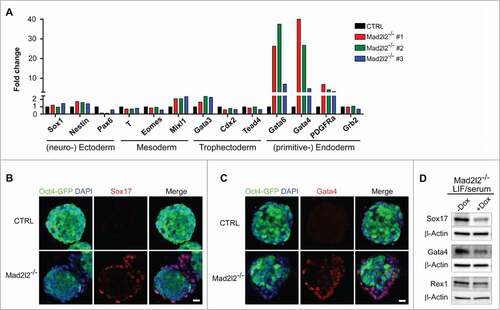
Figure 3. Adaptation of Mad2l2−/− ESC culture to LIF/2i inhibits differentiation. (A) Upon shifting from LIF/serum to LIF/2i, and further adaptation for 2–3 passages, differentiating cells in Mad2l2−/− culture disappear (red arrow in the left panel). (B) Western blot analysis of unsorted cells from LIF/serum culture (lanes #1 and 2), of FACS-sorted, Oct4-GFP positive cells from LIF/serum culture (lanes #3 and 4), and of LIF/2i-adapted ESCs (lanes #5 and 6). (C) Western blot analysis of pluripotency-related markers in ESCs from LIF/2i cultures. (D) LIF/2i-adapted Mad2l2−/− ESCs re-acquire the potential to generate chimeric pups (red arrow). A representative picture of 3 different litters obtained from embryo injection of each cell line is shown. (E) Western blot analysis of Oct4-GFP positive ESCs from LIF/serum cultures treated with the small molecule inhibitors CHIR 99021 (CH) and/or PD 0325901 (PD). Note that inhibition of the MAPK pathway by PD is sufficient to block primitive endoderm differentiation in Mad2l2−/− ESCs. (B–E) are the representative images of at least 3 independent biological replicates.
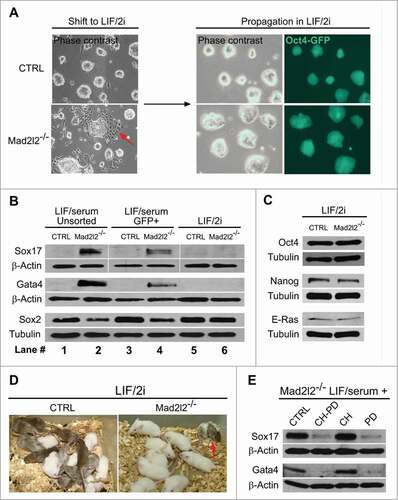
Culture in LIF/2i can block differentiation of Mad2l2−/− ESCs
The specification of primitive endoderm can be blocked by treatment of ex vivo-cultured murine blastocysts with 2 inhibitory chemicals, CHIR99021 (CHIR) and PD0352901 (PD) that block GSK3β and the MAPK pathway, respectively.Citation15,34 This causes an increase in the number of Nanog-expressing epiblast cells at the expense of Gata4-expressing primitive endoderm cells. The effect of these 2 inhibitors (2i) on the spontaneous differentiation of Mad2l2-deficient cells was investigated by shifting the culture conditions from LIF/serum to LIF/2i medium.Citation15,35 After 2–3 passages differentiated cells were no longer observed (), and Sox17 and Gata4 proteins were undetectable for up to more than 20 passages (, lanes 5 and 6). During the adaptation and further culture in LIF/2i conditions no appreciable levels of cell death were observed. Moreover, protein levels of Sox2, Oct4, E-Ras, and Nanog were restored to the control levels (). The developmental potential of knockout ESCs was tested by looking for chimera formation in vivo. In contrast to LIF/serum grown knockout ESCs (), Oct4-GFP-positive knockout cells from LIF/2i were able to contribute to the development of chimeric mice, though the level and the frequency of chimerism were lower than for control ESCs (; Table S4). These data suggest that upon culture in LIF/2i condition, knockout ESCs acquire, at least partially, the properties of the authentic pluripotent stem cells.
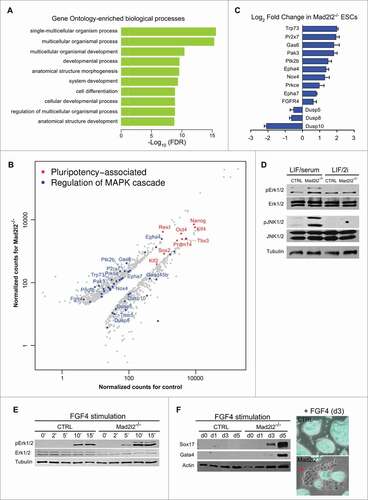
Figure 4 (See previous page). Genome-wide analysis of gene expression in LIF/2i Mad2l2−/− ESCs. (A) Identification of differentially expressed genes via R-package goseq from RNA-Seq experiments. The top 10 of 308 significantly enriched GO terms for the differentially expressed genes in Mad2l2−/− and CTRL ESCs are selected. Intriguingly, MAPK cascade gene ontology (GO) terms ′′regulation of MAPK cascade,′′ ′′positive regulation of MAPK cascade,′′ ′′regulation of fibroblast growth factor receptor signaling pathway,′′ and ′′fibroblast growth factor receptor signaling pathway′′ were also enriched (see Table S6). Normalization of counts and testing for differential expression were calculated via R-package DESeq. (B) Scatter plot of pluripotency-associated (in red) and differentially expressed MAPK transcripts (in blue). Normalized counts as CPM (counts per million mapped reads) were calculated via DESeq. Some genes associated to ′′regulation of MAPK cascade′′ are depicted. Pluripotency-associated genes are not differentially expressed except for Zfp42 (Rex1). (C) RT-qPCR analysis of gene expression for selected MAPK transcripts confirmed differential expressions seen by RNA sequencing. Note that positive components of the MAPK pathway are increased, whereas negative regulators such as the 3 dual specificity phosphatases are decreased in Mad2l2−/− ESCs. (D) Western blot analysis showed increased phosphorylation of Erk1/2 (pErk1/2) and JNK (pJNK1/2) proteins in LIF/serum Mad2l2−/− ESCs. The image is a representative of at 3 independent biological replicates of knockout line. (E) Western blot analysis of pErk1/2 levels upon short-term exposure of ESCs to FGF4 (3 ng/ml).Citation21 Note the earlier and stronger elevation of pErk1/2 in Mad2l2−/− samples upon stimulation by FGF4. (F) Left: western blotting for Sox17 and Gata4 primitive endoderm markers upon a continued exposure of ESCs to FGF4. In Mad2l2−/− ESCs, these markers were prominently upregulated already on day 3, while CTRL ESCs were refractory to differentiation even after 5 d. Right: a representative overlay (phase contrast and Oct4-GFP expression) of CTRL and Mad2l2−/− ESCs after 3 d of stimulation by FGF4 showed re-appearance of epithelial-like Oct4-GFP negative differentiating cells in knockout culture. (E and F), are the representative images of at least 3 independent biological replicates of FGF4 treatment of the Mad2l2 knockout line.
MAPK inhibition is sufficient to block differentiation of Mad2l2−/− ESCs
Oct4-GFP positive Mad2l2−/− cells growing in LIF/serum, were usually located in the middle of ESC colonies, and morphologically appeared undifferentiated (). However, they expressed significantly reduced levels of Oct4 and especially of Nanog proteins (). We asked whether these cells are primed for differentiation toward primitive endoderm. FACS-sorted Oct4-GFP positive Mad2l2−/− cells from LIF/serum culture still contained Sox17 and Gata4 proteins, and reduced levels of Sox2, though less prominent than unsorted cells containing more differentiated cells (, lanes 3–4 compared to 1–2). These observations indicate the existence of an Oct4-GFP positive cell population in LIF/serum, which is already deviating from an ESC identity toward a PrE fate, but not yet differentiated into a PrE phenotype.
To determine which inhibitor can effectively prohibit primitive endoderm differentiation, ESCs were treated with individual or combined inhibitors. To obtain a uniform population of less-differentiated cells, ESCs were FACS-sorted, in which a restrictive gating divided ESCs into 3 distinct populations based on Oct4-GFP expression: highly positive (Oct4-GFPhigh), weakly positive (Oct4-GFPlow), and negative cells (Oct4-GFPneg) (Fig. S2A). The negative population consisted mostly of differentiated or MEF feeder cells. Mad2l2−/− Oct4-GFPlow cells failed to form ESC colonies upon plating in either LIF/serum or LIF/2i, implying an irreversible differentiation toward primitive endoderm. In contrast, a majority of control Oct4-GFPlow cells formed normal ESC colonies within 3–4 d after plating (data not shown). Equal numbers of the FACS-sorted Oct4-GFPhigh cells from control and knockout cultures were plated on MEFs in LIF/serum medium overnight. The day after, the medium was replaced by LIF/serum and supplemented with CHIR, PD, or both. During the next 4–6 days, appearance of differentiating cells was monitored and western blot analysis was performed to analyze the expression of primitive endoderm markers, Sox17 and Gata4. Differentiating, epithelial-like cells reappeared in LIF/serum Mad2l2−/− ESC cultures, as well as in CHIR-treated cells, but not in 2i- or PD-treated cultures (Fig. S2B). A pronounced downregulation was observed in the levels of Sox17 and Gata4 proteins in protein extracts of Mad2l2−/− cultures treated with 2i or PD (). These data suggest that inhibition of the MAPK pathway is sufficient to rescue Mad2l2-deficient ESCs from deviating into PrE.
RNA expression profiles reveal impaired regulation of the MAPK pathway in undifferentiated Mad2l2−/− ESCs
Transcriptional profiles of Mad2l2−/− and control ESCs grown in LIF/2i were compared in order to understand the reasons for instability observed in LIF/serum conditions. Both knockout (line MPES#1) and control cells were first adapted for more than 10 passages in LIF/2i before RNA extraction. The expression profiles of wild type and Mad2l2-deficient ESCs were analyzed by deep sequencing in triplicates. A principal component analysis (PCA) of the data indicated that the 3 samples with the same genotype fall into a coherent group, and that both groups are significantly different (Fig. S3A). Gene ontology (GO) analysis of differentially expressed genes in knockout and control ESCs via the R-package goseqCitation36 showed a significant enrichment in developmental processes and differentiation ( and S3B, top 10 and 30 GO terms, respectively). Notably, expression levels of pluripotency associated genes like, Oct4, Nanog, Sox2, Klf4, and Tbx3 showed no differences, except for the upregulation of Zfp42 (Rex1), confirming our previous results (). Among the complete list of significantly enriched GO terms “regulation of MAPK cascade,” “positive regulation of MAPK cascade,” “regulation of fibroblast growth factor receptor signaling pathway“ and “fibroblast growth factor receptor signaling pathway” were identified (FDR-corrected p-value < 0.05; see also Tables S5 and S6). Genes associated with “regulation of MAPK cascade” () were either upregulated, including Trp73, Pr2x7, Gas6, Epha4 and 7, FGFR4, or downregulated, as for Dusp5, Dusp8, and Dusp10, the phosphatases that de-phosphorylate Erk and JNK kinases (). Interestingly, cell signaling-related terms, including “signal” and “phosphorylation” appeared among the enriched protein categories identified via DAVIDCitation37 (Fig. S3C). RT-qPCR analysis confirmed the differential expression of MAPK pathway genes in knockout and control LIF/2i ESCs (). Moreover, a similar, but not identical, pattern of gene expression misregulation could be observed in Oct4-GFP negative knockout LIF/serum ESCs in comparison to Oct-GFP positive ones; i.e., higher levels of FGFR4 and other positive regulators of MAPK were expressed in Oct4-GFP negative cells, whereas, lower levels of Dusp5, 10 were expressed in these cells compared to Oct4-GFP positive cells (Fig. S4).
To confirm the biological significance of these results, FACS-sorted Oct4-GFP positive LIF/serum Mad2l2−/− ESCs were treated with SU5402, a potent inhibitor of FGF receptor. Morphological analysis showed significant rescue of differentiation in SU5402-treated cultures (Fig. S2C).
Since the levels of positive and negative regulators of the MAPK pathway transcripts were up- and downregulated, respectively, in knockout ESCs, we hypothesized that this pathway might be hyperactive in Mad2l2−/− cells. First, immunoblotting of LIF/serum ESC extracts showed increased levels in knockout cells of the active transcription factors phospho-Erk1/2 (pErk1/2) and phospho-JNK1/2 (pJNK1/2), while their unphosphorylated, inactive forms (Erk1/2 and JNK1/2) appeared unchanged (). In LIF/2i, which contain the MEK inhibitor PD, levels of pErk1/2 and pJNK1/2 were in general decreased and showed no difference between wild type and knockout samples. Because knockout cells expressed higher levels of FGF receptor transcripts, we reasoned that they might be hypersensitive to FGF signaling, which would strongly induce the primitive endoderm lineage.Citation38 To address the immediate response of the cells to FGF4 stimulation, LIF/2i ESCs were released into N2B27 medium supplemented with CHIR, and FGF4, but without PD and LIF. In Mad2l2−/− cells, the level of pErk1/2 was elevated faster and stronger than in control cells (). Then, we investigated a long-term response of the ESCs to FGF4 stimulation by monitoring Sox17 and Gata4 protein levels. In Mad2l2−/− culture, they were detected as early as 3 d after FGF4 induction, whereas they were barely detectable in wild type cells, even after 5 d of FGF4 stimulation. Consistently, epithelial-like differentiating Oct4-GFP negative cells appeared back in FGF4-treated Mad2l2−/− ESCs, but not in controls (). In conclusion, Mad2l2−/− ESCs are deficient in the regulation of MAPK pathway transcripts, leading to a hyperactivity of the MAPK pathway, and consequently, to the increased susceptibility of Mad2l2−/− ESCs to FGF4 signaling and deviation toward primitive endoderm lineage.
Epigenetic changes in Mad2l2−/− ESCs
Since Mad2l2 is involved in the epigenetic reprogramming of PGCs,Citation24 we investigated the epigenetic configuration of Mad2l2−/− ESCs. In general, higher levels of suppressive H3K9me2 and H3K27me3 were detected in Mad2l2−/− ESCs in both LIF/serum and LIF/2i conditions (). This is in line with general elevation of these histone modifications during differentiation of ES cells.Citation6,7,39 High levels of H3K9me2 were particularly observed in the Oct4-GFP negative, differentiating cells in the periphery of knockout colonies (). Yet, increased levels of H3K9me2 and H3K27me3 were also detected in less differentiated FACS-sorted Oct4-GFP positive cells from Mad2l2−/− culture analyzed by protein gel blotting (). These results show an aberrant epigenetic configuration in knockout ESCs at a global level.
Figure 5 (See previous page). Epigenetic signature of Mad2l2 deficient cells. (A) Western blot analysis of histone modifications in CTRL and Mad2l2−/− ESCs in LIF/serum and LIF/2i culture conditions. Note to the consistent upregulation of histone marks in knockout cells. The image shows a representative blot of 3 independent biological samples of knockout line. (B) A representative immunocytochemistry analysis of H3K9me2 levels in ESCs. Differentiating Oct4-GFP negative cells in the periphery of Mad2l2−/− colonies harbor high levels of H3K9me2. Scale bar, 25 μm. (C) A representative protein gel blot analysis of histone modification in CTRL and Mad2l2−/− ESCs in FACS sorted Oct4-GFP positive ESCs from LIF/serum culture. Note the consistent upregulation of methylated histone 3 in knockout cells. (D) ChIP analysis of H3K27me3, Ezh2, and H3K9me2 deposition on Gata4, Sox17 and Nanog promoters in Oct4-GFP positive LIF/serum ESCs. 1% of pre-cleared sonicated chromatin was used as input. As the negative controls, mIgG, Normal mouse IgG, and rIgG, Normal rabbit IgG were used. ChIP analysis of β-Actin promoter shows no difference of histone marks between knockout and control LIF/serum ESCs. (E) Upper panel: G9a-inhibition hampers PrE differentiation of Mad2l2−/− ESCs as indicated by decreased levels of Sox17, Gata4 in western blot analysis. FACS sorted Oct4-GFP positive LIF/serum Mad2l2−/− ESCs were plated on fresh inactivated MEFs and treated for 4–5 d in LIF/serum (CTRL) or LIF/serum supplemented with G9a-inhibitor BIX01294 (+G9a-i). A significant decrease in H3K9me2 level confirms effective inhibition of G9a. Lower panel: The representative phase contrast and Oct4-GFP overlay of the cultures used for protein gel blotting. The red arrow points to the epithelial-like Oct4-GFP negative differentiated cells emerged in LIF/serum culture (CTRL) of Mad2l2−/− ESCs. (F) ChIP analysis of H3K27me3, Ezh2, and H3K9me2 deposition on FGFR4 and Dusp10 promoters in LIF/2i ESCs. 1% of pre-cleared sonicated chromatin was used as input. As the negative controls, mIgG, Normal mouse IgG, and rIgG, Normal rabbit IgG were used. ChIP analysis of β-Actin promoter shows no difference of histone marks between knockout and control LIF/2i ESCs. In (D and F), error bars represent mean ± SEM of 3 independent ChIPs. *,P< 0.05; paired Student's t test.
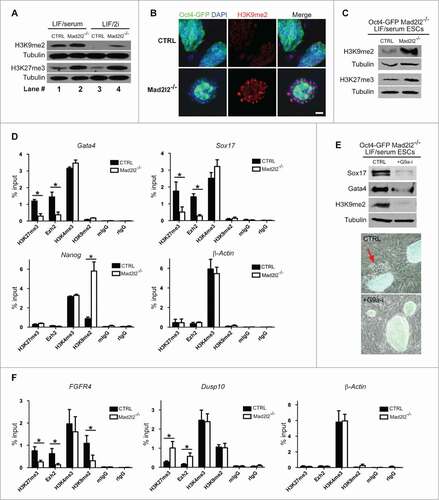
Next, we examined the distribution of these histone marks on Nanog and 2 PrE marker promoters, Gata4 and Sox17 in LIF/serum ESCs. Chromatin Immunoprecipitation (ChIP) analysis showed that H3K27me3, the repressive histone modification associated with bivalent loci, was decreased at Gata4 and Sox17 promoters in LF/serum Mad2l2−/− ESCs. However, no appreciable changes were observed in the deposition of H3K4me3 or H3K9me2 marks on these promoters (). Consistently, Gata4 and Sox17 promoters were also deprived of the polycomb protein Ezh2 in LIF/serum knockout samples. Together, these results point to a de-repression of Gata4 and Sox17 loci in Mad2l2 knockout cells. We further investigated the epigenetic configuration of the Nanog promoter. Mad2l2 was previously shown to bind directly to the histone methyltransferase G9a and thereby to be involved in H3K9me2 downregulation.Citation24 G9a is a barrier for reprogramming, is absent from the Nanog promoter in pluripotent ESCs, and is progressively deposited during differentiation.Citation40 ChIP analysis of the Nanog promoter showed a significant increase of H3K9me2 in the knockout samples (), suggesting the activity of histone methyltransferase G9a or a related enzyme. In contrast, levels of H3K4me3, H3K27me3 or Ezh2 were not changed on Nanog promoter. All ChIP experiments were performed on primitive endoderm-primed Oct4-GFP positive cells, which are the most undifferentiated cells in the knockout cultures. Therefore, the possibility of contamination with fully differentiated PrE cells can be ruled out. Furthermore, ChIP analysis of β-Actin promoter showed no obvious change between knockout and control samples, implying the selective effect of Mad2l2 loss on pluripotency- and PrE-related markers. To establish further functional connection between G9a and primitive endoderm differentiation of Mad2l2 knockout ESCs, we treated them with the selective G9a-inhibitor, BIX01294.Citation41 FACS-sorted Oct4-positive Mad2l2−/− ESCs were plated on fresh, inactivated MEFs, and were treated with LIF/serum medium supplemented with BIX01294 for 4–5 d. Whereas differentiation commenced in non-treated cells, G9a-inhibitor treatment hampered primitive endoderm differentiation as indicated by morphology and reduced levels of Sox17 and Gata4 (). The level of H3K9me2 in the BIX01294-treated ESCs was suppressed, as expected from a successful inhibition of G9a ().
Next, we examined the epigenetic signature of MAPK-associated loci in LIF/2i cells, especially those that were misregulated in Mad2l2 knockout cells (). Primers were designed to amplify a region in the potential promoter (−500 to +100 bp of Transcription Start Site, which were also enriched for H3K4me3 and H3K27me3 marks) of FGFR4 and Dusp10 (http://genome.ucsc.edu/)42. ChIP analysis of FGFR4 and Dusp10 promoters were performed in LIF/2i knockout and control cells. Decreased levels of H3K27me3, Ezh2, and H3K9me2, but not of H3K4me3, were observed on FGFR4 promoter of Mad2l2−/− ESCs (), suggesting the de-repression of FGFR4 expression that is in line with RNA-Seq analysis (). In contrast, elevated levels of H3K27me3 and Ezh2, but not of H3K9me2, repressed Dusp10 promoter in Mad2l2−/− LIF/2i ESCs. Consistently, treatment of Mad2l2−/− LIF/2i ESCs with G9a-inhibitor attenuated the expression of FGFR4, but had no effect on Dusp10 level (Fig. S5A). Finally, to establish a connection between epigenetic misregulation of MAPK genes and MAPK-hypersensitivity of Mad2l2-deficient ESCs, we utilized the FGF4-dependent primitive endoderm induction approach () in the presence of G9a-inhibitor. RT-qPCR analysis of Gata4 and Gata6 showed that G9a-inhibitor could hamper the primitive endoderm differentiation of FGF4-treated Mad2l2−/− LIF/2i ESCs in a dose-dependent manner (Fig. S5B).
In conclusion, we observed inappropriate epigenetic configurations at the Gata4, Sox17 and Nanog promoters (in LIF/serum) and at MAPK-associated loci (in LIF/2i) in Mad2l2−/− ESCs with respect to H3K27me3 and H3K9me2. These correspond to the observed hypersensitivity to FGF4/MAPK signaling, loss of pluripotency, and the differentiation into primitive endoderm. These phenotypes could be rescued, at least partially, by selective inhibition of the Mad2l2 interacting protein G9a.Citation24
Discussion
Pluripotency requires not only the presence of the core factors Oct4, Nanog, Sox2 and Klf4, but also their levels must be properly balanced.Citation43–Citation45 PrE formation from ESCs mimics closely the in vivo situation, when Nanog expressing segregates pluripotent epiblast cells from Gata4 expressing primitive endoderm cells. In vivo as well as in vitro, this process is under the control of FGF4, and requires the activation of Grb2-MAPK signaling in PrE precursorsCitation19,38,46,47 Normally, murine ESCs produce FGF4 in an autocrine fashion, however the MAPK pathway is controlled on multiple levels by diverse biochemical mechanisms in order to prevent differentiation.Citation48 For example, Myc/Max complexes can effectively suppress Erk activity by transcriptionally regulating 2 members of the dual-specificity phosphatase (DUSP) family, the negative MAPK regulators Dusp2 and 7. They bind and inactivate Erk1/2 by dephosphorylation of residues required for the catalytic activity.Citation49 Another example is Dusp5, which maintains Nanog and Oct4 expression in mouse ESCs by inhibition of the Erk pathway.Citation50 Recently it was shown that JNK1/2 signaling negatively regulates Klf4 activity.Citation51 JNK1/2 phosphorylates Klf4 and this inhibits Klf4 transcription and transactivation activity. Also pluripotency factors themselves are involved in the suppression of MAPK signaling. Thus, Oct4 negatively regulates Stk40 expression, which otherwise associates with Rcn2 to activate the Erk/MAPK pathway, and thereby induces ESC differentiation into extraembryonic endoderm lineages.Citation52
Mad2l2−/− ESCs grown in conventional LIF/serum condition, are instable and deviate spontaneously toward PrE fate, a process that is independent of apoptosis, but is caused by impaired regulation of MAPK transcripts as well as pluripotency associated genes. The small molecules PD and SU5402 inhibited FGF signaling very efficiently, and thus made the maintenance of undifferentiated Mad2l2−/− ESCs cultures possible. In LIF/serum, knockout ESCs did not produce more FGF4 or Grb2, but rather represent a hyperactive MAPK pathway indicated by elevated pErk1/2 and pJNK1/2 protein levels. In LIF/2i conditions, the undifferentiated ESC phenotype seems not to be maintained by the induction of death in differentiating knockout cells. Rather, the pluripotent cells are shielded from differentiation triggers by the blocking of the MAPK pathway, in particular of the MEK-inhibitor PD0325901.Citation48 Our gene expression profiles demonstrated that several components of the MAPK pathway were indeed affected by the absence of Mad2l2 even in undifferentiated ESCs adapted to LIF/2i. We observed that positive effectors of MAPK signaling were up-, and negative effectors were downregulated. How exactly, genetically or epigenetically, Mad2l2 regulates the expression of MAPK-associated genes need further investigation. Nevertheless, in the absence of Mad2l2, ESCs become hypersensitive to FGF4 stimulation. Expression of pluripotency associated genes, especially of Nanog, was also downregulated in LIF/serum Mad2l2−/− ESCs which would destabilize them for further differentiation.Citation53 Downregulation of Nanog in mouse and human ESCs leads either to primitive endoderm inductionCitation54,55 or induction of extraembryonic endoderm-associated genes GATA4, and GATA6, together with trophectoderm-associated genes CDX2, and GATA2.Citation56 However, in Mad2l2 deficient ESCs, suppression of the Nanog locus via increased deposition of H3K9me2 is accompanied by misregulation of MAPK genes and de-repression of the Gata4 locus that deviate Mad2l2−/− ESCs only to primitive endoderm, but not to trophectoderm. On the other hand, Rex1 expression was upregulated in Mad2l2−/− ESCs, which can be explained by its involvement in extraembryonic endoderm development. Indeed, Rex1−/− ESCs are unable to differentiate into PrE, suggesting that Rex1 implicate in PrE differentiation.Citation57 In this sense, Rex1 would no longer be considered as a pluripotency marker, but rather as an indicator of PrE differentiation.
If pluripotency is destabilized in murine ESCs by e.g., suboptimal treatment or medium, a differentiation pattern is usually observed. Similar features were reported previously for SOCS3- and MYC-knockout ESCs, as well as for PRDM14-knock down ESCs. Their colonies start to differentiate in the periphery, while cells in the middle, where they have piled up, initially remain pluripotent, as judged by morphology, division rate, and markers. Obviously, the cells in the periphery of the ESC colonies are more exposed to differentiating signals, or less supported by surrounding ESCs. The resulting cell-autonomous instabilities demonstrate that genes like Mad2l2, Prdm14, c-Myc, and SOCS-3 function to safeguard pluripotent stem cells from primitive endoderm differentiation.Citation29,58-60 Prdm1 (Blimp1) is crucial for PGCs, but not for ESCs.Citation61 Mad2l2 and Prdm14, on the other hand, are required for both the stable development of PGCs in early embryos, and for the propagation of ESCs in LIF/serum. However, while Prdm14 functions at least partially in DNA methylation via DNMTs,Citation21 our transcriptional profiling did not provide evidence for such a function of Mad2l2. Rather, it appears that Mad2l2 functions both in ESCs and PGC development by affecting specific epigenetic signatures.Citation24 Mad2l2 has been identified as an inhibitor of cyclin dependent kinase 1 (Cdk1) kinase activity.Citation24 In the present communication, we showed an increased activity of Erk1/2 kinase in the absence of Mad2l2, suggesting an inhibitory function of Mad2l2 on Erk1/2 in murine ESCs. In contrast, human ESCs (hESCs) were reported to phosphorylate Nanog via the selective kinases CDK1 and ERK2. Although the biological consequences of such phosphorylations need further investigation, they seem to indicate supportive effects of ERK and CDK1 on NANOG expression in hESCs.Citation62 In fact, hESCs are dependent on MAPK/ERK activation via bFGF and hESC self-renewal is impaired in the absence of FGF signaling.Citation63 In hESCs, ERK and its downstream effector ELK1 function to support self-renewal through activation of pluripotency related genes and/or inhibition of lineage specification genes.Citation64 This establishes a connection between CDK1, ERK and NANOG. Another link between Mad2l2 and MAPK signaling in human cells is established by binding of Mad2l2 to ELK-1, which promotes ELK-1 activation by c-Jun-dependent phosphorylation, and subsequently the upregulation of ELK-1 target genes.Citation26 The observation of a positive influence of Mad2l2 on MAPK signaling in human cells is in contrast to the negative effect of Mad2l2 on Erk and Cdk1 kinase activity, which we found in murine ESCs. It reflects the opposite requirements of murine and human ESCs, respectively for MAPK signaling.
Lethality was previously observed for mid-gestation, but not earlier, Mad2l2-deficient embryos.Citation24 This indicates that enough pluripotent cells in Mad2l2−/− blastocysts survive to support a further continuation of embryogenesis. Indeed, pluripotency is a transient state of epiblast cells that is immediately followed by differentiation and gastrulation. Moreover, contribution of residual maternal mRNA and protein is a possibility that cannot be completely ruled out. An intriguing question to ask is whether a similar phenotype to ESCs is observed during early in vivo development of Mad2l2 deficient embryos. Further studies are needed to unravel the PrE deviation of pluripotent epiblast cells in Mad2l2−/− embryos. It is well known that early murine embryos can accommodate a significant loss of cells. So if inner cell mass cells would be lost due to differentiation, we would not necessarily expect a complete discontinuation of embryogenesis. Rather, it would predict smaller animals, as were actually observed. We conclude that pluripotent inner cell mass cells in the blastocyst do not suffer substantially from the absence of Mad2l2, either because their growth conditions are optimal or because defects can be compensated.
Based on our previous findings we argue that H3K9 methyltransferase G9a is a direct target of Mad2l2, which becomes inhibited by protein-protein binding. In ChIP analysis of mutant ESCs, we observed a slight global increase of the H3K9me2 level, and a strong increase of H3K9me2 on the promoter of the pluripotency gene Nanog (in LIF/serum condition). The H3K27 methyltransferase Ezh2, on the other hand, can be protected from inhibitory phosphorylation by Mad2l2Citation24 via the blocking of Cdk1, a known inhibitory kinase for Ezh2. This effect was obvious on the bivalent Gata4 promoter, where the repressive H3K27me3 became strongly reduced in the absence of Mad2l2. The increased levels of both H3K9me2 and H3K27me3 may be explained by a general increase of suppressive histone marks in the chromatin during differentiation.Citation6,7,39 ChIP analysis of LIF/2i adapted knockout ESCs showed de-repression of FGFR4, the positive regulator of MAPK, through decreased promoter deposition of H3K27me3, Ezh2, and H3K9me2. In contrast, Dusp10 promoter (the negative regulator of MAPK) was repressed in knockout cells by increased levels of H3K27me3 and Ezh2. How Mad2l2 specifically recognizes or is differentially recruited to these loci needs further investigation. Recently, a crosstalk between H3K9 methyltransferases G9a and GLP with H3K27 methyltransferase Ezh2 was described.Citation65 According to this report, both G9a and GLP interact physically with Ezh2 in a DNA-independently manner. The enzymatic activity of G9a was shown to be essential for recruitment of Ezh2 to a set of developmental regulator genes and H3K27me3 establishment on their promoters. In such a context, Mad2l2 may facilitate a crosstalk between H3K9me2 and H3K27me3 in mouse pluripotent ESCs influencing expression of MAPK genes and thus inhibiting the PrE lineage. An involvement of Mad2l2 in epigenetic regulation is not as unlikely as it occurs at first sight. Components of DNA synthesis come together with epigenetic regulators at the replication fork, where the ring-shaped PCNA protein represents a functional platform. After certain types of DNA damage Mad2l2 (Rev7) is recruited to ubiquitinated PCNA as the accessory subunit of translesion DNA polymerase zeta. PCNA on the other hand also contacts G9a/GLP proteins, which transfer methyl groups onto DNA and histones. Thus, there is a scenario, where the discussed proteins come together and may become involved in each other's regulation.Citation66,67
The induction and maintenance of pluripotency has traditionally been interpreted to depend on the expression of core pluripotency factors. A more recent, not necessarily contradictory concept interprets pluripotency as an intricate balance between the activities of lineage specifying factors, which on their own would promote a specific developmental direction.Citation68 As proof of principle, it was shown that the trophectoderm specifier Gata3, in combination with the ectodermal specifier Geminin could efficiently reprogram fibroblasts to pluripotency.Citation69 Destabilization of the pluripotent status after deletion of a single gene may therefore point to a lineage specification role of the mutated gene. Applying this model to our results, the ablation of a germ cell critical gene like Mad2l2 could be interpreted as the cause for an imbalance of the metastable pluripotency equilibrium, where the primitive endoderm specifier Gata4 becomes de-repressed and dominant, leading to differentiation.
Materials and Methods
Ethics statement
All animal works have been conducted according to relevant national and international guidelines.
ESC establishment, culture and differentiation
E3.5 pre-implantation blastocysts from Oct4-GFP; Mad2l2+/− intercourses were harvested, plated on Mitomycin-C (Sigma)-inactivated mouse embryonic fibroblasts (MEFs) in N2B27 medium supplemented with 20% Fetal Bovine Serum (FBS, PAN), L-glutamine (2 nM, Invitrogen), sodium pyruvate (1 mM, Invitrogen), non essential amino acids (0.1 mM each, Invitrogen), 2-mercaptoethanol (100 μM, Invitrogen), penicillin/streptomycin (104 U/ml), LIF (1000 U/ml, Milipore), CHIR99021 (3 μM, Stemgent), and PD0325901 (1 μM, Merck).Citation15 At passage 3–4, cultures were continued to either KO-DMEM supplemented with LIF and serum (LIF/serum condition), or N2B27 medium supplemented with LIF, CHIR99021, PD0325901 (LIF/2i condition). For embryoid body differentiation, an ESC suspension (106 cells/ml) was cultured in bacterial culture dishes in DMEM supplemented with 20% serum, penicillin/streptomycin (104 U/ml), and sodium pyruvate (1 mM, Invitrogen). To investigate the effects of small molecule inhibitors on primitive endoderm differentiation of Mad2l2−/− ESCs, FACS sorted Oct4-GFP positive LIF/serum cells were plated overnight on fresh inactivated MEFs and treaded the day after with PD, CHIR, SU5402 (10 μM, Calbiochem), BIX01294 (4.1 μM, Sigma). For FGF4 stimulation, LIF/2i ESCs were trypsinized and single cells were plated on new gelatin-coated dished and treaded the day after with N2B27 medium supplemented with CHIR and FGF4 (3 ng/ml, Peprotech). A lentiviral vector–based, conditional gene expression system for drug-controllable expression of Mad2l2 for rescue experiments was generated and employed as described.Citation70 Mad2l2−/− ESCs were cultured in LIF/2i medium on MEFs for 24 h, were infected with lentivirus particles for 24 h, and then kept in the presence or absence, respectively, of 10 μg/ml doxycycline (Sigma). After 24 h medium was changed to either LIF/serum/doxycycline or LIF/serum. After three days of culture, cells were harvested in RIPA buffer for western blot analysis.
Immunocytochemistry
Table S1 describes the set of antibodies used in this study. Cells were washed in PBS, fixed for 20 min in paraformaldehyde, permeablized with 0.1% Tween 20, and blocked for 1 hour in 10% normal goat serum with 1% bovine serum albumin. Primary antibodies (in blocking solution) were applied overnight at 4°C before using appropriate secondary antibodies. The nuclei were counterstained with 4,6-Diamidin-2-phenylindol (DAPI, Vectashield).
Flowcytometry and FACS sorting
Cells were fixed for at least 30 min in 70% ethanol, stained with propidium iodide, treated for 30 min with RNase, and analyzed by flowcytometry (FACS Calibur). For sorting, single cell suspension of ESCs was prepared. MEFs were eliminated by differential attachment to gelatin. ESCs were washed with PBS, and then sorted by FACS Aria II (BD). Sorted cells were either cultured back on inactivated MEF feeders, or were lysed in RIPA buffer for protein gel blotting.
RT-qPCR
LIF/2i ESCs were trypsinized and washed twice with PBS. For LIF/serum ESC cultures, first MEFs and differentiated cells were eliminated by differential adhesion to gelatin-coated dishes. Then the total RNA was extracted with RNeasy kit (Qiagen), DNA was digested by DNaseI (Qiagen) and cDNA was synthesized from 1–2 μg RNA by reverse transcriptase (Omniscript, Qiagen) with a combination of random hexamere and oligo dT primers (Promega). Using pre-tested qPCR-grade set of primers (Table S3), 20–40 ng cDNA per reaction was amplified by KAPA SYBR® FAST qPCR Master Mix (KAPA biosystems) in real-time PCR with an Applied Biosystems 7300 Sequence Detection system. For each set of RT-qPCR primer, the efficiency (e) of amplification was measured through standard curves. The Ct values were determined using default threshold settings, after which the expression levels of genes in different samples were normalized to their corresponding GAPDH level. Then, fold changes in gene expression to control cells were calculated using e−ΔΔCt formula and plotted for knockout samples.
Western blotting
MEFs and differentiated cells were eliminated by differential adhesion to gelatin-coated dishes and the cells in the supernatant, which were mostly consisted of ESCs, were lysed in RIPA. Protein extracts were dissolved on SDS-PAGE gels and then transferred to the nitrocellulose membranes, blocked for 1 hour at room temperature with blocking solution (5% low-fat milk dissolved in PBS 0.1% Tween), and then incubated with primary antibodies (diluted in blocking solution) overnight at 4° C on shaking plate. Then, membranes were washed in PBS 0.1% Tween, incubated for 1–2 hour at room temperature with an appropriate HRP-conjugated secondary antibody. Finally, membranes were washed and developed following treatment with Pico chemiluminescent substrate (Thermo Scientific).
RNA-Sequencing
ESCs were adapted for more than 10 passages before RNA isolation. Library preparation for RNA-Seq was performed using the TruSeq Stranded Total RNA Sample Preparation Kit (Illumina, RS-122–2201) starting from 400 ng of total RNA. Accurate quantitation of cDNA libraries was performed by using the QuantiFluor™ dsDNA System (Promega). The size range of final cDNA libraries was determined applying the DNA 1000 chip on the Bioanalyzer 2100 from Agilent (280 bp). cDNA libraries were amplified and sequenced by using the cBot and HiSeq2000 from Illumina (SR; 1×50 bp;5- 6 GB ca. Thirty–35 million reads per sample). Sequence images were transformed with Illumina software BaseCaller to bcl files, which were demultiplexed to fastq files with CASAVA v1.8.2. Quality check was done via fastqc (v. 0.10.0, Babraham Bioinformatics). Sequences were aligned by Bowtie2 v2.0.2Citation71 to the Ensembl cDNA and non-coding RNA release 69 mouse reference genome. Counting the reads to each gene to the Ensembl gene annotation file (release 69) was done via HTSeq python scripts (0.5.3p9). Data was preprocessed and analyzed in the R/Bioconductor environment (2.10/2.15.2) loading DESeq, gplots, biomaRtCitation72 and goseq packages. After filtering the genes exceeding more than 20 counts for at least one sample, normalization, estimation of dispersions and testing for differentially expressed genes assuming negative binomial data distribution was computed via DESeq.Citation73 Candidate genes were filtered to a minimum of 2× fold change and FDR-corrected P value <0.05. For functional characterization of the differentially expressed transcripts, gene ontology (GO) enrichment analysis was conducted via goseq accounting for gene length bias and correcting for multiple testing.Citation36 Further functional association of candidate genes was performed with the webtool DAVID.
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed on 70–80 percent confluent LIF/serum ESC cultures following instruction manual for EZ ChIP chromatin immunoprecipitation kit (Upstate, Millipore). Briefly, single cell suspensions of ESCs were plated on gelatin-coated dishes to eliminate MEFs and differentiated cells. Equivalent of around 106 ESCs were used for each ChIP reaction. Single cells in suspension were fixed in 1% formaldehyde (Thermo Scientific) in PBS for 10 minutes. Unreacted formaldehyde was quenched afterwards with 125 mM glycine. Cells were lysed in 1% SDS buffer (1% SDS, 10mM EDTA, 50 mM Tris, pH 8.1). Then, cell lysates were sonicated in the Bioruptor XL sonicator (Diagenode) at 4°C to shear the chromatin and obtain 200–1000 bp DNA fragments. Chromatin samples were diluted in ChIP dilution buffer and pre-cleared by protein A/G agarose beads (Santa Cruz Biotechnology). Immunoprecipitation was performed using 5–10 μg of the appropriate antibodies (Table S2) with rotation at 4°C overnight, following by addition of Protein A/G agarose beads (Santa Cruz Biotechnology). Normal rabbit IgG (Upstate) and normal mouse IgG (Upstate) were served as negative controls. Beads were washed and protein/DNA complexes were eluted. Immunoprecipitated samples together with 1% input chromatin were subjected to reverse crosslinking overnight at 65°C in the presence of 200 mM NaCl. QIAquick PCR purification kit (Qiagen) was used to purify DNA samples, after which qPCR reactions were performed using indicated primers (Table S3).
Embryo injection and transfer
Mice were kept in standard IVC housing (Tecniplast Blue Line) with ad libitum breeding chow and water in 12L/12D from 6.00 a.m. to 6 p.m. To obtain host blastocysts for ESC injection, 4-week-old FVB/N females were superovulated with 7,5 IU PMSG at 12.30 p.m. and 5 IU HCG at 10.00 a.m. 2 d later. Mice were mated 4 hours after application of HCG. Blastula stage embryos were flushed at E3.5 from oviducts into standard M2 buffer (Sigma). Eight to 14 ES cells were injected in the space between zona pellucida and blastomeres. Manipulated embryos were transferred either the same day into oviducts of E0.5 CD1 surrogate females or after overnight culture in M16 buffer (Sigma) into uteri of E2.5 CD1 surrogate females. Recipients were treated with 4 mg/kg carprofen preoperatively to alleviate pain. Resulting pups were checked for appearance of chimerism at 5–14 d after the birth.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Tables.pdf
Download PDF (626.7 KB)Supplemental_Figures.zip
Download Zip (37.2 MB)Supplemental_Figure__and_Table_legends.docx
Download MS Word (119.6 KB)Acknowledgments
We thank P Rus for technical assistance, D Wollradt for maintenance of the mouse colonies. We thank GG Galli for technical support and A Klimke for discussions.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008; 132:567–82; PMID:18295576; https://doi.org/10.1016/j.cell.2008.01.015
- Yeo JC, Ng HH. The transcriptional regulation of pluripotency. Cell Res 2013; 23:20–32; PMID:23229513; https://doi.org/10.1038/cr.2012.172
- Denholtz M, Plath K. Pluripotency in 3D: genome organization in pluripotent cells. Curr Opin Cell Biol 2012; 24:793–801; PMID:23199754; https://doi.org/10.1016/j.ceb.2012.11.001
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 2007; 128:747–62; PMID:17320511; https://doi.org/10.1016/j.cell.2007.02.010
- Young RA. Control of the embryonic stem cell state. Cell 2011; 144:940–54.
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006; 10:105–16; PMID:16399082; https://doi.org/10.1016/j.devcel.2005.10.017
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 2010; 6:479–91; PMID:20452322; https://doi.org/10.1016/j.stem.2010.03.018
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 2006; 7:540–6; PMID:16723974; https://doi.org/10.1038/nrm1938
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 2007; 1:299–312; PMID:18371364; https://doi.org/10.1016/j.stem.2007.08.003
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315–26; PMID:16630819; https://doi.org/10.1016/j.cell.2006.02.041
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006; 8:532–8; PMID:16570078; https://doi.org/10.1038/ncb1403
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009; 460:118–22; PMID:19571885; https://doi.org/10.1038/nature08113
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003; 115:281–92; PMID:14636556; https://doi.org/10.1016/S0092-8674(03)00847-X
- Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A 2004; 101:6027–32; PMID:15075392; https://doi.org/10.1073/pnas.0401367101
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature 2008; 453:519–U5; PMID:18497825; https://doi.org/10.1038/nature06968
- Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J, et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 2013; 13:351–9; PMID:23850245; https://doi.org/10.1016/j.stem.2013.06.004
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 2012; 149:590–604; PMID:22541430; https://doi.org/10.1016/j.cell.2012.03.026
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 2010; 18:675–85; PMID:20412781; https://doi.org/10.1016/j.devcel.2010.02.012
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell 2006; 10:615–24; PMID:16678776; https://doi.org/10.1016/j.devcel.2006.02.020
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005; 122:947–56; PMID:16153702; https://doi.org/10.1016/j.cell.2005.08.020
- Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 Ensures Naive Pluripotency through Dual Regulation of Signaling and Epigenetic Pathways in Mouse Embryonic Stem Cells. Cell Stem Cell 2013; 12:368–82; PMID:23333148; https://doi.org/10.1016/j.stem.2012.12.012
- Leitch HG, Blair K, Mansfield W, Ayetey H, Humphreys P, Nichols J, Surani MA, Smith A. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development 2010; 137:2279–87; PMID:20519324; https://doi.org/10.1242/dev.050427
- Pirouz M, Klimke A, Kessel M. The reciprocal relationship between primordial germ cells and pluripotent stem cells. J Mol Med (Berl) 2012; 90:753–61; PMID:22584374; https://doi.org/10.1007/s00109-012-0912-1
- Pirouz M, Pilarski S, Kessel M. A critical function of Mad2l2 in primordial germ cell development of mice. PLoS genetics 2013; 9:e1003712; PMID:24009519; https://doi.org/10.1371/journal.pgen.1003712
- Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res 2008; 18:174–83; PMID:18157155; https://doi.org/10.1038/cr.2007.117
- Zhang L, Yang SH, Sharrocks AD. Rev7/MAD2B links c-Jun N-terminal protein kinase pathway signaling to activation of the transcription factor Elk-1. Mol Cell Biol 2007; 27:2861–9; PMID:17296730; https://doi.org/10.1128/MCB.02276-06
- Hong CF, Chou YT, Lin YS, Wu CW. MAD2B, a novel TCF4-binding protein, modulates TCF4-mediated epithelial-mesenchymal transdifferentiation. J Biol Chem 2009; 284:19613–22; PMID:19443654; https://doi.org/10.1074/jbc.M109.005017
- Szabo PE, Hubner K, Scholer H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev 2002; 115:157–60; PMID:12049782; https://doi.org/10.1016/S0925-4773(02)00087-4
- Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 2010; 7:343–54; PMID:20804970; https://doi.org/10.1016/j.stem.2010.06.023
- Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol 2011; 350:393–404; PMID:21146513; https://doi.org/10.1016/j.ydbio.2010.12.007
- Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci U S A 2010; 107:6364–9; PMID:20308546; https://doi.org/10.1073/pnas.0915063107
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 2008; 135:3081–91; PMID:18725515; https://doi.org/10.1242/dev.021519
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev 2010; 24:312–26; PMID:20123909; https://doi.org/10.1101/gad.1833510
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 2009; 136:3215–22; PMID:19710168; https://doi.org/10.1242/dev.038893
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 2011; 13:838–45; PMID:21685889; https://doi.org/10.1038/ncb2267
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010; 11:R14; PMID:20132535; https://doi.org/10.1186/gb-2010-11-2-r14
- Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4:P3; PMID:12734009; https://doi.org/10.1186/gb-2003-4-5-p3
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 2010; 137:715–24; PMID:20147376; https://doi.org/10.1242/dev.043471
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet 2009; 41:246–50; PMID:19151716; https://doi.org/10.1038/ng.297
- Yamamizu K, Fujihara M, Tachibana M, Katayama S, Takahashi A, Hara E, Imai H, Shinkai Y, Yamashita JK. Protein kinase A determines timing of early differentiation through epigenetic regulation with G9a. Cell Stem Cell 2012; 10:759–70; PMID:22704517; https://doi.org/10.1016/j.stem.2012.02.022
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 2007; 25:473–81; PMID:17289593; https://doi.org/10.1016/j.molcel.2007.01.017
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12:996–1006; PMID:12045153; https://doi.org/10.1101/gr.229102
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 2006; 38:431–40; PMID:16518401; https://doi.org/10.1038/ng1760
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol 2011; 13:490–6; PMID:21540844; https://doi.org/10.1038/ncb0511-490
- Radzisheuskaya A, Chia Gle B, dos Santos RL, Theunissen TW, Castro LF, Nichols J, Silva JC. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat Cell Biol 2013; 15:579–90; PMID:23629142; https://doi.org/10.1038/ncb2742
- Kuijk EW, van Tol LT, Van de Velde H, Wubbolts R, Welling M, Geijsen N, Roelen BA. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 2012; 139:871–82; PMID:22278923; https://doi.org/10.1242/dev.071688
- Kang M, Piliszek A, Artus J, Hadjantonakis AK. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development 2013; 140:267–79; PMID:23193166; https://doi.org/10.1242/dev.084996
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 2007; 134:2895–902; PMID:17660198; https://doi.org/10.1242/dev.02880
- Chappell J, Sun Y, Singh A, Dalton S. MYC/MAX control ERK signaling and pluripotency by regulation of dual-specificity phosphatases 2 and 7. Genes Dev 2013; 27:725–33; PMID:23592794; https://doi.org/10.1101/gad.211300.112
- Chen Q, Zhou Y, Zhao X, Zhang M. Effect of dual-specificity protein phosphatase 5 on pluripotency maintenance and differentiation of mouse embryonic stem cells. J Cell Biochem 2011; 112:3185–93; PMID:21732408; https://doi.org/10.1002/jcb.23244
- Yao K, Ki MO, Chen H, Cho YY, Kim SH, Yu DH, Lee SY, Lee KY, Bae K, Peng C, et al. JNK1 and 2 play a negative role in reprogramming to pluripotent stem cells by suppressing Klf4 activity. Stem cell research 2014; 12:139–52; PMID:24211391; https://doi.org/10.1016/j.scr.2013.10.005
- Li L, Sun L, Gao F, Jiang J, Yang Y, Li C, Gu J, Wei Z, Yang A, Lu R, et al. Stk40 links the pluripotency factor Oct4 to the Erk/MAPK pathway and controls extraembryonic endoderm differentiation. Proc Natl Acad Sci U S A 2010; 107:1402–7; PMID:20080709; https://doi.org/10.1073/pnas.0905657107
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol 2009; 7:e1000149; PMID:19582141; https://doi.org/10.1371/journal.pbio.1000149
- Hough SR, Clements I, Welch PJ, Wiederholt KA. Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of OCT4 and Nanog. Stem Cells 2006; 24:1467–75; PMID:16456133; https://doi.org/10.1634/stemcells.2005-0475
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003; 113:631–42; PMID:12787504; https://doi.org/10.1016/S0092-8674(03)00393-3
- Hyslop L, Stojkovic M, Armstrong L, Walter T, Stojkovic P, Przyborski S, Herbert M, Murdoch A, Strachan T, Lako M. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells 2005; 23:1035–43; PMID:15983365; https://doi.org/10.1634/stemcells.2005-0080
- Thompson JR, Gudas LJ. Retinoic acid induces parietal endoderm but not primitive endoderm and visceral endoderm differentiation in F9 teratocarcinoma stem cells with a targeted deletion of the Rex-1 (Zfp-42) gene. Mol Cell Endocrinol 2002; 195:119–33; PMID:12354678; https://doi.org/10.1016/S0303-7207(02)00180-6
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Structural Mol Biol 2011; 18:120–7; PMID:21183938; https://doi.org/10.1038/nsmb.2000
- Tsuneyoshi N, Sumi T, Onda H, Nojima H, Nakatsuji N, Suemori H. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem Biophys Res Commun 2008; 367:899–905; PMID:18194669; https://doi.org/10.1016/j.bbrc.2007.12.189
- Forrai A, Boyle K, Hart AH, Hartley L, Rakar S, Willson TA, Simpson KM, Roberts AW, Alexander WS, Voss AK, et al. Absence of suppressor of cytokine signalling 3 reduces self-renewal and promotes differentiation in murine embryonic stem cells. Stem Cells 2006; 24:604–14; PMID:16123385; https://doi.org/10.1634/stemcells.2005-0323
- Bao S, Leitch HG, Gillich A, Nichols J, Tang F, Kim S, Lee C, Zwaka T, Li X, Surani MA. The germ cell determinant Blimp1 is not required for derivation of pluripotent stem cells. Cell Stem Cell 2012; 11:110–7; PMID:22770244; https://doi.org/10.1016/j.stem.2012.02.023
- Brumbaugh J, Russell JD, Yu P, Westphall MS, Coon JJ, Thomson JA. NANOG Is Multiply Phosphorylated and Directly Modified by ERK2 and CDK1 In Vitro. Stem Cell Rep 2014; 2:18–25; PMID:24678451; https://doi.org/10.1016/j.stemcr.2013.12.005
- Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 2011; 8:326–34; PMID:21362572; https://doi.org/10.1016/j.stem.2011.01.001
- Goke J, Chan YS, Yan J, Vingron M, Ng HH. Genome-wide kinase-chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Mol Cell 2013; 50:844–55; PMID:23727019; https://doi.org/10.1016/j.molcel.2013.04.030
- Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, Margueron R, Ait-Si-Ali S. The Histone H3 Lysine 9 Methyltransferases G9a and GLP Regulate Polycomb Repressive Complex 2-Mediated Gene Silencing. Mol Cell 2014; 53:277–89; PMID:24389103; https://doi.org/10.1016/j.molcel.2013.12.005
- Takahashi A, Imai Y, Yamakoshi K, Kuninaka S, Ohtani N, Yoshimoto S, Hori S, Tachibana M, Anderton E, Takeuchi T, et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol Cell 2012; 45:123–31; PMID:22178396; https://doi.org/10.1016/j.molcel.2011.10.018
- Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature 2013; 498:70–4; PMID:23708966; https://doi.org/10.1038/nature12201
- Loh KM, Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 2011; 8:363–9; PMID:21474100; https://doi.org/10.1016/j.stem.2011.03.013
- Shu J, Wu C, Wu Y, Li Z, Shao S, Zhao W, Tang X, Yang H, Shen L, Zuo X, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell 2013; 153:963–75; PMID:23706735; https://doi.org/10.1016/j.cell.2013.05.001
- Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Method 2006; 3:109–16; PMID:16432520; https://doi.org/10.1038/nmeth846
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Method 2012; 9:357–9; PMID:22388286; https://doi.org/10.1038/nmeth.1923
- Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protocol 2009; 4:1184–91; PMID:19617889; https://doi.org/10.1038/nprot.2009.97
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106; PMID:20979621; https://doi.org/10.1186/gb-2010-11-10-r106
