ABSTRACT
SUMO modification is required for the kinetochore localization of the kinesin-like motor protein CENP-E, which subsequently mediates the alignment of chromosomes to the spindle equator during mitosis. However, the underlying mechanisms by which sumoylation regulates CENP-E kinetochore localization are still unclear. In this study, we first elucidate that the kinetochore protein Nuf2 is not only required for CENP-E kinetochore localization but also preferentially modified by poly-SUMO-2/3 chains. In addition, poly-SUMO-2/3 modification of Nuf2 is significantly upregulated during mitosis, which is temporally correlated to the kinetochore localization of CENP-E during mitosis. We further show that the mitotic defects in CENP-E kinetochore localization and chromosome congression caused by global inhibition of sumoylation can be rescued by expressing a fusion protein between Nuf2 and the SUMO-conjugating enzyme Ubc9 for stimulating Nuf2 SUMO-2/3 modification. Moreover, the expression of another fusion protein between Nuf2 and three SUMO-2 moieties (SUMO-2 trimer), which mimics the trimeric SUMO-2/3 chain modification of Nuf2, can also rescue the mitotic defects due to global inhibition of sumoylation. Conversely, expressing the other forms of Nuf2-SUMO fusion proteins, which imitate Nuf2 modifications by SUMO-2/3 monomer, SUMO-2/3 dimer, and SUMO-1 trimer, respectively, cannot rescue the same mitotic defects. Lastly, compared to Nuf2, the fusion protein simulating the trimeric SUMO-2 chain-modified Nuf2 exhibits a significantly higher binding affinity to CENP-E wild type containing a functional SUMO-interacting motif (SIM) but not the CENP-E SIM mutant. Hence, our results support a model that poly-SUMO-2/3 chain modification of Nuf2 facilitates CENP-E kinetochore localization and chromosome congression during mitosis.
Abbreviations: CENP-E, centromere-associated protein E; SUMO, small ubiquitin-related modifier; SIM, SUMO-interacting motif.
Introduction
Sumoylation, similar to ubiquitination and phosphorylation, has emerged as a major mechanism in regulating the cell cycle progression through mitosis [Citation1–3]. Small ubiquitin-related modifier (SUMO) proteins are covalently conjugated to lysine (K) residue(s) of protein targets through the E1-activating enzyme (SAE1/SAE2 heterodimer), the E2-conjugating enzyme (Ubc9), and multiple E3 ligases. As a reverse process of sumoylation, SUMOs are removed from their substrates by a group of SUMO-specific isopeptidases called SENPs in vertebrates [Citation4]. Although sumoylation frequently occurs at a lysine residue within a sumoylation consensus motif (Ψ-K-x-[D/E]) (Ψ: a hydrophobic residue; x: any residue; D/E: an acidic residue), SUMOs are also conjugated to a lysine residue within a non-consensus site [Citation5–7]. In contrast to a single SUMO protein in yeast and invertebrates, vertebrates express three SUMO proteins: SUMO-1, SUMO-2, and SUMO-3. In humans, SUMO-2 and SUMO-3 are ~95% identical to each other and therefore referred to as SUMO-2/3, but they share only ~45% sequence identity to SUMO-1. Similar to ubiquitin, SUMO can be conjugated to target proteins as a single monomer or as a poly-SUMO chain. Although both poly-SUMO-1 and poly-SUMO-2/3 chain modifications have been observed using in vitro sumoylation assays, poly-SUMO-2/3 chain modification of protein targets seems to be predominant in vivo [Citation8–10]. One explanation is that both SUMO-2 and SUMO-3, but not SUMO-1, contain a consensus sumoylation motif (10-VKTE-13) for the assembly of poly-SUMO-2/3 chains through the K11 residue [Citation9–11].
Our previous immunofluorescence microscopy analysis revealed that SUMO-1 and SUMO-2/3 display distinct subcellular localization during mitosis in mammalian cells [Citation12]. While SUMO-1 signals are present at the mitotic spindle, SUMO-2/3 signals are associated with centromeres and kinetochores during prophase and metaphase [Citation12]. As one of the largest protein complexes, the kinetochore contains over 100 different proteins and consists of the inner kinetochore and the outer kinetochore [Citation13,Citation14]. While the inner kinetochore proteins permanently associate with centromeric chromatin throughout the cell cycle, the outer kinetochore proteins temporally assemble onto the inner kinetochore during mitosis to mediate the kinetochore-microtubule attachment. Many different centromere/kinetochore proteins have been identified as SUMO-modified targets in various organisms ranging from yeast to humans [Citation2,Citation3,Citation12,Citation15–21].
The SUMO E2 enzyme Ubc9 and three E3 ligases (including PIAS3, PIASy, and Nup358/RanBP2) have been detected at kinetochores during mitosis [Citation17,Citation22,Citation23]. In budding yeast, inhibition of Ubc9 expression blocks the cell cycle progression at the G2/M phase with a defect in chromosome segregation [Citation24]. Consistent with this observation in budding yeast, global inhibition of sumoylation in mammalian cells by either overexpression of the SUMO isopeptidase SENP2 or RNAi depletion of Ubc9 leads to a prometaphase arrest and a defect in targeting the centromere-associated protein E (CENP-E) to kinetochores [Citation12]. In addition, SENP2 is associated with kinetochores during mitosis [Citation25], suggesting that SENP2 may play a critical role in downregulating levels of SUMO-2/3 modification in kinetochores.
As a kinetochore-associated and plus end-directed kinesin, CENP-E contains an N-terminal microtubule-binding motor domain, a long coiled-coil domain for dimerization, and a C-terminal tail domain (1958–2701 amino acids) for kinetochore localization [Citation26]. In addition, CENP-E is temporally present at the outer kinetochore from late prophase to late anaphase during mitosis [Citation27,Citation28]. CENP-E plays an essential role in chromosome congression by transporting chromosomes located near the spindle poles to the spindle equator, also called the metaphase plate, along the preexisting spindle microtubules [Citation29]. It has been shown previously that inhibition or depletion of CENP-E causes a pro-metaphase arrest with a subgroup of chromosomes clustered around the spindle poles [Citation30–32].
Our previous study elucidated that the C-terminal tail domain of CENP-E contains a SUMO-interacting motif (SIM) (2307–2311 amino acid) essential for its specific interaction with poly-SUMO-2/3 chains in vitro and for its association with kinetochores in vivo [Citation12]. In addition, we also found that two known CENP-E-interacting kinetochore proteins, Nuf2 [Citation33] and BubR1 [Citation26], are specifically modified by SUMO-2/3 in vivo [Citation12]. By forming the stable Hec1/Ndc80 complex with Hec1, Spc24, and Spc25 at the outer kinetochore, Nuf2 plays a critical role in kinetochore-microtubule attachment and chromosome congression during mitosis [Citation26,Citation34–36].
In this study, we tested the hypothesis that SUMO-2/3 modification of Nuf2 facilitates the kinetochore localization of CENP-E, which in turn mediates the chromosome alignment to the metaphase plate and the progression through mitosis. We first revealed that Nuf2 is necessary for CENP-E localization to the kinetochore and specifically modified by poly-SUMO-2/3 chains. We then provided multiple lines of evidence to support a model that poly-SUMO-2/3 modification of Nuf2 increases its interaction with CENP-E, resulting in CENP-E kinetochore localization, chromosome congression, and mitotic progression in mammalian cells.
Results
Nuf2 is required for CENP-E localization to kinetochores
The hypothesis that SUMO-2/3 modification of Nuf2 facilitates CENP-E localization to kinetochores is based on the assumption that Nuf2 itself is required for CENP-E localization to kinetochores. However, two previously published studies provided totally opposite findings on whether or not the CENP-E kinetochore localization depends on Nuf2 [Citation33,Citation34]. To solve this problem, we used RNA interference (RNAi) to inhibit Nuf2 expression by transfecting HeLa cells with Nuf2-specific siRNA 1 and siRNA 2, respectively. Our immunoblot analysis showed that levels of Nuf2 expression were almost completely inhibited in Nuf2 RNAi cells compared to control RNAi cells transfected with the scrambled siRNA (). We then performed immunofluorescence microscopy using mouse CENP-E antibody and human CREST antibody against CENP-A, CENP-B, and CENP-C for labeling centromeres and kinetochores. While nearly all the mitotic cells transfected with control siRNA (~95%) showed bright CENP-E foci at kinetochores, most of the mitotic cells transfected with Nuf2 siRNA 1 or 2 (~88% or ~75%) had no obvious CENP-E staining at kinetochores ().
Figure 1. Inhibition of Nuf2 expression by RNAi causes the defects in CENP-E localization to kinetochores and chromosome alignment to the metaphase plate. HeLa cells were transfected with control siRNA or Nuf2-specific siRNA 1 or 2 for 72 h. (a, b) The transfected cells were analyzed by immunoblotting with anti-Nuf2 and anti-tubulin antibodies (a) and by immunofluorescence microscopy with mouse anti-CENP-E antibody and human CREST antibody against the centromere/kinetochore proteins CENP-A, CENP-B, and CENP-C (b). Bar, 10 μm. (c–e) The portion (%) of mitotic cells without CENP-E staining at each stage of mitosis (n ≥ 100 mitotic cells for each siRNA) (c), the mitotic index (n ≥ 578 total cells for control siRNA, n ≥ 524 total cells for Nuf2 siRNA 1, and n ≥ 495 total cells for Nuf2 siRNA 2) (d), and the fraction (%) of mitotic cells present at each stage of mitosis (n ≥ 100 mitotic cells for each siRNA) (e) were determined by DAPI staining and immunofluorescence microscopy (Pro, prophase and prometaphase; Met, metaphase; A&T, anaphase and telophase). The plotted values indicate the means ± standard deviation (SD) from three independent experiments
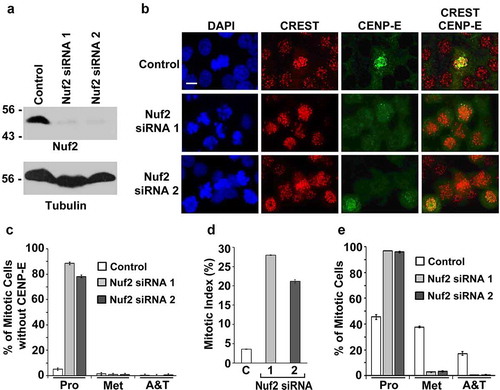
The absence of any evident CENP-E signals at kinetochores and in the cytosol in Nuf2 RNAi cells () raised a question of whether Nuf2 RNAi reduces levels of CENP-E expression. However, our immunoblot analysis indicated that levels of CENP-E expression were comparable between control and Nuf2 RNAi cells (Fig. S1A). We then examined whether the loss of CENP-E staining in Nuf2 RNAi cells was due to the extraction of the free or unbound cytosolic CENP-E from the cells treated with the PBS buffer containing the detergent Triton X-100 for cell permeabilization. To reduce the extraction of the cytosolic CENP-E during the cell permeabilization, we increased the time for cell fixation using 3.5% paraformaldehyde from 7 min () to 20 min (Fig. S1B). As shown in Fig. S1B, we observed an obvious CENP-E staining throughout the cytosol but no CENP-E foci at kinetochores during mitosis in most of the Nuf2 RNAi cells, whereas nearly all the control RNAi cells showed bright CENP-E foci at kinetochores but no evident staining in the cytosol during mitosis. Therefore, Nuf2 RNAi caused the mislocalization of CENP-E from kinetochores to the cytosol during mitosis.
Given that the localization of CENP-E to kinetochores is essential for its function in mediating both chromosome congression and mitotic progression [Citation31,Citation32], we investigated if the defect in CENP-E kinetochore localization caused by Nuf2 RNAi affects these two mitotic processes. We found that inhibition of Nuf2 expression by Nuf2-specific siRNA 1 or 2 significantly increased the mitotic index from ~3.5% in control RNAi cells to ~27% or ~20% (). While ~95% of the mitotic cells transfected with Nuf2-specific siRNA 1 or 2 were present at prophase and prometaphase (Pro), only ~45% of the mitotic cells transfected with the control siRNA were at Pro (). Therefore, RNAi-mediated inhibition of Nuf2 expression caused the defects in CENP-E kinetochore localization and chromosome congression during mitosis.
Upregulation of Nuf2 sumoylation at kinetochores by expressing the Nuf2-Ubc9 fusion protein rescues the mitotic defects caused by global inhibition of sumoylation
It has been shown previously that a linear fusion of the SUMO-conjugating enzyme Ubc9 to one of the known SUMO targets, including p53 and STAT1, dramatically increases their sumoylation in vivo [Citation37]. To test whether upregulation of Nuf2 sumoylation at kinetochores can rescue the mitotic defects in cells with global inhibition of sumoylation caused by overexpression of the SUMO-specific isopeptidase SENP2, we first generated a DNA construct encoding Flag-tagged Nuf2-Ubc9 fusion protein (Flag-Nuf2-Ubc9), in which Ubc9 wild type was fused to Nuf2 (). We also made a control plasmid encoding Flag-Nuf2-Ubc9D, in which Nuf2 was fused with a catalytically inactive Ubc9 dead mutant (Ubc9D) with the cysteine (C) 93 to alanine (A) mutation (C93A) [Citation16,Citation38] (). We confirmed that the constructs encoded the correct fusion proteins, including Flag-Nuf2, Flag-Nuf2-Ubc9D, and Flag-Nuf2-Ubc9, by transfection and immunoblot analysis ( and S2). Additionally, our immunofluorescence microscopy indicated that the three fusion proteins localized to kinetochores labeled by human CREST antibody (Fig. S3A). Lastly, global inhibition of sumoylation by SENP2 overexpression did not affect the kinetochore localization of the three fusion proteins during mitosis ().
Figure 2. The expression of the Nuf2-Ubc9 fusion protein rescues the defects in CENP-E kinetochore localization and chromosome congression caused by SENP2 overexpression. (a) The constructs encoding Flag-Nuf2, Flag-Nuf2-Ubc9D with catalytically dead Ubc9 mutant, and Flag-Nuf2-Ubc9 with Ubc9 wild type. (b) 293T cells were transfected with the indicated DNA constructs and analyzed by immunoblotting with anti-Flag antibody. (c–f) HeLa cells were co-transfected with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-Nuf2, Flag-Nuf2-Ubc9D, or Flag-Nuf2-Ubc9, for 48 h followed by immunofluorescence microscopy with anti-Flag and anti-Myc antibodies (c, d) or anti-Myc and anti-CENP-E antibodies (e, f) (Pro, prophase and prometaphase; Met, metaphase; Ana, anaphase; Ana/Tel, anaphase and telophase). Bar, 10 μm. (d, f) The portion (%) of mitotic cells with both Myc and Flag staining and present at Met and Ana/Tel (n ≥ 100 mitotic cells for each co-transfection) (d), and the fraction (%) of mitotic cells with both Myc and CENP-E staining (n ≥ 100 mitotic cells for each co-transfection) (f) were determined by DAPI staining and immunofluorescence microscopy. The plotted values are the means ± standard error of the mean (SEM) from three independent experiments (*: p < 0.05; **: p < 0.01; NS: not significant; Student’s t test)
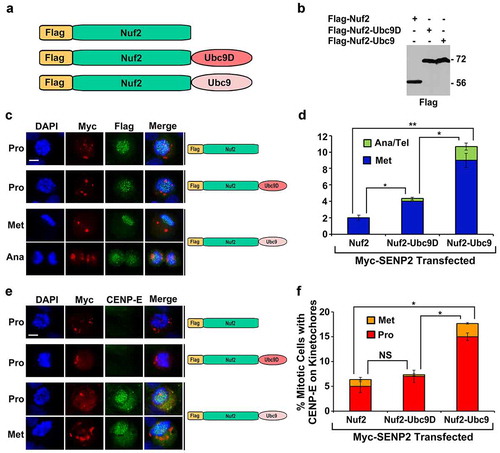
To test if Flag-Nuf2-Ubc9 can rescue the mitotic defects caused by SENP2 overexpression, we co-transfected HeLa cells with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-Nuf2, Flag-Nuf2-Ubc9D, or Flag-Nuf2-Ubc9, for 48 h followed by immunofluorescence microscopy using anti-Flag and anti-Myc antibodies (). We found that only a small percentage of mitotic cells with co-expression of Myc-SENP2 and Flag-Nuf2 (2%) or Flag-Nuf2-Ubc9D (4%) were present at metaphase, while the vast majority of the mitotic cells co-expressing Myc-SENP2 and Flag-Nuf2 (98%) or Flag-Nuf2-Ubc9D (96%) were at prophase and prometaphase (). Compared to only 2–4% of the control transfected mitotic cells at metaphase, a significantly higher percentage (11%) of the mitotic cells co-expressing Myc-SENP2 and Flag-Nuf2-Ubc9 were at metaphase and anaphase/telophase, which represented a rescue of the chromosome congression defect caused by SENP2 overexpression.
We then test whether Flag-Nuf2-Ubc9 rescues the chromosome congression defect in cells with Myc-SENP2 overexpression by targeting CENP-E to kinetochores. HeLa cells were co-transfected with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-Nuf2, Flag-Nuf2-Ubc9D, or Flag-Nuf2-Ubc9, followed by immunofluorescence microscopy using anti-Myc and anti-CENP-E antibodies. Compared to the co-expression of Flag-Nuf2 or Flag-Nuf2-Ubc9D with Myc-SENP2, the co-expression of Flag-Nuf2-Ubc9 with Myc-SENP2 caused a 2.5–3.0 times increase in the percentage of mitotic cells with an obvious CENP-E staining (). Hence, upregulation of Nuf2 sumoylation by Flag-Nuf2-Ubc9 rescued the mitotic defects in CENP-E kinetochore localization and chromosome congression caused by SENP2 overexpression.
Nuf2 is specifically modified by poly-SUMO-2/3 chains in vivo
Our previous study revealed that Nuf2 is specifically modified by SUMO-2/3 in vivo, and CENP-E contains poly-SUMO-2/3 chain-binding activity essential for its kinetochore localization [Citation12]. In addition, CENP-E is only temporally associated with kinetochores during mitosis, reaching its highest levels at prometaphase [Citation27,Citation28]. We thereby hypothesized that Nuf2 SUMO-2/3 modification is upregulated in mitotic cells compared to asynchronous cells, leading to a temporal recruitment of CENP-E to kinetochores during mitosis. To test this hypothesis, we transfected 293T cells with the construct encoding Flag-Nuf2 for 48 h and incubated the cells for 5 h in the absence (‒) or presence (+) of nocodazole, which was used for arresting or synchronizing the cells at prophase/prometaphase. We then performed immunoprecipitation using anti-Flag beads under stringent denaturing conditions followed by immunoblot analysis. Consistent with our previous finding [Citation12], Nuf2 was specifically modified by SUMO-2/3 instead of SUMO-1 (). The signal intensities of Flag-tagged Nuf2 (, left panel) and SUMO-2/3 modified Nuf2 (, right panel) were quantified with ImageJ (NIH). The relative levels of the SUMO-2/3 modified Nuf2 were generated by dividing the signal intensities of SUMO-2/3 modified Nuf2 with those of Nuf2 (). Based on three independent experiments, we found that the relative levels of Nuf2 SUMO-2/3 modification in mitotic cells were ~70% higher than those in asynchronous control cells (). We noticed that SUMO-2/3-modified Nuf2 exhibited high molecular weights from ~200 kDa to >300 kDa (, left panel). Due to the relatively small sizes of SUMO-2/3 (12 kDa) and Nuf2 (54 kDa), the large sizes of the (SUMO-2/3)n-Nuf2 conjugates indicated that each Nuf2 protein may be conjugated with about 10 to 20 moieties of SUMO-2/3. These 10–20 SUMO-2/3 moieties could be attached to Nuf2 in forms of monomeric SUMO-2/3 and/or poly-SUMO-2/3 chains.
Figure 3. Poly-SUMO-2/3 chain modifcation of Nuf2 is upregulated during mitosis and the expression of Nuf2-3× SUMO-2, but not Nuf2-3× SUMO-1, rescues the mitotic defects caused by SENP2 overexpression. (a) 293T cells were transfected with the construct encoding Flag-Nuf2 and cultured in the absence (–) or presence (+) of nocodazole followed by immunoprecipitation and immunoblot analysis. The high molecular weight bands of SUMO-2/3-modified Nuf2 were indicated by a bracket. (b) The relative levels of SUMO-2/3-modified Nuf2 immunoprecipitated from asynchronous cells and mitotic cells. (c) The in vitro sumoylation assays by incubating Nuf2 with SAE1/SAE2, Ubc9 and SUMO-1 or SUMO-2 followed by immunoblot analysis. (d, e) The in vitro sumoylation assays by incubating GST-tagged RanGAP1 NΔ419 fragment (d) or untagged Nuf2 (e) with SAE1/SAE2, Ubc9, and SUMO-1 wild type (WT) or mutant (K-less) followed by immunoblot analysis. The asterisk indicates the GST-Nuf2 fusion protein co-purified with the untagged Nuf2 protein (e). (f) The constructs encoding Flag-Nuf2, Flag-Nuf2-3× SUMO-1, and Flag-Nuf2-3× SUMO-2. (g, h) HeLa cells were co-transfected with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-Nuf2, Flag-Nuf2-3× SUMO-1, or Flag-Nuf2-3× SUMO-2. The portion (%) of mitotic cells with both Myc and Flag staining and present at metaphase (n ≥ 100 mitotic cells for each co-transfection) (g), and the fraction (%) of mitotic cells with both Myc and CENP-E staining (n ≥ 100 mitotic cells for each co-transfection) (h), were determined by DAPI staining and immunofluorescence microscopy (Pro, prophase and prometaphase; Met, metaphase). The plotted values are the means ± SEM from three independent experiments (*: p < 0.05; **: p < 0.01; ***: p < 0.001; NS: not significant; Student’s t test)
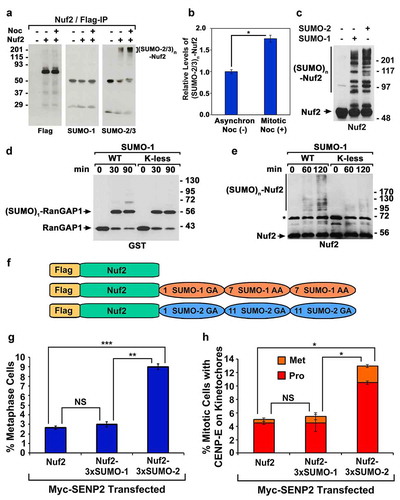
To examine the possibility that Nuf2 is modified by poly-SUMO chains, we performed in vitro Nuf2 sumoylation assays by incubating Nuf2 with SUMO-1 or SUMO-2 as well as the SUMO E1 (SAE1/SAE2) and E2 (Ubc9) enzymes followed by immunoblot analysis. In contrast to its SUMO-2/3-specific modification in vivo, Nuf2 was almost equally well modified by SUMO-1 and SUMO-2 in vitro (), suggesting that additional cellular proteins, such as Nuf2-specific SUMO E3 ligase(s), may determine the in vivo SUMO-2/3-specific modification of Nuf2. Similar to the in vivo SUMO-2/3 modified Nuf2 (), the in vitro SUMO-1 or SUMO-2 modified Nuf2 also contained the high molecular weight conjugates with sizes from ~200 kDa to >300 kDa, although they enclosed other low molecular weight conjugates with sizes from 90 kDa to ~200 kDa (). To achieve a better understanding of these high molecular weight Nuf2-SUMO-1 or Nuf2-SUMO-2 conjugates, we took advantage of the availability of SUMO-1 K-less mutant, in which all the lysine (K) residues were mutated to arginine (R) residues, resulting in its defect in forming poly-SUMO-1 chains. Using the well-defined SUMO target RanGAP1 known to be SUMOylated at a single K residue as a control substrate [Citation39,Citation40], we demonstrated that the recombinant SUMO-1 wild type (WT) and its K-less mutant had the comparable activity for RanGAP1 sumoylation (). We then performed the in vitro Nuf2 sumoylation assays using either SUMO-1 WT or its K-less mutant. In contrast to a faint smear of the SUMO-1 K-less mutant modified Nuf2 with sizes from ~90 kDa to ~130 kDa after the 120 min reaction, we observed a more robust smear of the SUMO-1 WT modified Nuf2 with sizes from ~90 kDa to >300 kDa. Hence, the high molecular weight bands of the (SUMO-2/3)n-Nuf2 conjugates with sizes ranging from ~200 kDa to >300 kDa likely represented poly-SUMO-2/3 chain-modified Nuf2 ().
To predict the potential sumoylation sites on Nuf2, we performed bioinformatic analysis of Nuf2 using two online programs, SUMOplot and GPS-SUMO [Citation41]. We identified six lysine (K) residues, K41, K165, K335, K348, K402, and K447, with the highest scores for Nuf2 sumoylation (Fig. S4A). We then generated the bacterial expression construct encoding the Nuf2-6× K/R mutant, in which all the six K residues were mutated to arginine (R) residues. Our in vitro sumoylation assays and immunoblot analysis indicated that Nuf2 wild type (WT) and Nuf2-6× K/R mutant were comparably modified by SUMO-2 in vitro (Fig. S4B), suggesting that the six lysine residues are not the major sites for Nuf2 sumoylation.
Trimeric SUMO-2 chain modified Nuf2, but not trimeric SUMO-1 chain modified Nuf2, rescues the mitotic defects caused by global inhibition of sumoylation
In this study, we found that overexpression of Flag-Nuf2-Ubc9, but not Flag-Nuf2-Ubc9D or Flag-Nuf2, rescued the mitotic defects caused by global inhibition of sumoylation (), and Nuf2 was preferentially modified by poly-SUMO-2/3 chain in vivo (). We then asked whether Flag-Nuf2-Ubc9 enhances SUMO-1 and/or SUMO-2/3 modification of Nuf2. To address this question, we transfected 293T cells with the construct encoding Flag-Nuf2 or Flag-Nuf2-Ubc9 and performed immunoprecipitation using anti-Flag beads followed by immunoblot analysis. We found that both Flag-Nuf2 and Flag-Nuf2-Ubc9 were specifically modified by SUMO-2/3 instead of SUMO-1 (Fig. S5). Although the amounts of Flag-Nuf2-Ubc9 pulled down by anti-Flag beads were only ~10% of those of Flag-Nuf2 (Fig. S5, left panel), levels of SUMO-2/3 modification on Flag-Nuf2-Ubc9 were almost comparable to those on Flag-Nuf2 (Fig. S5, right panel), suggesting that the fusion of Nuf2 to the SUMO-conjugating enzyme Ubc9 stimulated SUMO-2/3 modification of Nuf2. This result was consistent with the previous finding that the fusion of a known SUMO target, p53 or STAT1, with Ubc9 greatly increased levels of sumoylation on the corresponding protein target [Citation37].
Given that Nuf2 was modified by poly-SUMO-2/3 chains in vivo (), we thus tested whether the fusion protein imitating poly-SUMO-2/3 chain modified Nuf2 can rescue the mitotic defects caused by global inhibition of sumoylation. We generated two constructs encoding Flag-Nuf2-SUMO-1-SUMO-1-SUMO-1 (Flag-Nuf2-3× SUMO-1) and Flag-Nuf2-SUMO-2-SUMO-2-SUMO-2 (Flag-Nuf2-3× SUMO-2) to simulate poly-SUMO-1 and poly-SUMO-2 modified Nuf2, respectively (). While the SUMO-1 K7 residue represents a major site for poly-SUMO-1 chain formation [Citation8], the poly-SUMO-2/3 chain is formed through the K11 residue [Citation11]. The SUMO-1 trimer in Flag-Nuf2-3× SUMO-1 was generated by fusing the C-terminal ends of the first and second SUMO-1 moieties to the K7 residues of the second and third SUMO-1 moieties (). Similarly, the C-terminal ends of the first and second SUMO-2 in Flag-Nuf2-3× SUMO-2 were fused to the K11 residues of the second and third SUMO-2 ( and S6A). The same trimeric SUMO-2 fusion protein (3× SUMO-2) was used previously to screen human proteome microarrays and identified many poly-SUMO-2/3 chain-binding proteins [Citation42]. The peptide bonds between SUMO moieties in Flag-Nuf2-3× SUMO-1 and Flag-Nuf2-3× SUMO-2 are not cleavable by SUMO isopeptidases as the C-terminal double-glycine (GG) motifs of SUMO-1 and SUMO-2 are mutated to GA or AA (). Both Flag-Nuf2-3× SUMO-1 and Flag-Nuf2-3× SUMO-2 cannot be attached to protein targets as the third SUMOs lack the GG motif for sumoylation.
We found that Flag-Nuf2-3× SUMO-1 and Flag-Nuf2-3× SUMO-2 were expressed with the expected sizes (Figs. S2 and S6B) and targeted to kinetochores in the absence or presence of SENP2 overexpression (Figs. S3B and S6C). We then co-transfected HeLa cells with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-Nuf2, Flag-Nuf2-3× SUMO-1 or Flag-Nuf2-3× SUMO-2, followed by immunofluorescence microscopy. Only ~2.7% or ~3.0% of the mitotic cells co-expressing Myc-SENP2 and Flag-Nuf2 or Flag-Nuf2-3× SUMO-1 were present at metaphase with the rest mitotic cells arrested at prophase or prometaphase ( and S6C). In contrast, co-expression of Myc-SENP2 with Flag-Nuf2-3× SUMO-2 resulted in ~9.0% of the mitotic cells at metaphase, representing a ~3-fold increase in rescuing the chromosome congression defect ( and S6C). While only ~5.0% or ~5.5% of the mitotic cells co-expressing Myc-SENP2 and Flag-Nuf2 or Flag-Nuf2-3× SUMO-1 exhibited an obvious CENP-E staining at kinetochores, ~13.0% of the mitotic cells co-expressing Myc-SENP2 and Flag-Nuf2-3× SUMO-2 displayed a robust CENP-E staining at kinetochores ( and S6D). Therefore, overexpression of Flag-Nuf2-3× SUMO-2, a simulator of trimeric SUMO-2 modified Nuf2, rescued the mitotic defects in chromosome congression and CENP-E kinetochore localization caused by SENP2 overexpression.
Trimeric SUMO-2 chain modified Nuf2, but not monomeric or dimeric SUMO-2 modified Nuf2, rescues the mitotic defects caused by global inhibition of sumoylation
To test whether monomeric or dimeric SUMO-2 modified Nuf2 can also rescue the mitotic defects caused by global inhibition of sumoylation, we generated the constructs encoding Flag-Nuf2-SUMO-2 (Flag-Nuf2-1× SUMO-2) and Flag-Nuf2-SUMO-2-SUMO-2 (Flag-Nuf2-2× SUMO-2) (). Both Flag-Nuf2-1× SUMO-2 and Flag-Nuf2-2× SUMO-2 were expressed with the correct sizes (Figs. S2 and S7B) and localized to kinetochores during mitosis in the absence or presence of SENP2 overexpression (Figs. S3C and S7C). We then co-transfected HeLa cells with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-Nuf2, Flag-Nuf2-1× SUMO-2, Flag-Nuf2-2× SUMO-2, or Flag-Nuf2-3× SUMO-2, followed by immunofluorescence microscopy. While only ~1.5% of mitotic cells co-expressing Myc-SENP2 and Flag-Nuf2, Flag-Nuf2-1× SUMO-2, or Flag-Nuf2-2× SUMO-2 were observed at metaphase, ~7.5% of mitotic cells co-expressing Myc-SENP2 and Flag-Nuf2-3× SUMO-2 were present at metaphase ( and S7C). In contrast to only ~4.0‒5.0% of the mitotic cells with co-expression of Myc-SENP2 and Flag-Nuf2, Flag-Nuf2-1× SUMO-2 or Flag-Nuf2-2× SUMO-2 displaying an obvious CENP-E staining at kinetochores, ~12% of the mitotic cells with co-expression of Myc-SENP2 and Flag-Nuf2-3× SUMO-2 exhibited a strong CENP-E staining at kinetochores ( and S7D). Hence, a minimum length of trimeric SUMO-2 chain covalently conjugated to Nuf2 was required for rescuing the mitotic defects caused by SENP2 overexpression.
Figure 4. The expression of Nuf2-3× SUMO-2, but not Nuf2-1× SUMO-2 or Nuf2-2× SUMO-2, rescues the mitotic defects caused by global inhibition of sumoylation. (a) The constructs encoding Flag-Nuf2, Flag-Nuf2-1× SUMO-2, Flag-Nuf2-2× SUMO-2, and Flag-Nuf2-3× SUMO-2. (b, c) HeLa cells were co-transfected with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-Nuf2, Flag-Nuf2-1× SUMO-2, Flag-Nuf2-2× SUMO-2, or Flag-Nuf2-3× SUMO-2, followed by immunofluorescence microscopy. The percentage (%) of mitotic cells present at metaphase and with both Myc and Flag staining (n ≥ 100 mitotic cells for each co-transfection) (b), and the portion (%) of mitotic cells with both Myc and CENP-E staining (n ≥ 100 mitotic cells for each co-transfection) (c) were determined by DAPI staining and immunofluorescence microscopy (Pro, prophase and prometaphase; Met, metaphase). (d) HeLa cells were transfected with control or Ubc9 siRNAs for 72 h followed by immunoblot analysis. (e) HeLa cells were treated with Ubc9 siRNA for 24 h and then transfected with the construct encoding Flag-Nuf2 or Flag-Nuf2-3× SUMO-2 for 48 h followed by immunofluorescence microscopy with anti-Flag and anti-CENP-E antibodies. The percentage (%) of Flag-Nuf2 or Flag-Nuf2-3× SUMO-2 transfected mitotic cells with CENP-E staining at kinetochores was subtracted by that of untransfected mitotic cells with CENP-E staining, resulting in the % increase in mitotic cells with CENP-E staining (n ≥ 100 mitotic cells with or without Flag staining for each transfection). The plotted values are the means ± SEM from three independent experiments (*: p < 0.05; Student’s t test)
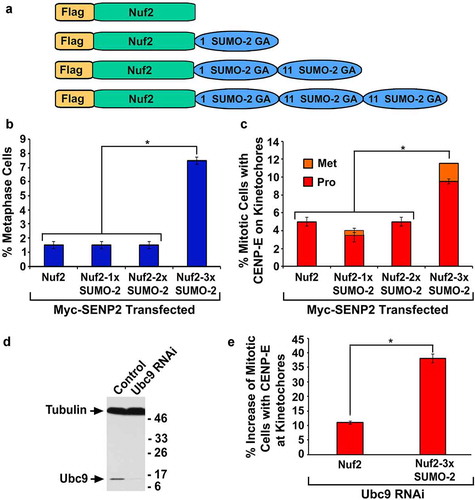
To investigate whether Flag-Nuf2-3× SUMO-2 can rescue the defect in CENP-E kinetochore localization caused by Ubc9 RNAi, we first showed that compared to control RNAi, Ubc9 RNAi greatly reduced levels of Ubc9 expression in HeLa cells (). We then transfected the Ubc9 RNAi cells with the construct encoding Flag-Nuf2 or Flag-Nuf2-3× SUMO-2 followed by immunofluorescence microscopy with antibodies against Flag and CENP-E. Compared to the Ubc9 RNAi cells with no Flag staining, the Flag-Nuf2 expression caused a ~10% increase in the mitotic cells with CENP-E staining at kinetochores, whereas the Flag-Nuf2-3× SUMO-2 expression resulted in a ~37% increase in the mitotic cells with CENP-E staining at kinetochores (). Hence, Flag-Nuf2-3× SUMO-2 was more effective than Flag-Nuf2 in rescuing the defect in CENP-E kinetochore localization caused by Ubc9 RNAi.
CENP-E has a higher binding affinity with trimeric SUMO-2 modified Nuf2 than with Nuf2
Based on our finding that Flag-Nuf2-3× SUMO-2 was more effective than Flag-Nuf2 in rescuing the defect in CENP-E kinetochore localization caused by global inhibition of sumoylation (), we hypothesized that compared to Nuf2, Nuf2-3× SUMO-2 has a higher-binding affinity with CENP-E. To test this hypothesis, we performed in vitro binding assays by immobilizing GST, GST-Nuf2, or GST-Nuf2-3× SUMO-2 on Glutathione beads. We then incubated the beads with CENP-E tail domain (CENP-Etail) wild type (WT) with a functional SUMO-interacting motif (SIM) or CENP-Etail SIM mutant (Mut) with a defect in interactions with poly-SUMO-2/3 chains [Citation12]. The relative levels of CENP-Etail were generated by dividing the signal intensities of CENP-Etail with those of GST, GST-Nuf2, and GST-Nuf2-3× SUMO-2, respectively (). Compared to GST-Nuf2, GST-Nuf2-3× SUMO-2 pulled down ~2.7-fold higher amounts of CENP-Etail WT, indicating that CENP-Etail WT had a significantly higher binding affinity with GST-Nuf2-3× SUMO-2 than with GST-Nuf2 (, left section). Conversely, we observed that compared to GST-Nuf2, GST-Nuf2-3× SUMO-2 only pulled down ~30% higher amounts of CENP-Etail Mut, which was not statistically significant (, right section). Moreover, GST-Nuf2-3× SUMO-2 pulled down ~3.2-fold higher amounts of CENP-Etail WT than those of CENP-Etail Mut, indicating that GST-Nuf2-3× SUMO-2 had a higher binding affinity with CENP-Etail WT than with CENP-Etail Mut (). Lastly, compared to GST, both GST-Nuf2 and GST-Nuf2-3× SUMO-2 pulled down significantly higher amounts of CENP-Etail WT or Mut (), which was consistent with the known interaction between Nuf2 and CENP-Etail [Citation33].
Figure 5. CENP-Etail has a higher-binding affinity with Nuf2-3× SUMO-2 than with Nuf2 both in vitro and in vivo. (a, b) Glutathione Sepharose beads were immobilized with GST, GST-Nuf2 or GST-Nuf2-3× SUMO-2 proteins and then incubated with 293T cell lysates expressing Flag-tagged CENP-Etail wild type (WT) or SIM mutant (Mut). The proteins on the beads were eluted and analyzed by immunoblotting with antibodies against GST (a) and Flag (b). (c) The relative levels of CENP-Etail WT or Mut pulled down by GST, GST-Nuf2, and GST-Nuf2-3× SUMO-2. The plotted values represent the mean values ± SEM from three independent experiments (*: p < 0.05; **: p ≤ 0.01; NS: not significant; Student’s t test). (d) 293T cells were co-transfected with two constructs encoding GFP-CENP-Etail WT and one of the Flag-tagged proteins, Flag-Nuf2 or Flag-Nuf2-3× SUMO-2, followed by co-immunoprecipitation (Co-IP) using anti-Flag beads and immunoblot analysis using anti-Flag and anti-GFP antibodies
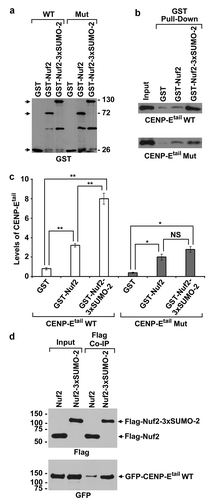
To test whether compared Nuf2, Nuf2-3× SUMO-2 has a higher binding affinity with CENP-Etail WT in vivo, we co-transfected 293T cells with two constructs encoding GFP-CENP-Etail WT and one of the Flag-tagged proteins, Flag-Nuf2 or Flag-Nuf2-3× SUMO-2, followed by co-immunoprecipitation (Co-IP) and immunoblot analysis. While the comparable amounts of Flag-Nuf2-3× SUMO-2 and Flag-Nuf2 were pulled down by anti-Flag beads (, upper panel), the amounts of GFP-CENP-Etail WT co-immunoprecipitated with Flag-Nuf2-3× SUMO-2 were at least 3-fold higher than those co-immunoprecipitated with Flag-Nuf2 (, lower panel). Hence, CENP-Etail WT had a higher binding affinity with trimeric SUMO-2 modified Nuf2 than with Nuf2 both in vitro and in vivo.
The mitotic defects caused by global inhibition of sumoylation can be rescued by trimeric SUMO-2 chain modified BubR1
Similar to Nuf2, the CENP-E interacting protein BubR1 is not only required for CENP-E kinetochore localization but also modified by SUMO-2/3 in vivo [Citation12,Citation26,Citation43,Citation44]. To test whether the BubR1-SUMO-2-SUMO-2-SUMO-2 (BubR1-3× SUMO-2) fusion protein that simulates trimeric SUMO-2 modified BubR1 can rescue the mitotic defects caused by SENP2 overexpression, we first showed that both Flag-BubR1 and Flag-BubR1-3× SUMO-2 were expressed with the correct sizes and associated with kinetochores during mitosis in the absence or presence of SENP2 overexpression ( and S8). We then co-transfected HeLa cells with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-BubR1 or Flag-BubR1-3× SUMO-2, followed by immunofluorescence microscopy (–c). Compared to only ~3.8% of the mitotic cells with co-expression of Myc-SENP2 and Flag-BubR1 observed at metaphase, ~13.2% of the mitotic cells with co-expression of Myc-SENP2 and Flag-BubR1-3× SUMO-2 were present at metaphase (). While ~24.2% of the mitotic cells co-expressing Myc-SENP2 and Flag-BubR1 had an obvious CENP-E staining at kinetochores, ~35.7% of the mitotic cells co-expressing Myc-SENP2 and Flag-BubR1-3× SUMO-2 exhibited a robust CENP-E staining at kinetochores (). Hence, the expression of Flag-BubR1-3× SUMO-2 also rescued the mitotic defects caused by SENP2 overexpression.
Figure 6. The expression of the BubR1-3× SUMO-2 fusion protein rescues the mitotic defects caused by SENP2 overexpression. (a) 293T cells were transfected with the constructs encoding Flag-BubR1 or Flag-BubR1-3× SUMO-2 followed by immunoblot analysis using anti-Flag and anti-SUMO-2 antibodies. (b-d) HeLa cells were co-transfected with two constructs encoding Myc-SENP2 and one of the Flag-tagged proteins, Flag-BubR1 or Flag-BubR1-3× SUMO-2, for 48 h and analyzed by immunofluorescence microscopy using anti-Myc and anti-CENP-E antibodies (b). Bar, 10 μm. The percentage (%) of mitotic cells with both Myc and Flag staining and present at metaphase (n ≥ 100 mitotic cells for each co-transfection) (c), and the fraction (%) of mitotic cells with both Myc and CENP-E staining (n ≥ 100 mitotic cells for each co-transfection) (d) were determined by DAPI staining and immunofluorescence microscopy (Pro, prophase and prometaphase; Met, metaphase). The plotted values are the means ± SEM from three experiments (***: p < 0.001; Student’s t test)
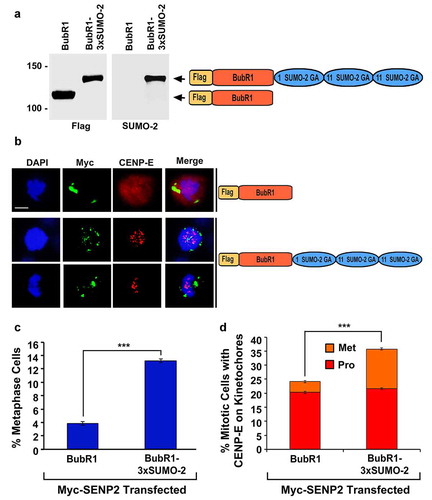
To test whether SUMO-2/3 modification of BubR1 is temporally upregulated during mitosis, we transfected 293T cells with the construct encoding Flag-BubR1 and then cultured the transfected cells in the absence or presence of nocodazole for arresting cells at mitosis followed by immunoprecipitation using anti-Flag beads and immunoblot analysis using antibodies against Flag, SUMO-1, and SUMO-2/3. We found that levels of BubR1 SUMO-2/3 modification were comparable between the nocodazole-treated mitotic cells and the untreated asynchronous cells (Fig. S9). Therefore, SUMO-2/3 modification of BubR1 was not upregulated during mitosis.
Discussion
Our results support a model that poly-SUMO-2/3 chain modification of Nuf2 enhances its interaction with CENP-E, leading to CENP-E kinetochore localization and CENP-E-mediated chromosome congression during mitosis. Trimeric SUMO-2/3 chains may represent the minimum length of poly-SUMO-2/3 chains conjugated on Nuf2 for an efficient recruitment of CENP-E to kinetochores during mitosis, because only Nuf2-3× SUMO-2, but not Nuf2-1× SUMO-2 or Nuf2-2× SUMO-2, can rescue the mitotic defect in CENP-E kinetochore localization in cells with global inhibition of sumoylation. It has been shown previously that poly-SUMO-2/3 chain signals are recognized by multiple SIMs present in various proteins, including the SUMO-targeted ubiquitin ligases (STUbLs) RNF4 and RNF111 [Citation45,Citation46]. Hence, CENP-E may contain multiple SIMs at its kinetochore-targeting tail domain (CENP-Etail) for its effective interactions with poly-SUMO-2/3 chain modified Nuf2 during mitosis. Notably, our bioinformatics analysis of CENP-Etail using the GPS-SUMO program [Citation41] identifies three SIMs (SIM1, SIM2, and SIM3) with high scores (Fig. S10). Our previous study reveals that SIM2 (2307–2311) is essential for CENP-E interaction with poly-SUMO-2/3 chains and localization to kinetochores [Citation12]. Here, we elucidate that SIM2 is also required for the high-affinity binding between CENP-Etail and trimeric SUMO-2/3 modified Nuf2. Additional studies are needed to determine whether SIM1 (2034–2038) and SIM3 (2439–2443) are critical for CENP-E interaction with poly-SUMO-2/3 modified Nuf2 and localization to kinetochores.
A growing number of outer kinetochore proteins have been identified as SUMO-2/3 substrates, including Nuf2, BubR1, CENP-E, Kif18A, NKAP, Mps1, and ANAPC4, in mammalian cells [Citation12,Citation18,Citation20,Citation47–50]. Similar to Nuf2, Kif18A is a known CENP-E interacting protein and modified by poly-SUMO-2/3 chain in vivo [Citation49,Citation51]. In this study, we demonstrate that either Nuf2-3× SUMO-2 or BubR1-3× SUMO-2, which simulates poly-SUMO-2/3 modified Nuf2 or BubR1, can rescue the defect in CENP-E kinetochore localization caused by global inhibition of sumoylation (). Therefore, multiple SUMO-2/3 modifications on a cluster of outer kinetochore proteins, such as Nuf2 and BubR1, may jointly contribute to the recruitment of CENP-E to kinetochores during mitosis. Consistent with this model, simultaneous SUMO modifications on a group of DNA repair proteins in response to DNA double-strand breaks synergistically stabilize interactions between these proteins and thus facilitate the repair process [Citation52]. Conversely, we observe a significant increase in SUMO-2/3 modification of Nuf2 during mitosis, whereas SUMO-2/3 modification of BubR1 is not upregulated during mitosis ( and S9). Given the temporal kinetochore association of CENP-E during mitosis [Citation27,Citation28], SUMO-2/3 modification of Nuf2 may play a more dominant role in targeting CENP-E to kinetochores than SUMO-2/3 modification of BubR1.
An intriguing question is how SUMO-2/3 modification of Nuf2 is temporally upregulated during mitosis for the timely recruitment of CENP-E to the kinetochore. To address this question, it would be necessary to identify and characterize the SUMO E3 ligase(s) and isopeptidase(s) responsible for upregulating and downregulating levels of Nuf2 SUMO-2/3 modification, respectively, during mitosis. The SUMO ligase PIASy is required for SUMO-2/3 modification of the centromere/kinetochore-associated Topo IIα during mitosis [Citation22,Citation53,Citation54]. In addition, the SUMO ligases, PIAS3 and Nup358/RanBp2, stimulate SUMO-2/3 modification of the centromere/kinetochore-associated chromosomal passenger complex (CPC) subunits, Aurora B and Borealin, respectively [Citation17,Citation55]. On the other hand, SENP2 is the sole SUMO isopeptidase that not only localizes to kinetochores during mitosis but also causes the mitotic defects in CENP-E kinetochore localization and chromosome congression when overexpressed in mammalian cells [Citation12,Citation25]. Moreover, the SUMO isopeptidase SENP3 deconjugates SUMO-2/3 from the CPC subunit Borealin during mitosis [Citation55]. Therefore, multiple SUMO ligases and isopeptidases may cooperatively regulate SUMO-2/3 modification of Nuf2 during mitosis.
Accumulating lines of evidence have shown that poly-SUMO-2/3 chain modification of various proteins often leads to RNF4/RNF111-mediated ubiquitylation and proteasomal degradation [Citation56,Citation57]. For example, poly-SUMO-2/3 modification of the centromere/kinetochore-associated proteins Mis18bp1 and CENP-I results in RNF4-mediated ubiquitylation and degradation [Citation19,Citation49,Citation58]. It would be interesting to examine whether the association of poly-SUMO-2/3 modified Nuf2 with CENP-E prevents RNF4/RNF111 from recognizing the poly-SUMO-2/3 chain signals on Nuf2 and thus inhibits RNF4/RNF111-mediated Nuf2 ubiquitylation and degradation. Consistent with this possibility, proteomic studies revealed that inhibition of proteasomal degradation in HeLa cells using the proteasome inhibitors dramatically increases levels of sumoylation on a variety of proteins, including Nuf2 and one of the best characterized RNF4/RNF111 targets called promyelocytic leukemia protein (PML) [Citation59]. It is well established that poly-SUMO-2/3 chain modification of PML leads to RNF4/RNF111-mediated ubiquitylation and degradation [Citation46,Citation60–63].
A novel strategy for cancer therapies is to target the SUMO pathway by small-molecule inhibitors against the sole SUMO E1 (SAE1/SAE2) or E2 (Ubc9) enzyme. At least three lines of evidence support this anticancer strategy. First, global inhibition of sumoylation in human cervical cancer HeLa cells by either SENP2 overexpression or Ubc9 RNAi causes the cell cycle arrest at mitosis followed by apoptosis [Citation12]. Second, RNAi depletion of the SUMO E1 enzyme subunit SAE2 or Ubc9 inhibits cancer cell proliferation in vitro and tumor growth in vivo [Citation64]. Third, sumoylation is required for tumorigenesis driven by Myc or Ras, which contributes to up to ~70% or ~50% of all human cancers [Citation65,Citation66]. Inhibition of sumoylation would be especially effective for treating cancers with Myc hyperactivation, Ras mutations, or upregulated sumoylation [Citation65–69]. A number of small-molecule inhibitors against SAE1/SAE2 (ML-792, ginkgolic acid, and COH000) or Ubc9 (spectomycin B1 and 2-D08) have been developed for therapeutic treatment of human cancers [Citation70–76].
Similar to SAE1/SAE2 and Ubc9, Nuf2 is also a potential anticancer target. Nuf2 is not only required for the cell cycle progression through mitosis but also overexpressed in many different types of human cancers [Citation77–83]. Furthermore, RNAi knockdown of Nuf2 expression inhibits cancer cell proliferation and tumor growth by triggering the cell cycle arrest and apoptosis [Citation78–80,Citation84,Citation85]. Based on our finding that Nuf2 sumoylation is critical for the mitotic progression by facilitating CENP-E kinetochore localization and chromosome congression, a combined targeting of Nuf2 and SAE1/SAE2 or Ubc9 by small-molecule inhibitors or siRNAs could be an effective approach for treating human cancers with both Nuf2 overexpression and Myc hyperactivation, Ras mutations, or upregulated sumoylation.
Materials and methods
Antibodies
We obtained antibodies from the following sources: anti-Nuf2 rabbit polyclonal antibody (pAb) (Bethyl Laboratories); anti-CENP-E mouse monoclonal antibody (mAb), Dr Tim Yen (Fox Chase Cancer Center); anti-CENP-E rabbit pAb, Dr Beth Weaver (University of Wisconsin–Madison); anti-SUMO-1 mouse mAb (21C7) [Citation86] and anti-SUMO-2/3 mouse mAb (8A2) [Citation12]; CREST human antibody against the centromere/kinetochore proteins CENP-A, CENP-B, and CENP-C, Dr William Brinkley (Baylor College of Medicine); anti-Flag mouse mAb (M2) (Sigma); anti-Ubc9 rabbit pAb (Abcam); anti-GST mouse mAb (Santa Cruz); anti-Myc (9E10) mouse mAb (Santa Cruz); anti-Myc rabbit pAb (Cell Signaling); anti-α-tubulin (DM1A) mouse mAb (Sigma); anti-GFP (GF28R) mouse mAb (UBPBio).
Plasmids and siRNAs
The DNA constructs encoding Flag-tagged Nuf2 fusion proteins were generated using pFlag-CMV vector, in which the Nuf2 coding sequence was inserted between ClaI and BamHI sites. To generate the pFlag-Nuf2-3× SUMO-1, pFlag-Nuf2-3× SUMO-2, and pFlag-Nuf2-Ubc9 constructs, the coding sequences of 3× SUMO-1, 3× SUMO-2, and Ubc9 were PCR amplified and subcloned between BamHI and SmaI sites in the pFlag-Nuf2 construct. The original constructs containing the coding sequences of 3× SUMO-1 and 3× SUMO-2 were a gift from Dr Michael Matunis (Johns Hopkins University). The pFlag-Nuf2-1× SUMO-2 and pFlag-Nuf2-2× SUMO-2 constructs were generated by site-directed mutagenesis using the pFlag-Nuf2-3× SUMO-2 construct as the template to introduce a stop codon at the end of the first and second SUMO-2 coding sequence, respectively. To generate pFlag-BubR1-3× SUMO-2 plasmid, the coding sequence of 3× SUMO-2 was subcloned into the pFlag-BubR1 plasmid [Citation12]. To produce the GST-tagged fusion proteins in bacteria, the coding sequences of Nuf2 and Nuf2-3× SUMO-2 were amplified by PCR and subcloned into the pGEX-6P-1 vector between SmaI and SalI sites. Using pFlag-Nuf2-Ubc9 as the template, pFlag-Nuf2-Ubc9D was generated by site-directed mutagenesis to convert the codon 93 encoding cysteine (C) to the codon encoding alanine (A). To knock down Nuf2 expression by RNA interference (RNAi), two small interfering RNAs (siRNAs) specific to Nuf2, siRNA 1 (5’-GCAUGCCGUGAAACGUAUAdTdT-3’) [Citation33,Citation34] and siRNA 2 (5’-GGCUUCUUACCAUUCAGCAdTdT-3’), and the scrambled control siRNA (5’-UUCUCCGAACGUGUCACGUdTdT-3’) [Citation87] were purchased from GE Dharmacon. HeLa cells were also transfected with Ubc9-specific siRNA for knockdown of Ubc9 expression [Citation12].
Cell culture and immunofluorescence microscopy
HeLa (human cervical cancer cells) and 293T (human embryonic kidney cells transformed with SV40 large T antigen) were grown in DMEM medium (Hyclone) supplemented with 1% Penicillin-Streptomycin and 10% fetal bovine serum (FBS). To analyze the expression of Flag-tagged fusion proteins, 293T cells were transfected with the corresponding construct using the calcium phosphate method [Citation88]. The transfected cells were lysed in 2× SDS sample buffer (2× SSB) followed by immunoblot analysis. To analyze the subcellular localization of Flag-tagged fusion proteins by indirect immunofluorescence microscopy, HeLa cells were grown to 70% confluency on coverslips and transfected with one of the constructs encoding a Flag-tagged fusion protein using Lipofectamine-Plus reagent (Invitrogen) for 48 h followed by fixation with 3.5% paraformaldehyde for 7 min and permeabilization with 0.2% Triton X-100 for 20 min. The fixed cells were first incubated with mouse anti-Flag mAb and human CREST antibody for 1 h and then with Alexa Fluor 488- and 594-conjugated secondary antibodies (Invitrogen) for 30 min. The stained cells were incubated with the mounting solution containing 4′,6-diamidino-2-phenylindole (DAPI) to stain DNA for 5 min and then analyzed by fluorescence microscope. In addition, HeLa cells were co-transfected with two constructs encoding Myc-tagged SENP2 and one of the Flag-tagged proteins for 48 h followed by immunofluorescence microscopy with rabbit anti-Myc antibody and mouse anti-Flag antibody or mouse anti-CENP-E antibody. The portion of the transfected cells at each stage of mitosis was determined by DAPI staining and immunofluorescence microscopy. The inverted Olympus IX81 fluorescence microscope with U-Plan S-Apo 60×/1.35 NA oil immersion objective was used to acquire images with the MicroSuite acquisition software (Olympus).
RNA interference
HeLa cells were transfected with one of the two Nuf2-specific siRNAs or a control siRNA for 72 h using Oligofectamine Transfection Reagent (Invitrogen). To check for the efficiency of RNAi-mediated knockdown of Nuf2, the transfected cells were lysed with 2× SSB and then analyzed by immunoblotting using anti-Nuf2 antibodies. To determine whether Nuf2 RNAi affects CENP-E kinetochore localization, HeLa cells were transfected with control or Nuf2-specific siRNAs for 72 h followed by immunofluorescence microscopy using human anti-CREST and mouse anti-CENP-E antibodies. The portion of the transfected cells at each stage of mitosis was determined by DAPI staining and immunofluorescence microscopy.
Immunoprecipitation
293T cells were transfected with the construct encoding Flag-tagged Nuf2 or BubR1 for 48 h followed by incubation in the presence or absence of nocodazole (100 ng/ml) for 5 h. All the immunoprecipitation experiments were performed under stringent denaturing conditions as described previously [Citation12]. In brief, the transfected cells were lysed in the lysis buffer containing protease inhibitors and 10 mM N-ethylmaleimide (NEM). The lysis buffer was obtained by diluting 2× SSB (5% SDS, 150 mM Tris-HCl, pH 6.7, 20% glycerol) with 1× RIPA buffer (20 mM HEPES, pH 8.0, 300 mM NaCl, 2 mM EDTA, 0.5% sodium deoxycholate, 1% Triton X-100, and 6% glycerol) in 1:4 ratio. The cell lysates were then sonicated and centrifuged at 14,000 rpm at 4°C for 10 min. The supernatant was incubated with anti-Flag M2 beads (Sigma) for 5 h at 4°C. The beads were washed five times in the wash buffer (20 mM HEPES, pH 8.0, 750 mM NaCl, 2 mM EDTA, 0.5% sodium deoxycholate, 1% Triton X-100, 6% glycerol, 0.1% SDS, and 10 mM NEM). The immunopurified proteins were eluted using 2× SSB followed by immunoblot analysis.
In vitro sumoylation assays
The in vitro sumoylation assays were performed at 37°C in 20 μl reactions containing 0.5 µg SAE1/SAE2, 0.3 µg Ubc9, 1 mM ATP, 10 mM phosphocreatine, 1.2 µg/ml inorganic pyrophosphatase, 40 U/ml creatine phosphokinase, 110 mM potassium acetate, 20 mM HEPES-KOH (pH 7.3), 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, and 1.0 µg Nuf2 or GST-RanGAP1 C-terminal fragment (NΔ419) in the absence or presence of 1.0 µg SUMO-1 WT, SUMO-1 K-less mutant, or SUMO-2.
In vitro and in vivo CENP-E binding assays
The GST, GST-Nuf2, and GST-Nuf2-3× SUMO-2 proteins were expressed in BL21 (DE3) cells and purified using Glutathione Sepharose beads (GE Healthcare). To produce Flag-tagged CENP-E tail domain (CENP-Etail) containing SIM wild type (WT) or mutant (Mut) [Citation12], 293T cells were transfected with the corresponding construct and then lysed in the lysis buffer containing 50 mM Tris-HCl (pH7.5), 0.2% NP-40, 10% Glycerol, 2 mM EDTA, 25 mM NaF, 50 mM NaCl, 10 mM NEM, and protease inhibitors. The 293T cell lysates were incubated with the Glutathione beads immobilized with GST, GST-Nuf2, or GST-Nuf2-3× SUMO-2 in the binding buffer consisting of the lysis buffer plus 0.02% NP-40 at 4°C for 5 h. The beads were washed with the buffer containing 50 mM Tris-HCl (pH 7.5), 0.02% NP-40, 10% glycerol, 2 mM EDTA, and 50 mM NaCl for five times. The proteins on the beads were eluted with 2× SSB and analyzed by immunoblotting with anti-GST and anti-Flag antibodies. The signal intensities of GST and Flag were quantified using ImageJ (NIH). Statistical analysis was performed using Student’s t-test for analyzing the data from three independent experiments.
The in vivo CENP-E binding assays or co-immunoprecipitation (Co-IP) experiments were performed by co-transfection of 293T cells with the constructs encoding GFP-CENP-Etail WT and Flag-Nuf2 or Flag-Nuf2-3× SUMO-2 using Lipofectamine 3000 (Invitrogen). The transfected cells were lysed with 1× RIPA buffer plus 0.1% SDS and protease inhibitors followed by sonication and centrifugation at 14,000 rpm at 4°C for 10 min. The supernatants were incubated with anti-Flag beads at 4°C for 5 h, and the beads were washed five times using 1× RIPA buffer plus 0.1% SDS and protease inhibitors. The proteins on the beads were eluted with 2× SSB followed by immunoblot analysis using anti-Flag and anti-GFP antibodies.
Supplemental Material
Download PDF (1,009 KB)Acknowledgments
We thank Drs Michael Matunis (Johns Hopkins University), William Brinkley (Baylor College of Medicine), Tim Yen (Fox Chase Cancer Center), and Beth Weaver (University of Wisconsin–Madison) for providing reagents, and Drs Micheal Yu and Paul Gollnick (SUNY Buffalo) for support on X-ray film processor. We also thank Jun Wan for assistance on the constructs encoding Flag-Nuf2-3× SUMO-2 and Flag-Nuf2-Ubc9, Michael Vespremi for support on microscopy, and Adam Lyle, Ellen Zhang, Lila Toczek, and Katie Bryant for comments on the manuscript. We acknowledge the reviewers for constructive suggestions and comments. This work was supported by SUNY Buffalo State Faculty Research Incentive Grant (XDZ) and American Cancer Society Institutional Research Grant 11-053-01-IRG (XDZ).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Abrieu A, Liakopoulos D. How does SUMO participate in spindle organization? Cells. 2019;8:801.
- Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5.
- Wan J, Subramonian D, Zhang XD. SUMOylation in control of accurate chromosome segregation during mitosis. Curr Protein Pept Sci. 2012;13:467–481.
- Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J Cell Sci. 2009;122:4249–4252.
- Bernier-Villamor V, Sampson DA, Matunis MJ, et al. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356.
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–12659.
- Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276(24):21664–21669.
- Pedrioli PG, Raught B, Zhang XD, et al. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat Methods. 2006;3:533–539.
- Tatham MH, Jaffray E, Vaughan OA, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374.
- Vertegaal AC. Small ubiquitin-related modifiers in chains. Biochem Soc Trans. 2007;35:1422–1423.
- Matic I, Van Hagen M, Schimmel J, et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144.
- Zhang XD, Goeres J, Zhang H, et al. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741.
- Cheeseman IM. The kinetochore. Cold Spring Harb Perspect Biol. 2014;6:a015826.
- Maiato H, DeLuca J, Salmon ED, et al. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477.
- Abrieu A, Kahana JA, Wood KW, et al. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102(6):817–826.
- Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487.
- Ban R, Nishida T, Urano T. Mitotic kinase Aurora-B is regulated by SUMO-2/3 conjugation/deconjugation during mitosis. Genes Cells. 2011;16:652–669.
- Li T, Chen L, Cheng J, et al. SUMOylated NKAP is essential for chromosome alignment by anchoring CENP-E to kinetochores. Nat Commun. 2016;7(1):12969.
- Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 2010;188(5):681–692.
- Restuccia A, Yang F, Chen C, et al. Mps1 is SUMO-modified during the cell cycle. Oncotarget. 2016;7(3):3158–3170.
- Yang F, Hu L, Chen C, et al. BubR1 is modified by sumoylation during mitotic progression. J Biol Chem. 2012;287:4875–4882.
- Agostinho M, Santos V, Ferreira F, et al. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 2008;68:2409–2418.
- Joseph J, Liu ST, Jablonski SA, et al. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617.
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373(6509):78–81.
- Cubenas-Potts C, Goeres JD, Matunis MJ. SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Mol Biol Cell. 2013;24:3483–3495.
- Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63.
- Cooke CA, Schaar B, Yen TJ, et al. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106(7):446–455.
- Yen TJ, Compton DA, Wise D, et al. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. Embo J. 1991;10:1245–1254.
- Kapoor TM, Lampson MA, Hergert P, et al. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311(5759):388–391.
- Putkey FR, Cramer T, Morphew MK, et al. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3(3):351–365.
- Schaar BT, Chan GK, Maddox P, et al. CENP-E function at kinetochores is essential for chromosome alignment. J Cell Biol. 1997;139(6):1373–1382.
- Wood KW, Sakowicz R, Goldstein LS, et al. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366.
- Liu D, Ding X, Du J, et al. Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J Biol Chem. 2007;282:21415–21424.
- DeLuca JG, Moree B, Hickey JM, et al. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555.
- Hori T, Haraguchi T, Hiraoka Y, et al. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci. 2003;116:3347–3362.
- McCleland ML, Gardner RD, Kallio MJ, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114.
- Jakobs A, Koehnke J, Himstedt F, et al. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Methods. 2007;4:245–250.
- Banerjee A, Deshaies RJ, Chau V. Characterization of a dominant negative mutant of the cell cycle ubiquitin-conjugating enzyme Cdc34. J Biol Chem. 1995;270:26209–26215.
- Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270.
- Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140(3):499–509.
- Zhao Q, Xie Y, Zheng Y, et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–330.
- Cox E, Hwang W, Uzoma I, et al. Global analysis of SUMO-binding proteins identifies SUMOylation as a key regulator of the INO80 chromatin remodeling complex. Mol Cell Proteomics. 2017;16:812–823.
- Legal T, Hayward D, Gluszek-Kustusz A, et al. The C-terminal helix of BubR1 is essential for CENP-E-dependent chromosome alignment. J Cell Sci. 2020;133:jcs246025.
- Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98.
- Sun H, Hunter T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem. 2012;287:42071–42083.
- Tatham MH, Geoffroy MC, Shen L, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546.
- Eifler K, Cuijpers SAG, Willemstein E, et al. SUMO targets the APC/C to regulate transition from metaphase to anaphase. Nat Commun. 2018;9:1119.
- Lee CC, Li B, Yu H, et al. Sumoylation promotes optimal APC/C activation and timely anaphase. Elife. 2018;7. DOI:10.7554/eLife.29539
- Liebelt F, Jansen NS, Kumar S, et al. The poly-SUMO2/3 protease SENP6 enables assembly of the constitutive centromere-associated network by group deSUMOylation. Nat Commun. 2019;10:3987.
- Yang F, Chen Y, Dai W. Sumoylation of Kif18A plays a role in regulating mitotic progression. BMC Cancer. 2015;15:197.
- Huang Y, Yao Y, Xu HZ, et al. Defects in chromosome congression and mitotic progression in KIF18A-deficient cells are partly mediated through impaired functions of CENP-E. Cell Cycle. 2009;8:2643–2649.
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151(4):807–820.
- Azuma Y, Arnaoutov A, Anan T, et al. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. Embo J. 2005;24:2172–2182.
- Diaz-Martinez LA, Gimenez-Abian JF, Azuma Y, et al. PIASgamma is required for faithful chromosome segregation in human cells. PLoS One. 2006;1:e53.
- Klein UR, Haindl M, Nigg EA, et al. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell. 2009;20(1):410–418.
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82(1):357–385.
- Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–568.
- Cuijpers SAG, Willemstein E, Vertegaal ACO. Converging small Ubiquitin-like modifier (SUMO) and Ubiquitin signaling: improved methodology identifies co-modified target proteins. Mol Cell Proteomics. 2017;16:2281–2295.
- Hendriks IA, Lyon D, Young C, et al. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325–336.
- Erker Y, Neyret-Kahn H, Seeler JS, et al. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol. 2013;33:2163–2177.
- Geoffroy MC, Jaffray EG, Walker KJ, et al. Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol Biol Cell. 2010;21:4227–4239.
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555.
- Zhang XW, Yan XJ, Zhou ZR, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–243.
- He X, Riceberg J, Pulukuri SM, et al. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS One. 2015;10(4):e0123882.
- Kessler JD, Kahle KT, Sun T, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335(6066):348–353.
- Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848.
- Eifler K, Vertegaal ACO. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. 2015;40:779–793.
- Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184–197.
- Subramonian D, Raghunayakula S, Olsen JV, et al. Analysis of changes in SUMO-2/3 modification during breast cancer progression and metastasis. J Proteome Res. 2014;13:3905–3918.
- Fukuda I, Ito A, Hirai G, et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009;16:133–140.
- He X, Riceberg J, Soucy T, et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol. 2017;13:1164–1171.
- Hirohama M, Kumar A, Fukuda I, et al. Spectomycin B1 as a novel SUMOylation inhibitor that directly binds to SUMO E2. ACS Chem Biol. 2013;8:2635–2642.
- Kim YS, Nagy K, Keyser S, et al. An electrophoretic mobility shift assay identifies a mechanistically unique inhibitor of protein sumoylation. Chem Biol. 2013;20(4):604–613.
- Li YJ, Du L, Wang J, et al. Allosteric inhibition of Ubiquitin-like modifications by a class of inhibitor of SUMO-activating enzyme. Cell Chem Biol. 2019;26(278–288):e276.
- Licciardello MP, Mullner MK, Durnberger G, et al. NOTCH1 activation in breast cancer confers sensitivity to inhibition of SUMOylation. Oncogene. 2015;34:3780–3790.
- Lv Z, Yuan L, Atkison JH, et al. Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme. Nat Commun. 2018;9:5145.
- Agarwal R, Narayan J, Bhattacharyya A, et al. Gene expression profiling, pathway analysis and subtype classification reveal molecular heterogeneity in hepatocellular carcinoma and suggest subtype specific therapeutic targets. Cancer Genet. 2017;216-217:37–51.
- Hayama S, Daigo Y, Kato T, et al. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 2006;66(21):10339–10348.
- Hu P, Chen X, Sun J, et al. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo. Biosci Rep. 2015;35(35). DOI:10.1042/BSR20140124
- Huang SK, Qian JX, Yuan BQ, et al. SiRNA-mediated knockdown against NUF2 suppresses tumor growth and induces cell apoptosis in human glioma cells. Cell Mol Biol (Noisy-le-grand). 2014;60:30–36.
- Shiraishi T, Terada N, Zeng Y, et al. Cancer/Testis Antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9:153.
- Sun ZY, Wang W, Gao H, et al. Potential therapeutic targets of the nuclear division cycle 80 (NDC80) complexes genes in lung adenocarcinoma. J Cancer. 2020;11:2921–2934.
- Xu W, Wang Y, Wang Y, et al. Screening of differentially expressed genes and identification of NUF2 as a prognostic marker in breast cancer. Int J Mol Med. 2019;44:390–404.
- Kaneko N, Miura K, Gu Z, et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun. 2009;390:1235–1240.
- Liu Q, Dai SJ, Li H, et al. Silencing of NUF2 inhibits tumor growth and induces apoptosis in human hepatocellular carcinomas. Asian Pac J Cancer Prev. 2014;15:8623–8629.
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470.
- Sekhri P, Tao T, Kaplan F, et al. Characterization of amino acid residues within the N-terminal region of Ubc9 that play a role in Ubc9 nuclear localization. Biochem Biophys Res Commun. 2015;458:128–133.
- Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res 1996;24: 596–601
