ABSTRACT
The heterochronic pathway in C. elegans controls the relative timing of cell fate decisions during post-embryonic development. It includes a network of microRNAs (miRNAs), such as let-7, and protein-coding genes, such as the stemness factors, LIN-28 and LIN-41. Here we identified the acn-1 gene, a homologue of mammalian angiotensin-converting enzyme (ACE), as a new suppressor of the stem cell developmental defects of let-7 mutants. Since acn-1 null mutants die during early larval development, we used RNAi to characterize the role of acn-1 in C. elegans seam cell development, and determined its interaction with heterochronic factors, including let-7 and its downstream interactors – lin-41, hbl-1, and apl-1. We demonstrate that although RNAi knockdown of acn-1 is insufficient to cause heterochronic defects on its own, loss of acn-1 suppresses the retarded phenotypes of let-7 mutants and enhances the precocious phenotypes of hbl-1, though not lin-41, mutants. Conversely, the pattern of acn-1 expression, which oscillates during larval development, is disrupted by lin-41 mutants but not by hbl-1 mutants. Finally, we show that acn-1(RNAi) enhances the let-7-suppressing phenotypes caused by loss of apl-1, a homologue of the Alzheimer's disease-causing amyloid precursor protein (APP), while significantly disrupting the expression of apl-1 during the L4 larval stage. In conclusion, acn-1 interacts with heterochronic genes and appears to function downstream of let-7 and its target genes, including lin-41 and apl-1.
Introduction
A fundamental feature of development is the relative timing of biological processes – whether proliferation, migration, or cell fate decisions. Caenorhabditis elegans (C. elegans) has proved immensely powerful in identifying genes and pathways involved in developmental timing due to the organism's discrete developmental stages and invariant cell lineages.Citation1 Many studies in C. elegans have focused on a pathway involved in developmental timing named the heterochronic pathway.Citation2-4 Heterochronic genes control the relative timing of development – the timing of developmental events relative to the developmental stage of the organism – and operate independent of spatial controls.Citation2,Citation4 Mutations in these genes lead to bypass (“precocious” phenotype) or repeat (“retarded” phenotype) of stage-specific cell fate decisions.Citation5 Analyses of these mutations, both alone and in combinations, have both revealed numerous genes that promote correct cell fate choices and elucidated how they genetically interact in the heterochronic pathway.Citation3,Citation4,Citation6
The key heterochronic genes responsible for controlling the timing pattern of stem-cell-like hypodermal seam cells in the later larval stages of C. elegans (L3 and L4) include the let-7 microRNA (miRNA), its family members (mir-48, mir-84, and mir-241), and its downstream interactor genes (hbl-1, lin-41, lin-42, and lin-29).Citation7-12 Here, we focus on let-7 and its two direct targets: lineage-abnormal 41 (lin-41), a Ring finger-B box-Coiled coil (RBCC) protein whose family members include several E3 ubiquitin ligases, and hunchback-like 1 (hbl-1), a Cys2-His2 zinc-finger transcription factor homologous to Drosophila melanogaster’s hunchback (hb) segmentation gene.Citation9-16 During the L4 stage, let-7 suppresses hbl-1 and lin-41 via miRNA-mediated post-transcriptional suppression mechanisms to promote the L4-to-adult transition.Citation9,Citation11,Citation12 Loss-of-function mutations in let-7 result in reiteration of L4 cell fates, leading to an extra round of seam cell division, a delay in seam cell terminal differentiation and adult alae production, and a transient increase in the apparent number of seam cells.Citation8 In contrast, mutations in the lin-41 or hbl-1 genes cause seam cells to skip the L4 cell divisions and terminally differentiate precociously, albeit with less than 100% penetrance.Citation9,Citation11,Citation12 Simultaneous knock-down of both genes enhances these precocious phenotypes, suggesting that LIN-41 and HBL-1 function in parallel.Citation9,Citation12 HBL-1 in turn inhibits transcription of let-7, forming a negative feedback loop between the adult- and larval-fate promoters.Citation17 hbl-1 is also suppressed by the let-7-family-member miRNAs mir-48, mir-84, and mir-241 in the seam cells during the L3 stage.Citation7.
While many key regulators of the heterochronic pathway are known, questions remain as to how the tightly timed expression patterns of these regulators promote stage-specific cell fate decisions. To answer these questions, several studies, including our own, have examined the roles of the downstream factors through which these regulators act in developmental timing. Of the several downstream genes of let-7, the amyloid precursor protein-like 1 (apl-1), is of particular interest.Citation18 apl-1 is a homologue of the human amyloid precursor protein (APP), a gene known to be associated with the progression of Alzheimer's disease.Citation19,Citation20 In C. elegans, apl-1 has been shown to be involved in neuronal plasticity, pharyngeal pumping, and molting.Citation21-23 Notably, Niwa et al. showed that apl-1 suppresses the let-7 retarded phenotypes and genetically interacts with hbl-1, lin-41, and lin-42.Citation18 In this study, we aimed to further elucidate the role of apl-1 in developmental timing in order to both gain insights into the mechanisms of the heterochronic pathway and potentially elucidate the function of an Alzheimer's disease-associated gene.
To this end, we identified a list of genes that could potentially interact with both apl-1 and other components of the heterochronic pathway. Human APP is intensely studied due to its connection with Alzheimer's disease, so there is a vast body of literature, including several genome-wide association studies (GWAS), on APP. We hypothesized that the C. elegans homologues of human genes that have been shown to interact with APP may in turn interact with the nematode apl-1 gene. Thus, we screened the C. elegans homologues of genes implicated in Alzheimer's disease for interactions with heterochronic genes.Citation24-37 Angiotensin-converting enzyme-like non-peptidase (acn-1), whose homologue ACE has been shown to interact with APP in GWAS, was one such gene. Here we identify acn-1 as a novel component of the heterochronic pathway. Specifically, we demonstrate that acn-1 suppresses let-7 phenotypes and enhances the phenotypes of apl-1 and hbl-1, though not lin-41. Conversely, loss of lin-41 but not hbl-1 alters the oscillatory pattern of acn-1 expression. This study suggests that acn-1 is a new factor involved in the developmental timing of seam cells in C. elegans. Moreover, as several studies have identified an association between mammalian ACE and Alzheimer's disease, our characterization of ACE's homologue in a model organism might prove useful in elucidating the nature of this association.
Results
Knockdown of acn-1 suppresses let-7(n2853) lethality
GWAS of Alzheimer's disease yielded 22 human genes with single nucleotide polymorphisms (SNPs) exhibiting statistically significant correlation with the disease phenotype.Citation24-26,Citation28-34,Citation37 An additional 23rd gene on our list was beta-secretase (BACE), which is known to cleave APP.Citation35,Citation36 Of these 23 human genes, 14 of them have C. elegans homologues as identified by the Basic Local Alignment Search Tool (BLAST) (). Due to constraints in RNAi construct availability and previously reported lethal effect of some RNAi constructs, we ultimately examined 24 C. elegans genes corresponding to 10 of these 14 human genes (). shows all genes that were examined here.
Figure 1. Knockdown of acn-1 partially suppresses let-7 lethality. This graph shows the result of an RNAi screen for let-7 suppressors. RNAi constructs targeting C. elegans homologues of the genes listed in the table in this figure were screened for suppression of the bursting-vulva phenotype of let-7(n2853) at the L4/adult transition. Except when noted, let-7(n2853) animals were placed on RNAi bacteria at L1. Animals marked with L2* were fed with empty vector RNAi clones for the first 16 hours before being moved to plates with the indicated RNAi constructs. Animals marked with L2** were fed with the OP50 strain of E. coli for 16 hours before being moved. Empty L4440 vectorCitation60 was used as a negative control; daf-12 RNAi was used as a positive control.Citation39
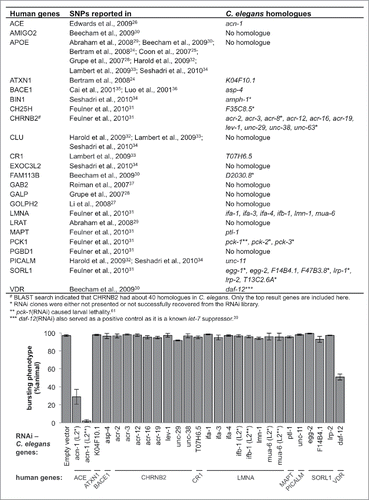
We hypothesized that some of these 24 C. elegans genes would interact with apl-1 and by extension the heterochronic genes with which apl-1 interacts. let-7 has one of the strongest mutant phenotypes of any heterochronic gene: loss of let-7 causes both retarded development of the seam cells and loss of vulval structure, resulting in lethality by bursting through the vulva soon after the L4/adult transition.Citation8 Identifying suppressors of let-7 mutant phenotypes is thus one of the most amenable ways to screen for novel heterochronic genes. For instance, loss of most of let-7's several downstream target genes suppresses both the vulval-related lethality and abnormal seam cell development phenotypes of let-7 mutants.Citation8,Citation9,Citation11,Citation12,Citation38,Citation39 In order to identify suppressors of the let-7 mutant phenotypes, we first examined whether RNAi knockdown of our candidate genes diminished the lethality caused by loss of let-7. For most RNAi constructs, let-7 mutant animals were fed E. coli carrying RNAi plasmids starting at the L1 larval stage; however, in order to bypass the early lethal effects of ifb-1 and mua-6 RNAi knockdowns, let-7(n2853) animals were fed the same RNAi construct-carrying bacteria starting from the L2 stage instead of L1. The majority of the RNAi constructs we screened resulted in more than 90% of animals bursting through the vulva, indicating no suppression of let-7 mutant phenotypes ().
acn-1 was the only gene whose knockdown appeared to suppress the let-7 bursting vulva phenotype. Animals fed with acn-1(RNAi) from the L1 stage mostly arrested at the L2-L3 stages and therefore never survived to the L4/adult transition during which bursting occurs. The early larval arrest phenotype observed with L1 RNAi feeding was phenocopied by the acn-1(null) mutant.Citation40 In order to overcome the problem of premature developmental arrest, we began feeding let-7(n2853) animals with acn-1(RNAi) starting at the L2 stage; as a result, more than 90% of animals survived to the L4/adult molt when they could be scored for bursting. Among these, less than 30% died by bursting (). The animals that survived appeared smaller than N2 wild-type animals but were able to lay eggs, confirming that they had reached the adult stage.
Knockdown of acn-1 suppresses let-7(n2853) retarded seam cell defects
Since bursting is a vulval phenotype, we next investigated whether knockdown of acn-1 affects development of the seam cells. While vulval cells undergo post-embryonic cell divisions, they do not divide during every larval stage as seam cells do; observing seam cells can therefore reveal more details of stage-by-stage development and heterochrony. let-7(n2853) mutants have retarded seam cell phenotypes: the seam cells do not stop dividing at the larval/adult transition and undergo terminal differentiation. This results in the transient detection of more than the normal 16 seam cells per side right after the L4/adult molt due to the presence of dividing daughter cells that should have already terminally differentiated;Citation18 additionally, adult alae are not completely produced at the young adult stage as normally observed in wild-type animals.Citation8 To visualize the seam cells in our knockout animals, we used a let-7(n2853);wIs79 strain (the wIs79 construct contains ajm-1::gfp;scm-1::gfp seam cell markers). Knocking down acn-1 eliminated the supernumerary seam cell divisions (scored as the normal number of 16 seam cells), suggesting that a few seam cells that may still undergo cell division due to let-7 mutation have stopped dividing when acn-1 was reduced (). Moreover, silencing acn-1 by RNAi also suppressed the retarded alae production phenotype caused by loss of let-7 (). Even though knocking down acn-1 restored some adult alae formation (‘any alae’ column), it disrupted the morphology of alae such that alae were not complete (‘>50% alae’ column) () – an observation consistent with previous studies.Citation40
Table 1. Knockdown of acn-1 partially suppressed the retarded phenotypes in seam cells of let-7 mutants. let-7 mutants have retarded seam cell phenotypes – displaying supernumerary divisions and transiently producing more than the normal 16 seam cells and delaying adult alae formation.Citation8 let-7(n2853);wIs79 animals were scored right after the L4/adult transition but before bursting. Knocking down acn-1 partially suppressed both phenotypes.
Loss of acn-1 enhances the loss-of-apl-1 suppressor phenotype and affects apl-1 expression
The let-7(n2853)-suppressing phenotypes of acn-1 knockdowns are similar to those of apl-1 mutants;Citation18 thus, we tested whether subjecting the animals to both RNAi simultaneously would enhance the heterochronic defects of the individual knockdowns. Since both apl-1(RNAi) and acn-1(RNAi) already suppressed the bursting vulva phenotype very effectively, any enhancement of this suppression could not be easily distinguished ( those labelled with ‘100%’); thus, we lowered the concentration of RNAi construct-carrying E. coli strain HT115 by mixing in E. coli containing only empty vector (L4440). We found that knocking down apl-1 and acn-1 together, at concentrations where knocking down either alone had little effect, suppressed the bursting vulva phenotype at a significantly higher rate (). Thus, apl-1 and acn-1 synergistically suppress the let-7 phenotype.
Figure 2. Loss of acn-1 enhanced the apl-1(RNAi) phenotype and affected apl-1 expression. (A) RNAi-mediated knockdown of acn-1 and apl-1 enhanced the suppression of the let-7 bursting-vulva phenotype. This effect was dose-dependent. A gradient of the quantity of RNAi was achieved by mixing the indicated percentage of tested RNAi construct with the corresponding amount of empty vector RNAi (experimental details in Materials and Methods). We observed a significant enhancement of the suppression phenotype in animals fed with a double RNAi construct (dbRNAi) targeting both acn-1 and apl-1 sequences. 10–19% of animals fed with dbRNAi stalled at the L1 or L2 stage and were excluded from the calculation of the percentage of animals exhibiting the bursting-vulva phenotype. The L1 and L2 labels designate the developmental stage at which animals were placed on RNAi bacteria. (B) Expression of the apl-1::gfp::unc-54 3′UTR construct in hypodermal seam cells at different developmental stages in nematodes fed empty vector (L4440) or acn-1 RNAi. Shown is the percentage of animals with seam cell GFP expression.
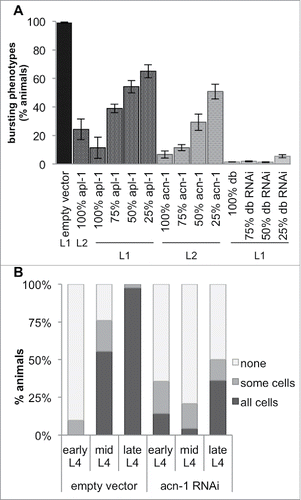
apl-1 is expressed from the L4 stage into adulthood,Citation18 while acn-1 is expressed in all larval stages and disappears during adulthood.Citation40 We thus tested whether acn-1 regulates apl-1 expression, employing wild-type animals with apl-1 promoter::gfp::unc-54 3’UTR constructs to measure apl-1 transcription. Only ∼10% of these animals expressed GFP at the early L4 stage, and when they did it was found in only a few seam cells (); expression of apl-1::gfp then increased during the mid-L4 stage to almost 100% of animals expressing it in all seam cells (). Knockdown of acn-1 disrupted this expression pattern (). Whereas about 75% and 100% of control animals expressed apl-1 at the mid- and late-L4 stages respectively, only about 20% and 50% of acn-1 knockdowns did the same (). This effect of acn-1 knockdown on apl-1 transcription parallels that of lin-41 silencing.Citation18
acn-1(lf) is not sufficient to cause heterochronic phenotypes
Since acn-1 is able to suppress let-7(n2853) retarded phenotypes, we hypothesized that acn-1(lf) might be sufficient to cause precocious phenotypes in hypodermal seam cells. However, this was not the case. acn-1 knockdown animals consistently failed to exhibit precocious hypodermal seam cell fusion or adult alae (data not shown). Therefore, acn-1 does not behave as a canonical heterochronic gene by itself.
Knockdown of acn-1 enhances the precocious phenotypes of hbl-1 mutants but not of lin-41 mutants
In wild-type animals, lateral hypodermal seam cells divide during the L1, L2, L3, and L4 stages before terminally differentiating and fusing together at the L4-adult molt. After fusion, these cells secrete a cuticular structure called the adult alae. Knockdown or mutation of hbl-1 results in precocious seam cell terminal differentiation and adult alae formation, albeit with less than 100% penetrance.Citation9,Citation12
Consistent with previous studies,Citation9,Citation12 we observed that 60% of hbl-1(mg285);wIs79(ajm-1::gfp;scm-1::gfp) animals exhibited at least one seam cell fusion event in L4; while 7% – 12% did so during the early and late L3 stage respectively (). On the other hand, in this background, 100% of acn-1(RNAi) knockdowns exhibited at least one precocious seam cell fusion event at the L4 stage (). More notably, 58% and 47% did the same at the late L3 and early L3 stages respectively () – the first time that precocious seam cell fusion has been reported in hbl-1(mg285) animals at a stage earlier than the L3 to L4 molt.
Table 2. Knockdown of acn-1 enhanced the precocious phenotypes of hbl-1(mg285) but did not affect those of lin-41(ma104).hbl-1(mg285) led to precocious hypodermal seam cell fusion and adult alae production though not with complete penetrance.Citation9,Citation12 RNAi knockdown of acn-1 enhanced these phenotypes further, promoting seam cell fusion and alae production as early as the L3 stage. lin-41(ma104) animals also exhibited precocious seam cell fusion and adult alae production during the L4 stage,Citation11 but knocking down acn-1 did not enhance these phenotypes.
Similarly, we also examined whether acn-1 knockdown enhanced the precocious adult alae production characteristic of hbl-1(mg285) mutants. Whereas 4% and 10% of hbl-1(mg285);wIs79 animals produced at least some alae at the early and late L3 stages respectively, 29% and 49% of acn-1 RNAi knockdowns did the same () – a marked increase due to acn-1 knockdown. Incidentally, the percentage of animals with any alae at late L3 was not significantly different from that at early L3.
These increases in the rates of both precocious seam cell fusion and alae production are statistically significant, as analyzed by Fisher's exact test (). These results clearly demonstrate that knockdown of acn-1 enhances the precocious phenotypes caused by loss of hbl-1.
Since hbl-1 and lin-41 are both targets of the let-7 miRNA, and since hbl-1 mutants exhibit enhanced penetrance of the precocious phenotypes of lin-41 mutants, it has been proposed that hbl-1 and lin-41 function in parallel.Citation9,Citation11,Citation12 Therefore, we investigated whether acn-1 could enhance lin-41 mutant phenotypes in the same manner that it did hbl-1 mutant phenotypes. We observed that lin-41(ma104);wIs79 animals exhibited precocious seam cell fusion and adult alae production, consistent with a previous report.Citation11 However, knockdown of acn-1 produced no increase in the percentage of lin-41 mutants exhibiting those phenotypes (). Thus, although knockdown of acn-1 enhances the precocious phenotypes of hbl-1 mutants, it does not have the same effect on lin-41 mutants.
The oscillatory expression pattern of acn-1 is influenced by lin-41 but not hbl-1
It has previously been reported that acn-1::gfp fusion reporter lines – which include the 5' UTR sequence, acn-1 coding sequence, and GFP – exhibit GFP expression in hypodermal seam cells starting during embryogenesis and continue until the young adult stage.Citation40 However, Frand et al. (2005) allude to unpublished data demonstrating that acn-1p::gfp-pest (a shorter-lived form of GFP) displays cyclic expression during all four molts.Citation41 In addition, two recent global studies of mRNA expression during C. elegans larval development demonstrate that acn-1 exhibits an oscillatory expression pattern.Citation42,Citation43 We confirmed this observation in wild-type animals by collecting RNA every 2 hours between the early L2 and early L3 stages, and measuring acn-1 transcription levels using qRT-PCR ( and ).
Figure 3. lin-41 influences acn-1’s oscillating expression pattern. (A and B) qRT-PCR of acn-1 mRNA from wild-type (N2) nematodes shows oscillating expression. These time points cover the period between early L2 and early L3 (the L2/L3 transition takes place between 18 and 20 hrs post-L1.) (A) hbl-1(mg285) mutation led to a slight rightward phase shift in the oscillation of acn-1 expression but no significant differences in any oscillation parameters, while (B) lin-41(ma104) mutation significantly dampened the amplitude of acn-1’s oscillations.
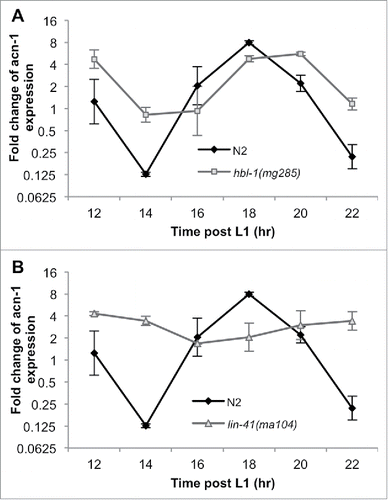
We then repeated this temporal acn-1 transcriptional analysis in hbl-1(mg285) animals and found that hbl-1 mutant animals also exhibited an oscillatory acn-1 expression pattern, albeit with some small differences in the oscillatory function parameters. Loss of hbl-1 did not affect the period of acn-1’s oscillations and the oscillations were only slightly phase-shifted to the right (p-value = 0.81) relative to those of wild-type animals (). Additionally, their amplitude was somewhat dampened (-0.87 ± 0.86 cycles, corresponding to a 0.65-fold change (p-value = 0.31) from that in wild-type animals) (). These changes observed were too small and variable to prove any functionally significant influence by hbl-1 on the expression profile of acn-1.
On the other hand, lin-41(ma104) mutant animals exhibited a significantly disrupted acn-1 expression pattern (). Though the mean acn-1 expression level remained relatively unchanged from that of wild-type animals, the amplitude of acn-1’s oscillations appeared to be greatly dampened (-1.73 ± 0.71 cycles, corresponding to a 0.29 fold change (p-value = 0.01) from that in wild-type animals) (), indicating that a functional copy of the lin-41 gene is necessary to maintain the normal oscillatory pattern of acn-1 expression. For other oscillation parameters, loss of lin-41 does not significantly affect the phase (slightly right-shifted) or the period of acn-1’s oscillations ().
In order to test whether acn-1 regulates expression of hbl-1 or lin-41 in a feedback loop, we treated wild-type animals with empty vector (L4440) and acn-1 RNAi constructs, collected them at 12 (L2), 21 (L3), and 29 (L4) hours post-treatment, and measured levels of hbl-1 and lin-41 mRNA using qRT-PCR. We found that the temporal expression levels of hbl-1 and lin-41 mRNAs remained unchanged in acn-1 knockdowns relative to those in control animals (data not shown). Therefore, it is unlikely that a feedback loop exists between acn-1 and either hbl-1 or lin-41.
Discussion
acn-1 genetically interacts with genes in the heterochronic pathway
In this study, we reveal several genetic interactions between acn-1 and certain heterochronic genes. Although knockdown of acn-1 does not on its own produce heterochronic phenotypes in the hypodermal seam cells, it partially suppresses the retarded phenotypes of let-7 mutants and enhances the precocious phenotypes of hbl-1 and apl-1 mutants. These results suggest that acn-1 promotes the larval identity of hypodermal seam cells, presumably in cooperation with other genes.
Since the acn-1 3' UTR contains no sequence with partial complementarity to let-7 or let-7 family member miRNAs (mir-48, mir-84, and mir-241), it is unlikely that acn-1 is a direct target of let-7 or its family members. Rather, acn-1 is likely an indirect target of let-7 that acts further downstream in the heterochronic pathway ().
Figure 4. A proposed model of the relationships between acn-1 and the heterochronic genes let-7, hbl-1, and lin-41.
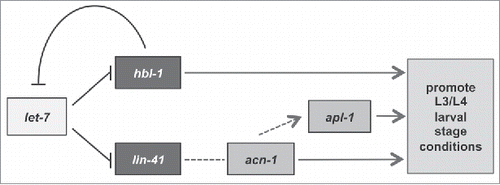
It has been proposed that hbl-1 and lin-41 function in parallel for three reasons: both genes are targets of let-7 and its family members, the majority of hbl-1 and lin-41 mutant phenotypes are identical, and knockdown of either gene by RNAi enhances the penetrance of the other's precocious phenotype.Citation9,Citation11,Citation12 In this study, we identify distinct features between hbl-1's and lin-41's interactions with acn-1. Knockdown of acn-1 enhances the precocious phenotypes of hbl-1 mutants but does not affect those of lin-41 mutants. Furthermore, whereas loss-of-function lin-41(ma104) mutations affect the oscillatory pattern of acn-1 expression, hbl-1(mg285) mutations do not. These data suggest that acn-1 may lie in the lin-41 arm of the pathway () and that lin-41 but not hbl-1 regulates acn-1 transcription in some way.
While mean acn-1 expression is not significantly diminished in lin-41(ma104) mutant animals, the amplitude of acn-1’s oscillations is considerably reduced. It is therefore unlikely that lin-41 functions in a straightforward way to promote or suppress acn-1 transcription. In a study of global mRNA expression, lin-41 was also shown to exhibit an oscillatory expression pattern with the same 8-hour period as that of acn-1, albeit with a much smaller amplitude (1.48 units) than that of acn-1 (3.94 units).Citation42 In wild-type nematodes during the L2-L3 sampling period, we observed some fluctuation in lin-41 expression, though not large enough to verify a pattern of oscillation (data not shown). On the other hand, hbl-1 is expressed at a steady level until it is down-regulated during the L3 stage.Citation9,Citation12,Citation42,Citation43 These findings indicate that lin-41 may play a role in regulating C. elegans’s cyclic progression through the larval stages,Citation42 and our results suggest that this regulation may at least partially occur through its interaction with acn-1. Such a model would explain why loss of lin-41 disrupts acn-1’s oscillatory expression pattern. However, it may also be the case that loss of lin-41 causes animals to develop less synchronously, thus making it difficult to fit a single sinusoidal curve to an asynchronous population.
A simple explanation of why knocking down acn-1 enhances the penetrance of the hbl-1(mg285) mutant's precocious phenotypes is that acn-1 functions in a pathway parallel to that of hbl-1 (), most likely the pathway regulated by lin-41. Such an indirect relationship between acn-1 and hbl-1 is supported by our observation that hbl-1(mg285) does not significantly affect the expression of acn-1. The slight change in acn-1 expression in hbl-1(mg285) mutants might have resulted from the negative feedback loop between hbl-1 and let-7,Citation17 which would indirectly affect lin-41 expression via let-7. Moreover, the hypothesis that acn-1 operates downstream of lin-41 is consistent with our findings that acn-1 knockdown does not enhance lin-41(ma104)’s heterochronic phenotypes in hypodermal seam cells as lin-41(ma104)’s seam cell phenotypes would accordingly already encompass the extent of acn-1 knockdown's phenotypes (). Thus, it appears that while both lin-41 and hbl-1 promote the same larval stage cell fate decisions, they may work through different genes to complement and achieve the same goal.
Heterochronic genes and molting
Previous studies have connected the process of molting to several genes that were originally shown to control the timing of hypodermal seam cell development. It has been demonstrated that mir-84 and let-7 promote the cessation of the molting cycle, and that several heterochronic genes – namely, lin-28, lin-14, lin-41, lin-42, and hbl-1 – are necessary to that end.Citation44 Two of the main signaling molecules that regulate molting (generating new cuticles and promoting ecdysis) are the nuclear hormone receptors, nhr-23 and nhr-25.Citation45-47 mir-84 and let-7 have been shown to require functional copies of nhr-23 and nhr-25 to terminate molting.Citation44 Moreover, knockdown of apl-1 was found to suppress the cuticle defect caused by the mir-48 and mir-84 mutations.Citation18 In addition, nhr-25 was shown to regulate apl-1 expression, and these two genes were found to act synergistically to cause a molting defect.Citation48
On the other hand, acn-1 was initially demonstrated to be necessary to promote the molting process.Citation40,Citation41 Feeding acn-1(RNAi) at the L1 larval stage leads to unsuccessful molting (generally at the L2/L3 molt).Citation40,Citation41 Furthermore, knocking down nhr-23 and nhr-25 reduces acn-1::gfp expression in the hypodermal seam cells.Citation40
In this study, we reveal novel interactions between the canonical heterochronic genes and acn-1, a gene that was previously thought to be just a molting gene. Indeed, acn-1 is involved not only in the molting cycle, which occurs at regular intervals during the progression of C. elegans through the larval stages, but also in cell fate decisions leading up to terminal differentiation of the hypodermal seam cells.
The interaction between human ACE and APP suggests a potential interaction between the C. elegans homologues, acn-1 and apl-1
It is well-established that mutations in the APP gene lead to early-onset Alzheimer's disease (AD).Citation20 However, the protein's normal function is less well understood. ACE has emerged as a gene linked to APP and AD. While the main function of the ACE protease is to cleave angiotensin I to produce the active form, angiotensin II, as part of the renin-angiotensin aldosterone system (RAAS) that regulates blood pressure,Citation49 several GWAS studies have associated ACE with APP and Alzheimer's Disease (for a review see Citationref. 50). Indeed, several point and insertion/deletion mutations in ACE1 exhibit high correlation with expression of the disease phenotype.Citation50,Citation51 Moreover, ACE has been shown to degrade APP-derived Aβ peptide in vitro.Citation52-54 Finally, administration of an ACE inhibitor was found to lead to improved learning and memory in mice.Citation55-57 Although these findings suggest a link between ACE, Alzheimer's disease, and APP processing, the exact mechanism of ACE’s contribution to Alzheimer's disease remains unclear.Citation27,Citation31,Citation57
In our exploration of the connection between the C. elegans homologues of ACE and APP we found them to form a mutual relationship in the heterochronic pathway. We found that loss of acn-1 enhanced apl-1(RNAi)’s suppression of the let-7 mutant phenotype. In addition, knockdown of acn-1 led to changes in the temporal expression pattern of apl-1. The C. elegans acn-1 protein lacks the key amino acids that bind zinc molecules in the metallopeptidase active site of ACE; consequently, acn-1 lacks ACE’s peptidase activity.Citation40 However, ACN-1's active sites may still bind peptides without cleaving them.Citation40 Here we show that acn-1 genetically and functionally interacts with apl-1 even without the metallopeptidase active site sequences. Given the complexity of the relationship between ACE1 and APP in humans, there might be several facets of their interactions that remain unknown. It would be interesting to further examine interactions between not only the human homologues, but between homologues in other model organisms as well.
Materials & methods
Nematode strains and culture
C. elegans were grown on standard nematode growth media (NGM) plates and fed with the OP50 strain of E.coli.Citation58 Nematodes were kept at either 20oC or 23oC. They were synchronized by bleaching (5% 5N NaOH, 10% bleach in M9 solution) and hatched in M9 without E. coli overnight before starvation-arrested L1 were plated for each experiment.
Strains used were: wild-type N2 Bristol, let-7(n2853),Citation8 let-7(n2853);wIs79(ajm-1::gfp;scm-1::gfp),Citation18 apl-1::gfp::unc-54 3’UTR,Citation18 wIs79(ajm-1::gfp;scm-1::gfp),Citation18 hbl-1(mg285),Citation9 and lin-41(ma104).Citation11 hbl-1(mg285);wIs79(ajm-1::gfp;scm-1::gfp) and lin-41(ma104);wIs79(ajm-1::gfp;scm-1::gfp) were created by R. Niwa. acn-1::gfp (rol-6 marker)Citation40 was kindly given to us by R. Elwyn Isaac.
RNAi experiments
Target genes were knocked down by feeding RNAi to the animals. All RNAi constructs were taken from the Ahringer lab's RNAi libraryCitation59 except for the apl-1;acn-1 RNAi construct, which was created by combining the two RNAi inserts in the library into a single construct via molecular cloning. All plasmids were expressed in the HT115 strain of E. coli. As a negative control, animals were fed E. coli containing the empty L4440 vector, the parental plasmid of other RNAi constructs.Citation60 These E. coli strains were selected on LB plates with ampicillin and tetracycline, grown in LB media with ampicillin, and induced with IPTG before being fed to the animals.Citation60 Except for acn-1(RNAi), synchronized nematodes were fed with RNAi bacteria starting from the L1 stage. Feeding animals with acn-1(RNAi) from the L1 stage resulted in arrested development at the L2/L3 molt (as also observed by Brooks et al.Citation40); therefore animals were fed with acn-1(RNAi) starting from the L2 stage.
In the vulva bursting assay, 100–200 let-7(n2853) mutants were placed on each RNAi plate. Each data point shown is the average across 3 plates of the percentage of animals that died by bursting.
In the experiment using different dilutions of RNAi, we first adjusted the concentration of each RNAi construct with LB media to control for the concentration of bacteria, as determined by optical density at 600nm (OD600). We used that starting concentration of RNAi bacteria as ‘100% RNAi’. The RNAi dilutions labeled as ‘75%, 50%, and 25%’ were diluted with E. coli containing the empty vector RNAi (L4440) at the same OD600 at ratios of 3:1, 1:1, and 1:3 of specific RNA:L4440 respectively.
Observation of animals by microscopy
Nematodes were anatomically staged based on the length of the gonad.Citation9,Citation11 Early, mid, and late L4 animals were categorized by whether their gonadal distal arms were 0 – ¼, ¼ – ½, or >½ the length of the proximal arms respectively. Early L3 animals were categorized both by the size of the animals to ensure that it has passed the L2/L3 molt and by a gonad length not significantly greater than that of the L2's gonad; late L3 animals were categorized by gonads approaching the gonadal turns. Animals were observed using an Axioplan II microscope (Carl Zeiss) equipped with an AxioCam MRm CCD camera (Carl Zeiss).
In and , animals were examined for alae production (either any at all or covering >50% of the length of the animal) and at least partial seam cell fusion.
The apl-1::gfp::unc-54 3’UTR was strongly expressed in the head region, so it was difficult to distinguish seam cells expression there.Citation18 In , what we described as GFP expressed in “all cells” refers to all seam cells that could be distinguished, excluding those in the head region.
RNA extraction, Reverse Transcription, and Quantitative RT-PCR
Animals were synchronized by bleaching (see ‘nematode strains and culture’). Starvation-arrested L1 animals were placed on NGM plates with OP50 and kept at 23oC for the amount of time specified before sample collection. RNA extraction was done using a mirVana miRNA Isolation Kit (ThermoFisher, https://www.thermofisher.com/order/catalog/product/AM1560). RNA samples were treated with Turbo DNase (ThermoFisher, https://www.thermofisher.com/order/catalog/product/AM1907). cDNA synthesis was performed according to the protocol for SuperScript III Reverse Transcriptase (ThermoFisher, https://www.thermofisher.com/order/catalog/product/18080044). We used SYBR Green-based real-time PCR for expression analysis (Roche, https://shop.roche.com/shop/products/lightcycler14301-480-sybr-green-i-master and the LightCycler 480 Instrument; https://shop.roche.com/shop/products/lightcycler14301-480-instrument-ii). 2.5 ng of cDNA was used per reaction. Primers used for qRT-PCR include: CTGCTGGACAGGAAGATTACG and CTCGGACATTCTCGAATGAAG for cdc-42; GTTCCCGTGTTCATCACTCAT and ACACCGTCGAGAAGCTGTAGA for pmp-3; GTCGCTTCAAATCAGTTCAGC and GTTCTTGTCAAGTGATCCGACA for Y45F10D.4;Citation61 GACAAGTCCTCCCTTTCCATC and GACAAGTCCTCCCTTTCCATC for acn-1; GCAGTTCACAAATTTCATCCGG and CTCGCCATCTTTCTCCATTGC for hbl-1; and CGGCTCCGTATTCGTCTCT and GATCGTCCCACTGATTTGGC for lin-41.
Fit of the acn-1 oscillatory pattern to a sinusoidal curve
Total RNA was collected every 2 hrs starting at t = 12 hrs and ending at t = 22 hrs post-hatching, and acn-1 expression levels at each time point were measured using quantitative RT-PCR. ddCt values were set to a sinusoidal function of time of the form y = A*sin(B*t + C) + D in MatLab to determine the accuracy of the fit. Next, confidence intervals for each parameter were obtained using Bayesian statistical inference. A random walk was performed using the “Just Another Gibbs Sampling” (JAGS) package in R to obtain 100,000 sets of parameter values, from which statistical values could be inferred. All parameters were assumed to have uniform prior distributions, and the distribution of real data values about the fit was assumed to be normal.
Abbreviations
| acn-1 | = | angiotensin-converting enzyme-like, non-peptidase-1 |
| ACE | = | angiotensin-converting enzyme |
| APP | = | amyloid precursor protein |
| miRNA | = | microRNA |
Disclosure of potential conflicts of interest
The authors have no conflict of interest.
Acknowledgments
The authors thank R. Elwyn Isaac and Ryusuke Niwa for C. elegans strains; and Alexandre de Lencastre and Sachi Inukai for critical reading of this manuscript.
Funding
F.J.S. was supported by a grant from the NIH (R01AG033921).
References
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110-56. doi:10.1016/0012-1606(77)90158-0. PMID:838129
- Slack F, Ruvkun G. Temporal Pattern Formation by Heterochronic Genes. Annu Rev Genet. 1997;31:611-34. doi:10.1146/annurev.genet.31.1.611. PMID:9442909
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol [Internet]. 2002;18:495-513. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12142272. doi:10.1146/annurev.cellbio.18.012502.105832
- Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol [Internet]. 2007;17:R425-34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17550772. doi:10.1016/j.cub.2007.03.043
- Ambros V, Horvitz HR. Heterochronic Mutants of the Nematode Caenorhabditis elegans. Science (80- ). 1984;226:409-16. doi:10.1126/science.6494891
- Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma [Internet]. 2009;118:405-18. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19340450. doi:10.1007/s00412-009-0210-z
- Abbott AL, Alvarez-saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA Family Members mir-48 , mir-84 , and mir-241 Function Together to Regulate Developmental Timing in Caenorhabditis elegans. Dev Cell. 2005;9:403-14. doi:10.1016/j.devcel.2005.07.009. PMID:16139228
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901-6. doi:10.1038/35002607. PMID:10706289
- Lin S, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C . elegans hunchback Homolog , hbl-1 , Controls Temporal Patterning and Is a Probable MicroRNA Target. Dev Cell. 2003;4:639-50. doi:10.1016/S1534-5807(03)00124-2. PMID:12737800
- Vella MC, Choi E-Y, Lin S-Y, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3’UTR. Genes Dev. 2004;18:132-7. doi:10.1101/gad.1165404. PMID:14729570
- Slack F, Basson M, Liu Z, Ambros V, Horvitz H, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell [Internet]. 2000;5:659-69. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10882102. doi:10.1016/S1097-2765(00)80245-2
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell [Internet]. 2003;4:625-37. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12737799. doi:10.1016/S1534-5807(03)00127-8
- Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol [Internet]. 1999;205:240-53. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9917360. doi:10.1006/dbio.1998.9096
- Lehmann R, Nüsslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119:402-17. doi:10.1016/0012-1606(87)90045-5. PMID:3803711
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn E a, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol [Internet]. 2009;11:1411-20. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19898466. doi:10.1038/ncb1987
- Schulman BRM, Liang X, Stahlhut C, Delconte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and seural tube closure. Cell cycle. 2008;7:3935-42. doi:10.4161/cc.7.24.7397. PMID:19098426
- Roush SF, Slack FJ. Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Dev Biol. 2009;334:523-34. doi:10.1016/j.ydbio.2009.07.012. PMID:19627983
- Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer's amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev Biol. 2008;315:418-25. doi:10.1016/j.ydbio.2007.12.044. PMID:18262516
- Daigle I, Li C. apl-i , a Caenorhabditis elegans gene encoding a protein related to the human beta-amyloid protein precursor. Proc Natl Acad Sci U S A. 1993;90:12045-9. doi:10.1073/pnas.90.24.12045. PMID:8265668
- Rossor MN, Newman S, Frackowiak RSJ, Lantos P, Kennedy AM. Alzheimer's disease families with amyloid precursor protein mutations. In: Annals of the New York Academy of Sciences. 1993. page 198-202.
- Ewald CY, Li C. Understanding the molecular basis of Alzheimer's disease using a Caenorhabditis elegans model system. Brain Struct Funct. 2010;214:263-83. doi:10.1007/s00429-009-0235-3. PMID:20012092
- Zambrano N, Bimonte M, Arbucci S, Gianni D, Russo T, Bazzicalupo P. feh-1 and apl-1, the Caenorhabditis elegans orthologues of mammalian Fe65 and beta-amyloid precursor protein genes, are involved in the same pathway that controls nematode pharyngeal pumping. J Cell Sci [Internet]. 2002;115:1411-22. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11896189
- Hornsten A, Lieberthal J, Fadia S, Malins R, Ha L, Xu X, Daigle I, Markowitz M, O'Connor G, Plasterk R, et al. APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc Natl Acad Sci U S A. 2007;104:1971-6. doi:10.1073/pnas.0603997104. PMID:17267616
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BMM, Hooli B, Divito J, Ionita I, et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623-32. doi:10.1016/j.ajhg.2008.10.008. PMID:18976728
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson J V, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, et al. A High-Density Whole-Genome Association Study Reveals That APOE Is the Major Susceptibility Gene for Sporadic Late-Onset Alzheimer's Disease. J Clin Psychiatry. 2007;614-9.
- Edwards TL, Pericak-Vance M, Gilbert JR, Haines JL, Martin ER, Ritchie MD. An association analysis of Alzheimer disease candidate genes detects an ancestral risk haplotype clade in ACE and putative multilocus association between ACE, A2M, and LRRTM3. Am J Med Genet Part B. 2009;150B:721-35. doi:10.1002/ajmg.b.30899. PMID:19105203
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol [Internet]. 2008;65:45-53. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17998437
- Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, et al. Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum Mol Genet [Internet]. 2007;16:865-73. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17317784. doi:10.1093/hmg/ddm031
- Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC, et al. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics. 2008;1:44. doi:10.1186/1755-8794-1-44. PMID:18823527
- Beecham GW, Martin ER, Li Y-J, Slifer M a, Gilbert JR, Haines JL, Pericak-Vance M a. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. 2009;84:35-43. doi:10.1016/j.ajhg.2008.12.008. PMID:19118814
- Feulner TM, Laws SM, Friedrich P, Wagenpfeil S, Wurst SHR, Riehle C, Kuhn K a, Krawczak M, Schreiber S, Nikolaus S, et al. Examination of the current top candidate genes for AD in a genome-wide association study. Mol Psychiatry [Internet]. 2010;15:756-66. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19125160. doi:10.1038/mp.2008.141
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088-93. doi:10.1038/ng.440. PMID:19734902
- Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094-9. doi:10.1038/ng.439. PMID:19734903
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith A V, Carassquillo MM, Lambert JC, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832-40. doi:10.1001/jama.2010.574. PMID:20460622
- Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, Wong PC. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci [Internet]. 2001;4:233-4. Available from: https://doi.org/10.1038/85064. doi:10.1038/85064
- Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, Kha H, Zhang J, Gong Y, et al. Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci [Internet]. 2001;4:231-2. Available from: https://doi.org/10.1038/85059. doi:10.1038/85059
- Reiman EM, Webster J a, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson J V, Hu-Lince D, Huentelman MJ, et al. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54:713-20. doi:10.1016/j.neuron.2007.05.022. PMID:17553421
- Lall S, Grün D, Krek A, Chen K, Wang Y-L, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol [Internet]. 2006;16:460-71. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16458514. doi:10.1016/j.cub.2006.01.050
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell [Internet]. 2005;8:321-30. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15737928. doi:10.1016/j.devcel.2004.12.019
- Brooks DR, Appleford PJ, Murray L, Isaac RE. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J Biol Chem [Internet]. 2003;278:52340-6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14559923. doi:10.1074/jbc.M308858200
- Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi:10.1371/journal.pbio.0030312
- Hendriks G-J, Gaidatzis D, Aeschimann F, Großhans H. Extensive oscillatory gene expression during C. elegans larval development. Mol Cell [Internet]. 2014;53:380-92. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24440504.
- Kim DH, Grün D, van Oudenaarden A. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat Genet. 2013;45:1337-44. doi:10.1038/ng.2763. PMID:24036951
- Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development [Internet]. 2006;133:4631-41. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17065234. doi:10.1242/dev.02655
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:7360-5. doi:10.1073/pnas.131171898. PMID:11416209
- Asahina M, Ishihara T, Jindra M, Kohara Y, Katsura I, Hirose S. The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells. 2000;5:711-23. doi:10.1046/j.1365-2443.2000.00361.x. PMID:10971653
- Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259-72. doi:10.1006/dbio.2000.9679. PMID:10772806
- Hada K, Asahina M, Hasegawa H, Kanaho Y, Slack FJ, Niwa R. The nuclear receptor gene nhr-25 plays multiple roles in the Caenorhabditis elegans heterochronic gene network to control the larva-to-adult transition. Dev Biol. 2010;344:1100-9. doi:10.1016/j.ydbio.2010.05.508. PMID:20678979
- Carey RM. Newly discovered components and actions of the renin-angiotensin system. Hypertension [Internet]. 2013;62:818-22. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24082058. doi:10.1161/HYPERTENSIONAHA.113.01111
- Kehoe PG, Russ C, McIlory S, Williams H, Holmans P, Holmes C, Liolitsa D, Vahidassr D, Powell J, McGleenon B, et al. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat Genet [Internet]. 1999;21:71-2. Available from: https://doi.org/10.1038/5009. doi:10.1038/5009
- Lehmann DJ, Cortina-Borja M, Warden DR, Smith AD, Sleegers K, Prince JA, van Duijn CM, Kehoe PG. Large meta-analysis establishes the ACE insertion-deletion polymorphism as a marker of Alzheimer's disease. Am J Epidemiol [Internet]. 2005;162:305-17. Available from: http://aje.oxfordjournals.org/content/162/4/305.short. doi:10.1093/aje/kwi202
- Hu J, Igarashi A, Kamata M, Nakagawa H. Angiotensin-converting enzyme degrades Alzheimer amyloid beta-peptide (A beta ); retards A beta aggregation, deposition, fibril formation; and inhibits cytotoxicity. J Biol Chem. 2001;276:47863-8. doi:10.1074/jbc.M104068200. PMID:11604391
- Toropygin IY, Kugaevskaya E V, Mirgorodskaya OA, Elisseeva YE, Kozmin YP, Popov IA, Nikolaev EN, Makarov AA, Kozin SA. The N-domain of angiotensin-converting enzyme specifically hydrolyzes the Arg-5-His-6 bond of Alzheimer's Abeta-(1-16) peptide and its isoAsp-7 analogue with different efficiency as evidenced by quantitative matrix-assisted laser desorption/ionization tim. Rapid Commun Mass Spectrom [Internet]. 2008;22:231-9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18085519. doi:10.1002/rcm.3357
- Sun X, Becker M, Pankow K, Krause E, Ringling M, Beyermann M, Maul B, Walther T, Siems WE. Catabolic attacks of membrane-bound angiotensin-converting enzyme on the N-terminal part of species-specific amyloid-β peptides. Eur J Pharmacol. 2008;588:18-25. doi:10.1016/j.ejphar.2008.03.058. PMID:18495113
- Wang J, Ho L, Chen L, Zhao Z, Zhao W, Qian X, Humala N, Seror I, Bartholomew S, Rosendorff C, et al. Valsartan lowers brain beta-amyloid protein levels and improves spatial learning in a mouse model of Alzheimer disease. J Clin Invest. 2007;117:3393-402. doi:10.1172/JCI31547. PMID:17965777
- Mogi M, Li JM, Tsukuda K, Iwanami J, Min LJ, Sakata A, Fujita T, Iwai M, Horiuchi M. Telmisartan prevented cognitive decline partly due to PPAR-gamma activation. Biochem Biophys Res Commun. 2008;375:446-9. doi:10.1016/j.bbrc.2008.08.032. PMID:18715543
- Kehoe PG, Miners S, Love S. Angiotensins in Alzheimer's disease – friend or foe? Trends Neurosci [Internet]. 2009;32:619-28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19796831. doi:10.1016/j.tins.2009.07.006
- Brenner S. The Genetics of Caenorhabditis elegans. Genetics. 1974;77:71-94. PMID:4366476
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature [Internet]. 2003;421:231-7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12529635. doi:10.1038/nature01278
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene [Internet]. 2001;263:103-12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11223248. doi:10.1016/S0378-1119(00)00579-5
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:9. Available from: https://doi.org/10.1186/1471-2199-9-9. doi:10.1186/1471-2199-9-9. PMID:18211699
