ABSTRACT
Everolimus (EV), a rapamycin analogue mTOR inhibitor, is used in the clinic to treat Estrogen positive (ER+) breast cancer in order to avoid the resistance to hormonotherapy. Here, we investigated whether EV efficacy varied according to administration timing by using the ER+ breast cancer cell line MCF-7 as model system. Our results showed that instead of apoptosis, EV induced a G0/G1 phase blockage of MCF-7 cells. Following serum shock, MCF-7 cells displayed a statistically significant 24h rhythm of mammalian target of Rapamycin (mTOR) activity, but perturbed circadian clock genes oscillations. Interestingly, the different delivery schedule of EV presented different efficacy in G0/G1 phase blockage in serum shocked MCF-7 cells. Moreover, serum shock induced also a circadian-like oscillation in expression or activity of several important G1 phase progression proteins, such as Cyclin D1 and phosphorylated Retinoblastoma protein (RB). Inhibition mTOR activity by EV reduced Cyclin D1 and Cyclin D3 protein level as well as RB phosphorylation level. Taken together, the results indicated that serum shock synchronization induced a circadian oscillation in mTOR activity in MCF-7 cells, which rhythmically regulated the synthesis or phosphorylation of key G1 progression proteins, such as Cyclin D1 and phosphorylated RB, ultimately resulting in different G0/G1 blockage efficiency according to different EV administration timing.
Introduction
Breast cancer is the most invasive female cancer, accounting for 25% of all cancers in women and resulting in 1.68 million cases and 522,000 deaths worldwide in 2012.Citation1 Nearly 70% of breast cancers express estrogen-receptors (ER+) and hence benefit from anti-estrogen therapies. Recent therapeutic progress has led to combine anti-estrogens with Everolimus (EV), an inhibitor of the mammalian target of rapamycin (mTOR) pathway/signaling, in order to avoid or reduce resistance to estrogen receptor targeted drugs.Citation2–6
The mTOR is a serine/threonine protein kinase in the phosphatidylinositol 3-kinase-related kinase protein family. mTOR participates at least the formation of two distinct multi-protein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which play important roles in cell growth, cell proliferation, metabolism and survival. EV binds to its protein receptor FK506 binding protein 12 (FKBP12) to form a complex, which inhibits the activity of mTORC1. One of the most important roles of mTORC1 is to promote protein synthesis by phosphorylating the eukaryotic initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) and the p70 ribosomal S6 kinase 1 (S6K1). The phosphorylation of 4E-BP1 inhibits the interaction between 4E-BP1 and eIF4E, finally promotes cap-dependent translation. The phosphorylation of S6K1 by mTORC1 increases mRNA biogenesis, promotes cap-dependent translation and elongation, and the translation of ribosomal proteins etc.Citation7–9 mTOR signaling is commonly deregulated in most human cancers. Hence, it is considered as a target for anti-cancer treatments, especially for kidney and neuroendocrine malignancies, as well as ER+ breast cancers.Citation2–9
Furthermore, mTOR activity displays a circadian (about 24 hours) variation in different mouse tissues and cells, including heart, liver, spleen, adipose tissue and brain (as cerebellum, frontal cortex and suprachiasmatic nucleus) etc., as well as in primary lung fibroblasts and renal carcinoma cell RenCa etc.Citation10–15 Circadian cycles exist in most living organisms and coordinate organismal behavior, physiology and metabolism with environmental cycles during the course of every 24 hours. In mammals, circadian rhythms are systemically coordinated by the hypothalamic suprachiasmatic nuclei.Citation16 At cellular level, circadian rhythm is generated through interwoven transcription and translation feedback loops, involving about 15 clock genes and proteins, including the core molecular Bmal1, Clock, Cryptochromes (Cry1 and Cry2), Periods (Per1, Per2 and Per3) etc.Citation17 In brief, the transcriptional activator complex of Bmal1/Clock or Bmal1/Npas2 activates the transcription of their targets Cry1, Cry2, Per1, Per2 and Per3, these Cry and Per proteins in turn repress their own transcription through their interactions with the Clock-Bmal1 heterodimer. The Bmal1/Clock heterodimer also activates the transcription of clock genes Rev-Erb α/β and Ror α/β/γ, which respectively repress and activate Bmal1 transcription.Citation17 Such rhythmical genetic circadian clock results that about 3%-10% of genes show one rhythmic expression, which are involved in major cells activity, as proliferation, metabolism, senescence, apoptosis and DNA damage response etc.Citation18–20
These circadian rhythms can further modify both tolerability and efficacy of drugs, resulting in dosing time-dependent pharmacology and effects. Recent experimental data using target anticancer agents have demonstrated the importance of dosing time as well as drug dose in pharmacological effects determination.Citation18–20 Clinical trials have further shown that the circadian timing of chrono-modulated chemotherapy could improve tolerability up to 5-fold and nearly double efficacy as compared to constant rate or oppositely-timed infusions.Citation19 Preliminary clinical data with EV indeed suggested that morning oral dosing could reduce side-effects in patients with metastatic breast cancers.Citation21 Moreover, in corresponding to circadian activation of mTOR in RenCa tumor mass, EV dosing time influence the survival rate of RenCa-bearing mice.Citation14 All these findings led us to investigate EV efficacy according to dosing-time in synchronized human MCF-7 cell cultures, as a model of ER+ human breast cancer.
Materials and methods
Cell culture and serum shock synchronization
MCF-7(ATCC® HTB-22™, LGC Standards SARL, France) cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with GlutaMAX (GIBCO, Life Technology, CA, USA) and 10% fetal bovine serum (FBS, Hyclone, UT, USA). MCF-10A (ATCC® CRL-10317™, LGC Standards SARL, France) cells were cultured as previously described.Citation20 For western-blot, qRT-PCR and flow cytometry analysis, MCF-7 cells were seeded into 6-well plates, allowed to reach 15%-20% confluence in exponential growth phase and then synchronized with a serum shock as previously described.Citation22,23 Subsequently, the cells were returned to their usual culture condition and collected at indicated times. The first sample (Time 0: T0) was taken just after the serum shock completion. During bioluminescence recordings, cells were cultured in phenol red-free media supplemented 1mM luciferin (Promega, WI, USA). For these recordings, MCF-7 and MCF-10A cultures were tested with starting confluence of 10–15%, 20–25% or 30–40% and recorded for at least three days.
EV treatment
EV powder (ApexBio Technology, TX, USA) was dissolved in dimethyl sulfoxide (DMSO, Sigma, MO, USA) and to constitute a 10mM stock solution. EV was added in synchronized MCF-7 cell culture at indicated times (T0, T12 or T24) at a final concentration of 1µM and an equivalent volume of DMSO was added in the control sample (CTRL). The cells were collected for western-blot and cytometry analysis 24 h after EV exposure onset.
Western-blot
MCF-7 cells were washed once with PBS and lysed in 2X Laemmli buffer (100 mM Tris, pH 6.8, 20% glycerol, 4% SDS, 0.05% bromophenol blue, and 10 mM DTT). The lysates were then boiled 5 minutes and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel. After transfer, nitrocellulose membranes were blotted with the following antibodies: Anti-Actin (A3854, Sigma, MO, USA); Anti-HSC70 (SPA-815, Stressgen, CA, USA); Anti-Cyclin D1 (556470, BD Bioscience, NJ, USA); anti-Bmal1 (ab3350), anti-Cry2 (ab38872) and anti-Per2 (ab179813) were from ABCAM (Cambridge, UK); Anti-Rev-ERB α (#13418), Anti-Phospho-S6 ribosomal protein (Ser240/244) (#5456), Anti-S6 ribosomal protein (#2317), Anti-Cyclin D3 (#2936), Anti-P21 Waf1/Cip1 (#2947) were from Cell Signaling Technology(MA, USA). Primary antibodies were detected with appropriate secondary antibodies conjugated with horseradish peroxidase and the membranes were developed with an enhanced chemiluminescence (ECL) system (ECL detection kit, Thermofisher, MA, USA). The results were quantified by Image J, Actin or HSC70 was used as control for protein loading.
Quantitative real time PCR (qRT-PCR)
Cells were collected at indicated times, then total RNA was extracted as previously described.Citation24,25 Reverse transcription was performed with Superscript II RT-kit (Invitrogen, CA, USA). Quantitative real time PCR was performed with LightCycler 480 using LightCycler 480 SYBR Green I master kit (Roche, Bâle, Switzerland). Primers used for gene amplification were previously described.Citation25 Primers for Bmal1 were as following, Fwd: AAGGATGGCTGTTCAGCACATGA; Rev: CAAAAATCCATCTGCTGCCCTG. Primers for 36B4 were as following, Fwd: AATCCCTGACGCACCGCCGTGATG, Rev: TGGGTTGTTTTCCAGGTGCCCTCG. Hybridization temperature for all primers was 60°C. The relative quantification of target RNA by using 36B4 as a reference was computed with the Relquant software (Roche, Bâle, Switzerland), which is based on the 2(–ΔΔT) method.
Flow cytometry analysis
Cells were trypsinized by Trypsin-EDTA (ThermoFisher, MA, USA), then fixed with ice-cold 70% ethanol and afterwards washed twice in ice-cold PBS with centrifugation at 300g for 10 minutes. The cells were then incubated with anti-Phospho-S6-APC (#14733) (Cell Signaling, MA, USA) and anti-Ki67-FITC (BD Biosciences, NJ, USA) in a PBS solution containing 0.5% BSA and 2mM EDTA for 30 min at 4°C. After being washed once with PBS, cells were suspended in PBS containing 50ug/ml propidium iodide (PI) and 20ug/ml RNaseA (Sigma, MO, USA) and incubated overnight at 4°C. A LSR Fortessa™ cell analyzer (BeTon Dickinson, NJ, USA) was used for flow cytometry analysis.
Generation of bioluminescence reporter cell lines and bioluminescence imaging
The lentiviral circadian bioluminescence reporter constructs pLV7-Bsd-p(Bmal1)-dLuc (p(Bmal1)-dLuc) was a kind gift of A.C Liu.Citation26 Lenti-viral particles were produced by transient transfection with TransIT-LT (Mirus-Bio, WI, USA) in HEK293FT cells (Thermo Fisher, Paisley, UK) according to the manufacturer's instructions. pWPI (Addgene plasmid# 12254, MA, USA) was used as transfection control. For target cell transduction, a 10 cm dish of 50% confluent MCF-7 or MCF-10A cells were incubated with 1ml filtered viral supernatant in the presence of 8 µg/ml polybrene (SIGMA, MO, USA) for 6 hours. Three days after viral transduction, infected cells were selected by adding Blasticidin (2.5 µg/ml to MCF-7 and 6 µg/ml to MCF-10A) to the culture medium for at least 1 week.
The LumiCycle (Actimetrics, IL, US) luminometer was used for bioluminescence recording of serum shocked MCF-7 p(Bmal1)-dLuc and MCF-10A p(Bmal1)-dLuc cells in 35 mm dishes at 37°C with 5% CO2. The bioluminescence signal was recorded every 10 min for at least 3 days. The LumiCycle Analysis program was used to obtain baseline-subtracted data.
Statistical analysis
For flow cytometry analysis, statistically significant differences were determined by two-way ANOVA or by t-test by using GraphPad Prism version 6.0. Error bars represent standard error of the mean (SEM) or standard deviation (SD) of independent experiments. For western-blot or qRT-PCR, time-dependent differences were first statistically validated with one-way ANOVA. If the time-dependent differences is significant, the results were then analyzed by Cosinor algorithm based on Fourier transformed analysis for sinusoidal function using SPSS to determine if the variation corresponds to a sinusoidal waveform over the circadian period. The Cosinor analysis provided the following parameters estimates: mesor (rhythm-adjusted mean), amplitude (difference between mesor and maximum of best-fitting cosine function), and acrophase (time of maximum in best-fitting cosine function). These parameters are reported with their respective 95% confidence limits. For bioluminescence recording, the circadian period and the statistical significance of the computed period are determined by Periodogram analysis.
Results
mTOR activity displayed a rhythmic variation after serum shock
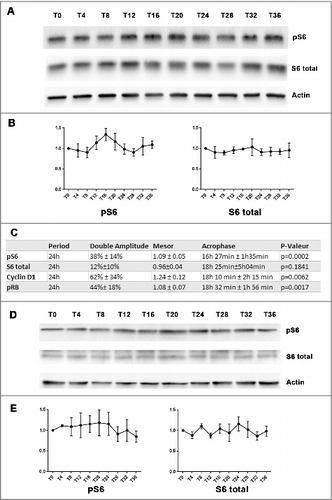
An about 24 hours (24h) oscillatory pattern in phosphorylated S6 ribosome protein was found in MCF-7 cell populations after serum shock ( and ). As one of the main mTOR effectors, Ribosomal protein S6 phosphorylation level was used as a proxy for mTOR activity.Citation8–9 In three independent experiments, phosphorylated S6 ribosome protein gradually increased from T8, reached a peak between T12 to T20, then decreased toward a nadir around T24 to T28, and rose again at T36 (). Time-dependent differences were statistically validated with one-way ANOVA (p = 0.03). A statistically significant 24h oscillation in phosphorylated S6 ribosome protein was further suggested by Cosinor analysis (p = 0.0002), with a double-amplitude of 38% ± 14% relative to the rhythm-adjusted mean (Mesor), and an Acrophase occurring at 16h27min ± 1h35min (). However, no apparent time dependent differences were observed for total S6 protein expression (one-way ANOVA, NS; Cosinor, NS; ). In contrast, unsynchronized MCF-7 cells did not show significant time-dependent differences in neither phosphorylated S6 nor total S6 protein expression (one-way ANOVA, NS; and >E)
Figure 1. Circadian rhythm in mTOR activity after serum shock (A and B): Western-blot revealed a rhythmic change of mTOR activity in MCF-7 cell after serum shock (3 independent experiments). MCF-7 cells were harvested every 4h after serum shock for 36h. The first time point (T0) was taken just after the serum shock. The phosphorylation level of S6 was used to evaluate mTOR activity. Actin was used as a control of protein loading to calculate phosphorylated S6 and total S6 level at every T time. (C): The table resumed Cosinor analysis which revealed a statistically significant 24h oscillation in phosphorylated S6 ribosome protein, Cyclin D1 and phosphorylated RB protein, but not in S6 protein. (D and E): Western-blot showed mTOR activity in MCF-7 cell without synchronization (3 independent experiments). The non-synchronized MCF-7 cells were harvested every 4h for 36h as the synchronized MCF-7 cells. Actin was used as a control of protein loading to calculate phosphorylated S6 and total S6 level at every T time.
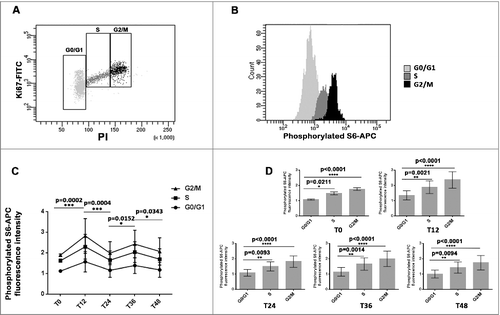
The oscillatory pattern in mTOR activity was consistent in every cell cycle phase
The use of the tri-staining method (Ki67 antibody, PI and phosphorylated ribosomal protein S6 antibody) for cytometry analysis, enabled us to assess mTOR activity dynamics in distinct cell cycle phases ( and ). A consistent oscillatory pattern in S6 phosphorylation level was found in G0/G1, S and G2/M phase cells that persisted for two consecutive 24 h cycles after serum shock (). Higher phosphorylated S6 expression occurred at T12 and T36, in alternation with lower ones at T0, T24 and T48. Irrespective of sampling time, phosphorylated S6 levels were higher in S and G2/M phase cells compared to G0/G1 phase cells ().
Figure 2. The oscillatory pattern in mTOR activity was consistent in every cell cycle phase (A and B): The combination of anti-piS6-APC, Ki67-FITC and PI in cytometry analysis allowed to determine mTOR activity of cells in different cell cycle phases. The combination of Ki67 and PI allowed to determine cell cycle phase distribution and mTOR activity is indicated by phosphorylated S6 level. (C): Cytometry analysis revealed an oscillatory pattern of mTOR activity in every cell cycle phase of MCF-7 cell. The first time point (T0) was taken just after the serum shock. Cells were then harvested at indicated times after synchronization. The Phospho-S6-APC level of every cell cycle phase (G0/G1, S, G2/M) at every T time (T0, T12, T24, T36 and T48) was calculated from 6 independent experiments. (D): The cells in S and G2/M phases presented one higher S6 phosphorylation levels compared to cells in G0/G1 phase at each sampling time T.
Lack of evident circadian oscillation of Bmal1 expression in serum shocked MCF-7 cells
Previous study on Bmal1−/− mice revealed the implication of Bmal1 in mTOR signaling regulationCitation11; we therefore probed Bmal1 expression in serum shock MCF-7 cells to check if there existed one circadian oscillation pattern in Bmal1expression.
First, real-time bioluminescence of MCF-7 p(Bmal1)-dLuc and MCF-10A p(Bmal1)-dLuc reporter cells were recorded over three days. MCF-10A p(Bmal1)-dLuc reporter cells which used as positive control displayed a statistically significant rhythm in Bmal1 promotor activation (p<0.05, Periodogram analysis, ). However, MCF-7 p(Bmal1)-dLuc did not show one evident rhythm in Bmal1 gene expression (p>0.05, NS for three conditions, The circadian period and the statistical significance of the computed period are determined by Periodogram analysis, ).
Figure 3. Lack of evident circadian oscillation of Bmal1 expression in serum shocked MCF-7 cells (A): Average real-time bioluminescence traces of serum shocked MCF-7 p(Bmal1)-dLuc or MCF-10A p(Bmal1)-dLuc reporter cell lines. Three different cell confluences were used at the beginning of record, condition (1) was around 10–15%, condition (2) was around 20–25% and condition (3) was around 30–40%. The plotted data represent bioluminescence (photon count/sec) against time (hours). For details see Material and Methods. The circadian period and the statistical significance of the computed period are determined by Periodogram analysis. MCF-10A p(Bmal1)-dLuc reporter cells presented one significant rhythm of Bmal1 promotor activation, condition: (1) p<0.048, Period = 20.5h; (2) p<0.048, Period = 21.5 h; (3) P<0.0005, Period = 21.7 h. However, MCF-7 p(Bmal1)-dLuc cells did not show any consistent oscillation in Bmal1 expression (p>0.05, NS for every confluence condition). (B): Western-blot analysis did not revealed an obvious oscillation in Bmal1 protein expression oscillation in serum shocked MCF-7 cells. Bmal1 protein expression was analyzed every 4 h after serum shock for 36 h. Actin was used as a control of protein loading. (C): qRT-PCR analysis revealed that MCF-7 cells did not display an obvious rhythm in Bmal RNA expression. Bmal1 RNA expression was analyzed every 4 h after serum shock for 36h. 36B4 was used as reference for relative quantification of target gene mRNA.
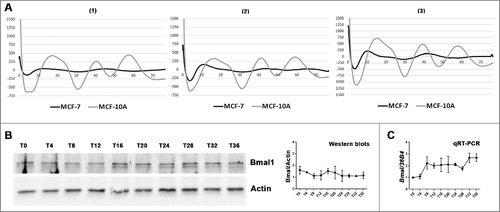
The lack of detectable Bmal1 expression oscillation was confirmed in serum shocked MCF-7 cells at mRNA and protein levels by qRT-PCR and western-blot respectively (). One-way ANOVA ruled out statistically significant time dependent differences in Bmal1 mRNA and protein expressions.
Moreover, no obvious time related oscillatory pattern was found in both mRNA and protein expression of other clock genes (Per2, Rev-Erbα and Cry2) in serum shocked MCF-7 cells over 36 hours (data not shown).
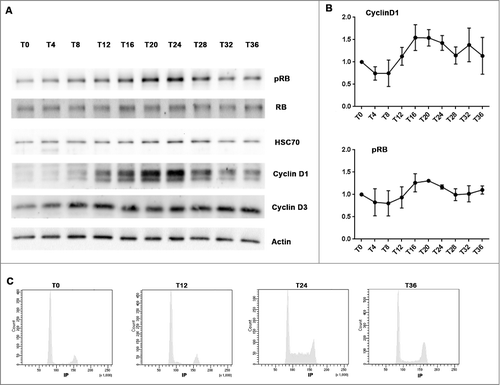
Serum shock induced rhythmic Cyclin D1 and pRB expressions and synchronized cell cycle in MCF-7 cells
Although no rhythmic mRNA and protein expression of the core circadian genes could be detected following a serum shock, Cyclin D1 and phosphorylated RB (pRB), two key G1 phase proteins displayed oscillatory patterns, with a period around 24 h ( and ). Time-dependent differences were statistically validated with one-way ANOVA (Cyclin D1, p = 0.0003; pRB, p = 0.04). Cosinor analysis further validated a 24h period (Cyclin D1, p = 0.0062; pRB, p = 0.0017 ) and revealed a double-amplitude of 62% ± 34% relative to mesor and an acrophase occurred at 18 h10 min ± 2 h 15 min for Cyclin D1; for the pRB protein rhythm, the double-amplitude relative to mesor is 44% ± 18% and the acrophase was located at 18 h32 min ± 1h56 min (). The 24h oscillation in Cyclin D1 and pRB expressions indicated that serum shock induced cell cycle synchronization. Cytometry analysis further confirmed that an accumulation of cells in G1/G0 phase at T0 which then released into S at T24 ().
Figure 4. Serum shock induced Cyclin D1 and pRB rhythmic oscillations and synchronized cell cycle in MCF-7 cells (A and B): Western-blot revealed the rhythmic expression of Cyclin D1 and phosphorylated RB in serum shocked MCF-7 cell. MCF-7 cells were harvested every 4h after serum shock for 36h. The first time point (T0) was taken just after the serum shock. Actin and HSC70 were used as protein loading control to calculate Cyclin D1 and phosphorylated RB expression level at every T time (3 or 4 independent experiments). HSC70 was used as a control of protein loading to calculate pRB, Cyclin D1 and Cyclin D3 expression level at every T time. (C): Cytometry analysis revealed that an accumulation of cells in G1/G0 phase at T0 induced by serum shock which then released into S at T24.
Everolimus inhibited MCF-7 cells proliferation without inducing apoptosis
Using phosphorylated S6 antibody and PI double labelling flow cytometry, similar dose-response curves of EV on the inhibition of S6 phosphorylation were found in every cycle phase of MCF-7 cells. A steeper slope of the dose-response relation was shown in S and G2/M phase cells (). Maximum inhibition was reached at 1µM EV and plateaued thereafter, since a similar effect was found with 10µM EV. Such dose-dependency was further confirmed by western-blot analysis for phosphorylated S6 protein level irrespective of cell cycle phase (). Therefore, an EV concentration of 1µM was selected for further experiments.
Figure 5. EV inhibited MCF-7 cells proliferation without inducing apoptosis (A): Flow cytometry analysis revealed dose-dependent inhibition of S6 phosphorylation level in different cell cycle population. (B): Western-blot result revealed dose-dependent inhibition of S6 phosphorylation level by EV. (C): Annexin V and PI analysis by cytometry revealed that EV did not induce apoptosis or cell death of MCF-7 cells. (D): Ki67 and PI analysis by cytometry showed that EV inhibited MCF-7 cells proliferation and induced a significant increase of G0/G1 and G2/M phases cells after 48 h incubation.
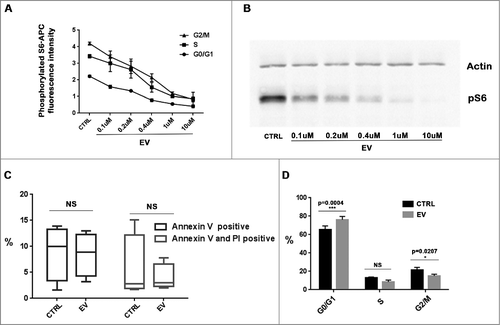
First, we investigated EV induced apoptosis and cell cycle arrest 48h after exposure onset. EV did not increase the proportion of apoptotic or dead cells compared to controls, as assessed with Annexin V positive cells or Annexin V and PI double positive cells, respectively (). However, EV inhibited MCF-7 cells proliferation, as indicated by a significant increase in G0/G1 phase and decrease in G2/M phase cells (). Thus, G0/G1 phase blockage was used as the main endpoint when assessing anti-proliferative effects of EV according to drug timing.
Time dependent anti-proliferative effect of EV in serum shocked MCF-7 cells.
In order to imitate different clinical dosing time (one time every day, morning or evening), serum shocked MCF-7 cells were exposed to EV for 24h under three different time schedules: T0 to T24, T12 to T36 or T24 to T48 respectively. G0/G1 blockage was used to determine EV anti-proliferative effect (). EV exposure from T0 to T24 or exposure from T24 to T48 induced a more evident G0/G1 blockage compared to EV exposure from T12 to T36 in serum shocked MCF-7 cells ( and ). In matching up to the control, EV exposure from T0 to T24 and from T24 to T48 increased respectively 27.5%±5.5% and 17.3%±3.6% G0/G1 phase cells. However, G0/G1 phase cell is only increased 7.19%±2.5% when MCF-7 cell was treated from T12 to T36. The time dependent efficacy of EV in serum shocked MCF-7 cells is statistically validated for T0-T24 VS T12-T36 (p = 0.0022) and T24-T48 VS T12-T36 (p = 0.0186). However, there is no significant difference for T0-T24 VS T24-T48. ().
Figure 6. Time-dependent anti-proliferative effect of EV in serum shocked MCF-7 cells. (A): After serum shock, cell cycles of EV treated MCF-7 cells and their controls under three different time schedules (T0-T24, T12-T36 or T24-T48) were analyzed by cytometry. (B): Graphical resumed from cell cycle cytometry analysis illustrated the different cell cycle phase distributions of T0-T24, T12-T36 or T24-T48 EV treated serum shocked MCF-7 cells compared to their control (5 independent experiments). The whiskers go down to the smallest value and up to the largest, the line in the box is plotted at the median. (C): Histograms depicting the relative changes in cell cycle phase distributions according to EV timing schedule. In matching up to the control, the proportion of G0/G1 phase cells were increased much more evident when EV exposure from T0 to T24 or from T24 to T48 in comparison with EV exposure from T12 to T36. (D): Graphic resumed from the cytometry analysis revealed S6 phosphorylation level was inhibited by EV with different dosing times in serum shocked MCF-7 cell. (E): Western-blot results showed S6 phosphorylation level was inhibited by EV with different dosing times in serum shocked MCF-7 cell.
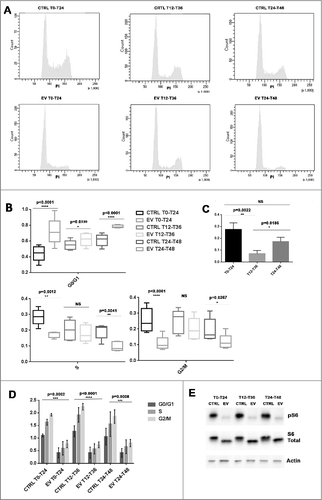
The increase in the proportion of G0/G1 phase cells matched significant decreases in both S and G2/M phase cells when EV dosing is started at T0 or T24. However, EV exposure starting at T12 did not significantly reduce the proportion of S or G2/M cells compared to controls (). Hence, the blockage efficacy was largely higher following drug exposure at either T0-T24 or T24-T48 compared to T12-T36.
The cytometry and western-blot analysis revealed that after a 24h incubation with 1uM EV, S6 phosphorylation was strongly inhibited in the three different conditions (T0-T24, T12-T36 or T24-T48). ().
EV inhibited Cyclin D1 and Cyclin D3 expressions as well as RB phosphorylation
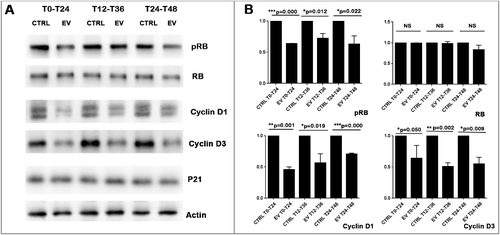
Western-blot analysis revealed that inhibition of mTOR activity by EV further decreased Cyclin D1 and Cyclin D3 protein levels (). Importantly, inhibition of mTOR by EV did not decrease RB expression but only its phosphorylation. As Cyclin D and phosphorylated RB (pRB) play an essential role in G1 phase progression, EV induced diminution of Cyclin D and pRB provided a potential explanation of the G0/G1 arrest induced by EV.
Figure 7. EV inhibited Cyclin D1 and Cyclin D3 expression as well as RB phosphorylation (A):Western-blot results show that different 24h exposure schedules of EV inhibit the protein expression of phosphorylated RB, Cyclin D1, and Cyclin D3, but not of total RB and P21. Actin was used as protein loading control to calculate RB, pRB, Cyclin D1 and Cyclin D3 expression levels. (B): Three independent western-blot results were quantified by Image J. Statistical analysis (T-test) revealed that EV induced significant diminution of Cyclin D1, Cyclin D3 and RB phosphorylation level but not the total RB expression level.
Discussion
Here we tried to understand how and why the administration time of Everolimus could affect its anti-proliferation efficacy by using ER+ breast cancer cell line MCF-7 as a model.
We found that mTOR activity, the target enzyme of EV, displayed a 24 h rhythmic pattern in serum shocked MCF-7 cells. EV dose dependently arrested MCF-7 cells in G0/G1 phase without eliciting detectable apoptosis. This result was consistent with previous results suggesting that several pan-mTOR inhibitors induced also cell cycle arrest but not cell death.Citation27 Interestingly, the extent of EV induced G0/G1 blockage varied significantly according to EV administration time in the serum shocked MCF-7 cells. Normally, serum shock is used to synchronize both the cell cycle and the circadian clock, two essential biological oscillators which are all around 24 h.Citation28,29 Especially, these two process share some common elements in biochemical and molecular regulation, like hormones, growth factor, E-box and bHLH transcription factors and can be reset by light or serum response, etc.Citation23,Citation28 The conservation of essential elements for the molecular regulation of circadian and cell cycle underlies similarities in the dedicated molecular mechanisms, which allow both the ordered progression along cell cycle and the maintenance of circadian rhythm. Recent studies revealed also possible molecular interaction between the endogenous circadian clock and cell cycle.Citation30–34 However, circadian rhythms are often altered in breast cancer patientsCitation35–37 as well as in breast tumors and breast cancer cells lines.Citation22,Citation38–42 Alterations and desynchronization of molecular clock machinery on genetic and epigenetic level were observed in more aggressive breast cancer and those lacking estrogen receptor. Numerous studies have suggested that there are no mRNA circadian-like oscillations of canonical circadian genes in many breast cancer cell lines, either in ER+ cells (such as MCF-7 or T47D) or in ER- cells (such as HS578T and MDA-MB231). However, cultured human mammary epithelial cells maintain an inner circadian clocks.Citation22,Citation41,42 Using luciferase reporter technology, our results revealed also serum shock induced the rhythmical expression of Bmal1 promotor in non-tumorigenic epithelial cell line MCF-10A, supporting it was endowed with functional circadian clocks. But ER+ MCF-7 cell did not display an evident circadian oscillations of core clock genes expression under the examination of luciferase reporter technology, qRT-PCR and western-blot. However, after a serum shock, MCF-7 cell exhibited surprisingly a rhythmic pattern of mTOR activity as well as a synchronized cell cycling and a significant 24h rhythms in G1 phase progression proteins, such as Cyclin D1 and phosphorylated RB.
Increasing mTOR activity was shown to drive cell cycle progression and increase cell proliferation mainly thanks to its effect on protein synthesis. Indeed, it was reported that mTOR activity regulated Cyclin D1 translation through phosphorylating S6 kinase or 4E-BP1.Citation43,44 Similarly, Rapamycin, an EV analog, decelerated the accumulation of Cyclin D1 during p21 induced cell cycle arrest.Citation45 Our results revealed mTOR inhibition by EV could decrease Cyclin D1 and Cyclin D3 protein expression, but not alter P21 expression. Interestingly, the mTOR activity peak started at T12, slightly before the Cyclin D1 expression peak (T16), suggesting that induction of Cyclin D1 expression after mTOR activation takes several hours. Usually, Cyclin D-Cdk4/6 dimer phosphorylates RB protein in G1 phase cell, which releases transcription factor E2F from phosphorylated Rb and become active to drive G1 to S phase transition. As RB is one of the main substrates of Cyclin D-Cdk4/6 dimer, it is reasonable that Cyclin D1 expression and RB phosphorylation presented a similar oscillation tendency and arrive to their peak at the same time. Moreover, EV induced only a decreased RB phosphorylation level but not total RB expression, which suggested that mTOR is not directly involved in RB expression. The decreased RB phosphorylation could be due to the diminution of Cyclin D expression. Taken together, our results suggested that the oscillation of mTOR activity might induce the oscillation of expression or phosphorylation of some G1 progression proteins, which in turn could couple it to cell cycle progression. In view of the rhythmic activation of mTOR, when EV was added at different times on serum shocked MCF-7 cells which had different mTOR activity level, different G1 progression proteins levels and therefore different cell cycle progression probabilities, it was reasonable the cells presented varied EV sensitivity.
The optimal cancers therapy should be to minimize the toxicity of drug to the host but maximize its effect on the tumor. But unfortunately, in most cancers the clinicians have to apply higher toxicity to gain good efficacy. Based on our results and a growing literature on chronotherapy,Citation18–20 drug dosing timing has the potential to become a critical factor, but currently its importance is underestimated. Chronotherapy aims to design optimal drug dosing schedules based on physiological circadian rhythms, which could increase its efficacy and reduce its toxicity.Citation16–18 The in vitro MCF-7 model investigated here provides useful hints toward EV chronotherapy in human ER+ breast cancer. A striking finding was that chrono-efficacy of EV seemed not require canonical circadian rhythm of core molecular clocks. Moreover, exogenous signals to synchronize the molecular clocks also coordinate and couple rhythmic mTOR activity and cell cycling with about 24h periods. These results suggested the potential importance of clock gene independent rhythms in chronotherapy. Clock gene independent rhythms appear pervasively in biology, as shown with the highly conserved Peroxiredoxins biochemical circadian clock, and the cell autonomous 12h clock in mouse liver, which coordinated ER and mitochondrial functions independently from the genetic circadian clock.Citation46,47 Even in MCF-7 cells who did not possess one net clock gene rhythm, the serum shock could still induce rhythmic mRNA expressions of more than 400 genes.Citation41 Thus, in chronotherapy, how to take into account not only clock gene dependent rhythms but also clock gene independent rhythms will be a new potential clue for the future. Remarkably, the xenografting of MCF-7 cells into nude rats could re-establish a circadian pattern in Bmal1 transcription, suggesting that the importance of host circadian time cues in the circadian coordination of cancer cells in vivo.Citation42 Thus, if it is possible to re-establish cancer cell's rhythm by reinforcing the breast cancer patients’ rhythms which are often deregulated during cancer progression, how to take in consideration the interaction between cancer cells and health host cells also need to be considered for jointly optimizing EV chronotherapy effects.
The chrono-efficacy of EV found here provided a new perspective for cancer chronotherapy, which could be potentially applied to other anticancer agents.
Conflict-of-interest disclosure
The authors declare no competing financial interests.
Acknowledgments
This work is supported by INSERM, by University Paris 11 and by the Agence Nationale de la Recherche (Hyclock ANR-14-CE9-0011-01). Yuan Zhang is supported by the China Scholarship Council. Eva Hadadi is supported by the Vaincre le Cancer NRB association. Robert Dallmann acknowledges support from the Royal Society (RG160266).
References
- World Health Organization. World Cancer Report 2014; pp. Chapter 1.1. ISBN 92-832-0429-8.
- Lumachi F, Brunello A, Maruzzo M, Basso U, Basso SM. Treatment of estrogen receptor-positive breast cancer. Curr Med Chem. 2013;20(5):596–604. doi:10.2174/092986713804999303. PMID:23278394.
- Steelman LS, Martelli AM, Cocco L, Libra M, Nicoletti F, Abrams SL, McCubrey JA. The therapeutic potential of mTOR inhibitors in breast cancer. Br J Clin Pharmacol. 2016;82(5):1189‐212. doi:10.1111/bcp.12958. PMID:27059645.
- Hortobagyi GN. Everolimus plus exemestane for the treatment of advanced breast cancer: a review of subanalyses from BOLERO-2. Neoplasia. 2015;17:279‐88. doi:10.1016/j.neo.2015.01.005. PMID:25810012.
- Piccart M, Hortobagyi GN, Campone M, Pritchard KI, Lebrun F, Ito Y, Noguchi S, Perez A, Rugo HS, Deleu I, et al. Everolimus plus exemestane for hormone-receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: overall survival results from BOLERO-2. Ann Oncol. 2014;25:2357‐62. doi:10.1093/annonc/mdu456. PMID:25231953.
- Baselga J, Campone M, Piccart M, Burris HA 3rd, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, et al. Everolimus in postmenopausal hormone receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520‐29. doi:10.1056/NEJMoa1109653. PMID:22149876.
- Xu K, Liu P, Wei W. mTOR signaling in tumorigenesis. Biochimica et Biophysica Acta. 2014;1846(2):638‐54. doi: 10.1016/j.bbcan.2014.10.007. PMID:25450580.
- Laplante M, Sabatini DM. mTOR signaling at a glance J Cell Sci. 2009;122(Pt 20):3589‐94. doi: 10.1242/jcs.051011. PMID: 19812304.
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926‐45. doi:10.1101/gad.1212704. PMID:15314020.
- Khapre RV, Patel SA, Kondratova AA, Chaudhary A, Velingkaar N, Antoch MP, Kondratov RV. Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging (Albany NY). 2014;6(8):675‐89. doi:10.18632/aging.100686. PMID:25239872.
- Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, Kondratov RV BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY). 2014;6(1):48‐57. doi:10.18632/aging.100633. PMID:24481314.
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, Sonenberg N. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron. 2013;79(4):712‐24. doi:10.1016/j.neuron.2013.06.026. PMID:23972597.
- Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181:79‐88. doi:10.1016/j.neuroscience.2011.03.005. PMID:21382453.
- Okazaki H, Matsunaga N, Fujioka T, Okazaki F, Akagawa Y, Tsurudome Y, Ono M, Kuwano M, Koyanagi S, Ohdo S. Circadian regulation of mTOR by the ubiquitin pathway in renal cell carcinoma. Cancer Res. 2014;74(2):543‐51. doi:10.1158/0008-5472.CAN-12-3241. PMID:24253377.
- Drägert K, Bhattacharya I, Hall MN, Humar R, Battegay E, Haas E Basal mTORC2 activity and expression of its components display diurnal variation in mouse perivascular adipose tissue. Biochem Biophys Res Commun. 2016:22;473(1):317‐22. doi:10.1016/j.bbrc.2016.03.102. PMID: 27016480.
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69(6):1583‐6. doi:10.1073/pnas.69.6.1583. PMID:4556464.
- Takahashi JS Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164‐79. doi:10.1038/nrg.2016.150. PMID:27990019.
- Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci. 2013;119:221‐82. doi:10.1016/B978-0-12-396971-2.00009-9. PMID:23899600.
- Dallmann R, Okyar A, Lévi F. Dosing-Time Makes the Poison: Circadian Regulation and Pharmacotherapy. Trends Mol Med. 2016;22(5):430‐45. doi:10.1016/j.molmed.2016.03.004. PMID:27066876.
- Lévi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377‐421. doi:10.1146/annurev.pharmtox.48.113006.094626. PMID:20055686.
- Giacchetti S, Li XM, OZTURK N, Cuvier C, Machowiak J, Arrondeau J, Chang-Marchand Y, Espié M, Okyar A, Lévi F Consistent dosing-time dependent tolerability of everolimus (EV) in a pilot study in women with metastatic breast cancer (MBC) and in a mouse chronopharmacology investigation Abstract of San Antonio Breast Cancer Symposium 2016 Abstract N° 850329.
- Rossetti S, Esposito J, Corlazzoli F, Gregorski A, Sacchi N. Entrainment of breast (cancer) epithelial cells detects distinct circadian oscillation patterns for clock and hormone receptor genes. Cell Cycle. 2012;11(2):350‐60. doi:10.4161/cc.11.2.18792. PMID:22193044.
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929‐37. doi:10.1016/S0092-8674(00)81199-X. PMID:9635423.
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156‐9. doi:10.1016/0003-2697(87)90021-2. PMID:2440339.
- Ballesta A, Dulong S, Abbara C, Cohen B, Okyar A, Clairambault J, Lévi F. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput Biol. 2011;7:e1002143. doi:10.1371/journal.pcbi.1002143. PMID:21931543.
- Ramanathan C, Khan SK, Kathale ND, Xu H, Liu AC. Monitoring cell-autonomous circadian clock rhythms of gene expression using luciferase bioluminescence reporters. J Vis Exp. 2012;27(67):pii:4234. doi: 10.3791/4234. PMID: 23052244.
- Leontieva OV, Blagosklonny MV Gerosuppression by pan-mTOR inhibitors Aging (Albany NY). 2016;30; 8(12):3535‐51. doi:10.18632/aging.101155. PMID:28077803.
- Canaple L, Kakizawa T, Laudet V. The days and nights of cancer cells. Cancer Res. 2003;63(22):7545‐52. PMID:14633665.
- Goldbeter A, Gérard C, Gonze D, Leloup J-C, Dupont G Systems biology of cellular rhythms, FEBS Lett. 2012;586(18):2955‐65. doi:10.1016/j.febslet.2012.07.041. PMID:22841722.
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255‐9. doi:10.1126/science.1086271. PMID:12934012.
- Gréchez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F The circadian clock component Bmal1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535‐4542. doi:10.1074/jbc.M705576200. PMID:18086663.
- Fu L, Pelicano H, Liu J, Huang P, Chi Lee C The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111: 41‐50. doi:10.1016/S0092-8674(02)00961-3. PMID:12372299.
- Pérez-Roger I, Solomon DLC, Sewing A, Land H Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27Kip1 binding to newly formed complexes. Oncogene. 1997;14:2373‐81. doi:10.1038/sj.onc.1201197. PMID:9188852.
- Feillet C, Krusche P, Tamanini F, Janssens RC, Downey MJ, Martin P, Teboul M, Saito S, Lévi F, Bretschneider T, et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc Natl Acad Sci U S A. 2014;111(27):9828‐33. doi:10.1073/pnas.1320474111. PMID:24958884.
- Cash E, Sephton SE, Chagpar AB, Spiegel D, Rebholz WN, Zimmaro LA, Tillie JM, Dhabhar FS Circadian disruption and biomarkers of tumor progression in breast cancer patients awaiting surgery. Brain Behav Immun. 2015;48:102‐14. doi:10.1016/j.bbi.2015.02.017. PMID:25728235.
- Sephton SE, Sapolsky RM, Kraemer HC, Spiegel DJ Diurnal cortisol rhythm as a predictor of breast cancer survival. Natl Cancer Inst. 2000: 21;92(12):994‐1000. doi:10.1093/jnci/92.12.994. PMID:10861311.
- Touitou Y, Lévi F, Bogdan A, Benavides M, Bailleul F, Misset JL Rhythm alteration in patients with metastatic breast cancer and poor prognostic factors. J Cancer Res Clin Oncol. 1995;121(3):181‐8. doi:10.1007/BF01198101. PMID:7713990.
- Blakeman V, Williams JL, Meng QJ, Streuli CH Circadian clocks and breast cancer. Breast Cancer Res. 2016;18(1):89. doi:10.1186/s13058-016-0743-z. PMID:27590298.
- Reszka E, Przybek M Circadian Genes in Breast Cancer. Adv Clin Chem. 2016;75:53‐70. doi:10.1016/bs.acc.2016.03.005. PMID:27346616.
- Cadenas C, van de Sandt L, Edlund K, Lohr M, Hellwig B, Marchan R, Schmidt M, Rahnenführer J, Oster H, Hengstler JG Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle. 2014;13(20):3282‐91. doi:10.4161/15384101.2014.954454. PMID:25485508.
- Gutiérrez-Monreal MA, Treviño V, Moreno-Cuevas JE, Scott SP Identification of circadian-related gene expression profiles in entrained breast cancer cell lines. Chronobiol Int. 2016;33(4):392‐405. doi:10.3109/07420528.2016.1152976. PMID:27010605.
- Xiang S, Mao L, Duplessis T, Yuan L, Dauchy R, Dauchy E, Blask DE, Frasch T, Hill SM Oscillation of clock and clock controlled genes induced by serum shock in human breast epithelial and breast cancer cells: regulation by melatonin. Breast Cancer (Auckl). 2012;6:137‐50. doi: 10.4137/BCBCR.S9673. PMID:23012497.
- Koziczak M, Hynes NE Cooperation between fibroblast growth factor receptor-4 and ERBB2 in regulation of cyclin D1 translation. J Biol Chem. 2004;279(48):50004‐11. doi:10.1074/jbc.M404252200. PMID:15377668.
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477‐80. doi:10.1038/nature03205. PMID:15690031.
- Leontieva OV, Lenzo F, Demidenko ZN, Blagosklonny MV Hyper-mitogenic drive coexists with mitotic incompetence in senescent cells, Cell Cycle. 2012: 11:24:4642‐9; doi:10.4161/cc.22937.
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459‐64. doi: 10.1038/nature11088. PMID:22622569.
- Zhu B, Zhang Q, Pan Y, Mace EM, York B, Antoulas AC, Dacso CC, O'Malley BW. A Cell-Autonomous Mammalian 12 hr Clock Coordinates Metabolic and Stress Rhythms, Cell Metab. 2017;25(6):1305‐19. e9. doi: 10.1016/j.cmet.2017.05.004.
