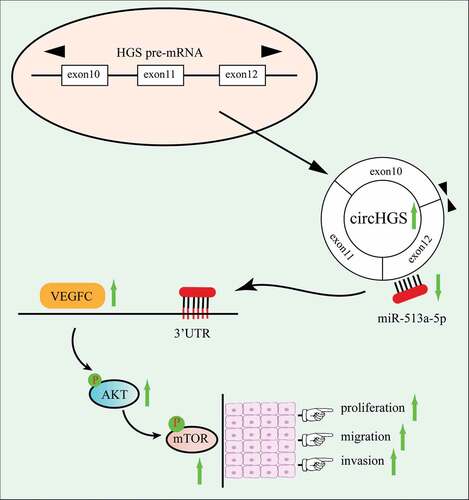
ABSTRACT
Bladder cancer (BCa) is a malignant tumor that occurs in the bladder mucosa with high mortality. Circular RNAs (circRNAs), as newly discovered noncoding RNAs, are associated with the occurrence and development of BCa. However, the effects of circRNAs in BCa have not been fully elucidated. Through the GEO (Gene Expression Omnibus) database, an abnormally expressed circular RNA, circHGS (hsa_circ_0004721), was first identified in BCa. qRT – PCR was performed to measure the expression of circHGS in BCa tissues and cells. The intracellular localization of circHGS was detected by nucleocytoplasmic separation experiment and fluorescence in situ hybridization assay. In vitro experiments were conducted to detect the effects of circHGS on cell cycle, proliferation, migration and invasion. The correlations between miR-513a-5p and circHGS or VEGFC were confirmed by dual-luciferase reporter assay, qRT – PCR and western blot. The role of circHGS in vivo was verified by xenograft tumor mice model. In this study, we clarified the roles and potential mechanism of circHGS in BCa. CircHGS, originating from the HGS gene, is upregulated in BCa tissues compared to normal tissues. Moreover, the expression of circHGS in BCa was positively associated with tumor grade and pathological T stage. Functionally, silencing of circHGS apparently suppressed cell cycle, proliferation, migration and invasion, but circHGS overexpression showed the opposite result. In vivo experiments also suggested that knockdown of circHGS suppressed tumor growth. Mechanistically, circHGS functions as a sponge of miR-513a-5p to elevate VEGFC expression and activate the AKT/mTOR signaling pathway, ultimately promoting BCa progression. Our findings indicated that circHGS promotes BCa progression via the miR-513a-5p/VEGFC/AKT/mTOR pathway and can be a promising therapeutic target of BCa.
Introduction
Bladder cancer (BCa) is a kind of aggressive malignant tumor of the human urinary system with high morbidity and mortality [Citation1,Citation2]. It has been reported that approximately 573,000 BCa cases were diagnosed and 213,000 BCa patient deaths occurred worldwide in 2020 [Citation3]. According to the depth of tumor growth in the bladder wall, BCa consists of roughly two major groups, nonmuscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC), of which NMIBC patients account for approximately 80% of all diagnosed BCa patients [Citation4,Citation5]. To date, tobacco smoking is still considered the principal risk factor for BCa patients [Citation6]. Moreover, multiple studies have identified genetic alterations in BCa, which is considered an early adverse event of BCa [Citation4]. Depending on the different conditions of the patients, surgery, radiotherapy, chemotherapy and immunotherapy are usually performed to treat BCa cases [Citation1,Citation7]. Nevertheless, a considerable number of BCa patients still have a poor prognosis, and the 5-year survival rate is not optimistic [Citation8]. In addition, the biomarkers used for BCa early diagnosis are insufficient [Citation5]. These deficiencies prompted us to find novel diagnostic markers and therapeutic targets for BCa.
Recently, with the development of high-throughput sequencing technology, considerable circular RNAs (circRNAs) have been identified [Citation9,Citation10]. CircRNAs, mostly generated from the back-splicing of exons of precursor mRNA, are a new kind of covalently circular noncoding RNA without 5’ caps and 3’ poly (A) tail structures [Citation11]. Benefiting from their continuous loop structure, circRNAs are protected from the degradation of RNase R [Citation12,Citation13]. Thus, circRNAs are more stable in the cytoplasm than corresponding linear mRNAs. Moreover, circRNAs have attracted great attention because of their special biological roles, such as acting as miRNA sponges, binding to RNA-binding proteins, regulating transcription and participating in protein translation [Citation13,Citation14].
Various studies have demonstrated that circRNAs are abnormally expressed in BCa and are closely correlated with the occurrence of BCa [Citation9]. For example, circRNA hsa_circ_0014130 is upregulated in BCa and mainly acts as a sponge of miR-132-3p to play oncogenic roles in BCa [Citation15]. Moreover, more circRNAs, such as circEHBP1 [Citation16], circNUDT21 [Citation17], and circ_0000658 [Citation18], have been identified to be dysregulated in BCa. Therefore, circRNAs are promising biomarkers and valuable therapeutic targets for BCa.
In our study, we first identified an upregulated circRNA, circHGS (hsa_circ_0004721), in BCa and aimed to investigate the effects of circHGS on cell cycle, proliferation, migration and invasion of BCa. Loss-of-function experiments showed that knockdown of circHGS markedly inhibited the cell cycle, proliferation, migration and invasion of BCa cells. In vivo experiments also indicated that silencing of circHGS suppressed tumor growth. Moreover, we found that circHGS enhanced BCa proliferation, migration and invasion via the miR-513a-5p/VEGFC/AKT/mTOR axis. Generally, our study showed that circHGS plays an oncogenic role in BCa and has the potential to be a novel target for BCa therapy.
Materials and methods
Cell culture
Human BCa cells (T24, UM-UC-3 and 5637) and human uroepithelial immortalized SV-HUC-1 cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). T24 and 5637 cells were cultivated in RPMI 1640 medium (Gibco, USA) containing 10% fetal bovine serum (FBS, TransStem), and UM-UC-3 cells were cultivated in DMEM medium (Gibco, USA) containing 10% FBS. SV-HUC-1 cells were cultured in F-12K medium with 10% FBS. All cells were cultured at 37°C in a 5% CO2 incubator.
Patient specimens
Thirty pairs of BCa tissues and matched nontumor tissues were harvested from patients who underwent radical cystectomy at Affiliated Hospital of Guangdong Medical University from February, 2021 to August, 2022. Twenty-six additional BCa tissues were collected from patients who underwent transurethral resection of bladder tumor or partial cystectomy at Affiliated Hospital of Guangdong Medical University from October, 2020 to August, 2022. The pathological types were confirmed by two professional pathologists. Tissue samples were subsequently stored at −80°C for later experiments. Written informed consent was acquired from all patients, and this study was approved by the Ethics Committee of Affiliated Hospital of Guangdong Medical University.
RNA extraction and qRT – PCR
Total RNA of tissues and cells was extracted with TRIzol Reagent (Invitrogen, USA) following the manufacturer’s instructions. Then, RNA was reverse transcribed into cDNA with a PrimeScriptTM RT Reagent Kit with gDNA Eraser (TaKaRa, Japan). TB Green Premix Ex TaqTM (TaKaRa, Japan) was utilized for qRT – PCR following the manufacturer’s protocol. All primers were designed and synthesized by Shanghai Sangon Biotech. GAPDH and U6 were utilized as internal references, and all primer sequences are shown in Supplementary Table S2. All assays were independently repeated three times.
Isolation of cytoplasmic and nuclear RNAs
Cytoplasmic and nuclear RNAs were isolated from BCa cells using a Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Canada) according to the manufacturer’s protocol. qRT – PCR was conducted to measure the abundance of circHGS.
Actinomycin D assays
For the actinomycin D assays, BCa cells were seeded into six-well plates in advance and then treated with actinomycin D (Sigma, USA) at a concentration of 2 μg/ml for 4 h, 8 h, and 12 h. The cells were collected at the corresponding time points, and the expression levels of circHGS and HGS mRNA were investigated by qRT – PCR. The assays were independently repeated three times.
Fluorescence in situ hybridization
Cy3-labeled circHGS and 18S probes were designed and synthesized by GenePharma (Suzhou, China). The subcellular localization of circHGS in BCa cells was identified by fluorescence in situ hybridization assays. After costaining nuclei with Hoechst 33,342, images were obtained with a confocal microscope (Leica, Germany). The assays were independently repeated three times.
Cell transfection
A total of 2 × 105 T24 cells or 1.5 × 105 UM-UC-3 cells were seeded into six-well plates. The next day, according to the manufacturer’s protocol, circHGS siRNA, miR-513a-5p mimics and the corresponding control (GenePharma, China) were transfected into BCa cells with LipofectamineTM RNAiMAX Reagent (Invitrogen, USA). The circHGS overexpression plasmid using the GV486 vector (Genechem, China) and the corresponding control were transfected into BCa cells with X-treme GENE HP DNA (Roche, Switzerland). qRT – PCR was performed to quantify the transfection efficiency.
Lentivirus infection
Based on the circHGS gene sequences, three RNAi target sequences used to knock down circHGS expression and a negative control sequence were designed (Genechem, China). According to the manufacturer’s instructions, we used packaged lentivirus with GV493 as a vector to infect BCa cells and selected cells twice with puromycin at 3 μg/ml for 48 h. Cells were divided into control, LV-circRNA-RNAi-1, LV-circRNA-RNAi-2 and LV-circRNA-RNAi-3 groups. Then, qRT – PCR was performed to measure the infection efficiency. The assays were independently repeated three times.
CCK8 assay
BCa cells were treated with circHGS siRNA or circHGS overexpression plasmid. Twenty-four hours later, the cells were seeded into 96-well plates. CCK8 (Dojindo, Japan) assays were performed to detect cell proliferation ability at 0, 1, 2, 3, 4, and 5 d, and the absorbance was detected at 450 nm.
Flow cytometry assay for cell cycle
BCa cells were harvested and adjusted cell number to 1 × 106. Then, BCa cells were fixed with cold 70% ethanol for 1 hour at room temperature and washed with cold PBS twice. Next, BCa cells were resuspended with 500 μl PI/RNase staining solution (Sungene biotech, China) and incubated for 30 minutes at room temperature, protected from light. The cell cycle result was analyzed by flow cytometry and all assays were independently repeated three times.
Colony formation assay
For the colony formation assay, transfected BCa cells were seeded into six-well plates (500 cells per well) and grown in the incubator for 1–2 weeks. Then, the cells were treated with 4% paraformaldehyde and crystal violet staining solution, and stained cell colonies were counted. All assays were independently repeated three times.
Wound-healing assay
BCa cells were seeded into six-well plates one day in advance. When the cells grew to 90% of the area of the six-well plate, a 200 μl pipette tip was utilized to make wounds in the center of the six-well plate, and the cells were cultivated using serum-free medium. Cell migration pictures were taken with a cell imaging system (EVOS XL Core, Invitrogen) at 0, 12, 24 h, 36 h and the cell migration rates were calculated. The assays were independently repeated three times.
Transwell migration and invasion assay
For the Transwell migration assays, 8 × 104 T24 cells or UM-UC-3 cells suspended in 200 μl serum-free medium were added to the upper chamber, and 600 μl medium containing 20% serum was added to the lower chamber of a 24-well plate. After culturing for 18 h for T24 cells and 24 h for UM-UC-3 cells in the incubator, the cells were fixed with 4% paraformaldehyde and stained with crystal violet staining solution. Then, 3 fields of view were randomly selected to be photographed and counted. For the Transwell invasion assays, the upper chambers containing Matrigel (Corning, USA) were used for invasion assays, and the remaining experimental steps were the same as the migration assay. All assays were independently repeated three times.
Bioinformatics analysis
The online databases CircInteractome, miRanda and RNAhybrid were used to predict miRNAs that bind to circHGS. TargetScan, micro-T, PITA, miRmap and miRNANet databases were used to filter the potential target genes of miR-513a-5p.
Dual-luciferase reporter assay
The wild-type sequences of circHGS (circHGS-wt) and VEGFC 3’-UTR (VEGFC-wt) capable of binding to miR-513a-5p were inserted into GP-miRGLO plasmids. Then, the corresponding binding sequences were mutated (circHGS-mut and VEGFC-mut) and inserted into the GP-miRGLO plasmids to construct dual-luciferase reporter plasmids (GenePharma, China). Then, miR-513a-5p mimics and plasmids were cotransfected into BCa cells. Twenty-four hours later, firefly luciferase and Renilla luciferase were detected by using a Dual-Lumi™ Luciferase Assay Kit (Beyotime, China), and the assays were independently repeated three times.
Western blot
Proteins were obtained from BCa cells by using a mixture of RIPA lysis buffer (Beyotime, China) and PMSF protease inhibitor (Solarbio, China). Then, 5×loading buffer (Beyotime, China) was added, and the proteins were heated and denatured. The protein samples were separated on SDS – PAGE gels (Beyotime, China) and transferred to PVDF membranes (Bio – Rad, USA). Then, the PVDF membranes were incubated with the corresponding primary antibody overnight at 4°C and chemically exposed the next day. The primary antibodies used were as follows: Cyclin D1 (26939-1-AP, Proteintech), CDK4 (11026-1-AP, Proteintech), CDK6 (14052-1AP, Proteintech), VEGFC (67116-1-lg, Proteintech), phospho-mTOR (Ser2448) (67778-1-lg, Proteintech), mTOR (66888-1-lg, Proteintech), phospho-AKT (Ser473) (66444-1-lg, Proteintech), AKT (60203-2-lg, Proteintech), GAPDH (10494-1-AP, Proteintech), and β-actin (66009-1-lg, Proteintech). All assays were independently repeated three times.
Animal experiment
Four-week-old female BALB/c nude mice were randomly divided into a control group and a LV-circHGS-RNAi-2 group (5 per group). Lentivirus-infected UM-UC-3 cells were injected subcutaneously into the right backs of nude mice (8 million cells per nude mouse). After the tumors grew, the longest and widest diameters of the tumors were detected every week. The tumor volumes were calculated as (length × width2)/2. After 4 weeks, the nude mice were sacrificed, and tumors were excised and weighed. The animal experiment was conducted in accordance with the guidelines of the Laboratory Animal Ethics Committee of Guangdong Medical University.
Statistical analysis
The experimental data are shown as the mean ± standard deviation (SD). Student’s t test was performed to compare differences between groups. A chi-square test was conducted to assess the relationship between circHGS expression and clinicopathological features. The experimental data were analyzed by SPSS 21.0 software, and GraphPad Prism 8.0.1 software was utilized for statistical graphs. A P value <0.05 was considered statistically significant.
Results
Identification and characteristics of circHGS
To clarify the relationship between circRNAs and BCa, we analyzed the circRNA expression profiles of three pairs of BCa tissues and matched normal bladder tissues from the GEO database (GSE97239). In BCa samples, we identified 5 upregulated circRNAs that are derived from exons and available in the circBase database ( and Supplementary Table S1). Among these differentially expressed circRNAs, qRT – PCR results showed that the expression of circHGS in 30 pairs of BCa tissues was significantly higher than that in matched nontumor tissues (). Moreover, the levels of circHGS in BCa cells were elevated compared to human immortalized normal urothelium cells SV-HUC-1 (). In addition, circHGS expression was positively correlated with tumor grade and pathological T stage, but not with gender, age, tumor size, and lymph node metastasis (). According to the circBase database, circHGS (circBase ID: hsa_circ_0004721) was generated from the head-to-tail splicing of exons 10, 11 and 12 of HGS (). To examine the stability of circHGS, we collected total RNA from BCa cells after actinomycin D treatment at 4 different time points. The results showed that the half-life of circHGS exceeds 12 h, and the corresponding HGS mRNA is only 4 hours (), indicating that circHGS has better stability than HGS mRNA. Furthermore, nucleocytoplasmic separation experiments and fluorescence in situ hybridization assays showed that circHGS mainly exists in the cytoplasm of BCa cells ().
Figure 1. Identification of circHGS. (a) Five upregulated circRnas were screened from the GEO database (GSE97239) in circRNA expression profiles of three pairs of BCa tissues and paired adjacent normal bladder tissues. (b) the expression of circHGS in 30 BCa tissues and paired normal tissues. (c) the expression of circHGS was elevated in BCa cells compared with SV-HUC-1 cell. (d) CircHGS is generated by back-splicing of exons 10, 11 and 12 of HGS. (e) qRT – PCR was performed to measure relative circHGS and HGS mRNA levels at 4 different time points in BCa cells treated with actinomycin D. (f) qRT – PCR was used to detect the expression of circHGS in the cytoplasm and nucleus of BCa cells; GAPDH and U6 were used as internal references. (g) FISH experiments were conducted to confirm the subcellular localization of circHGS in T24 and UM-UC-3 cells, and nuclei were stained with Hoechst 33,342. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01.
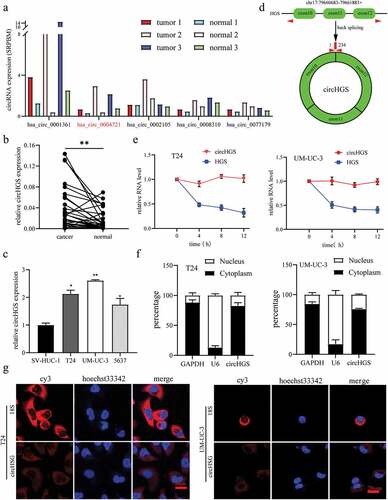
Table 1. Relationship between circHGS expression and clinical characteristics in BCa patients.
Knockdown of circHGS inhibited BCa cell proliferation and cell cycle progression, but circHGS overexpression showed the opposite results
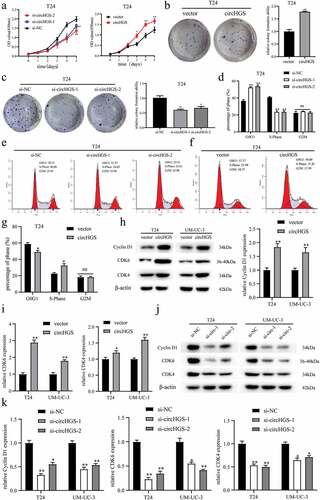
To evaluate the biological roles of circHGS in BCa, two circHGS siRNA and circHGS overexpression plasmids were designed and transfected into UM-UC-3 and T24 cells to establish circHGS-knockdown and circHGS-overexpressing BCa cells. Then, the transfection efficiencies (Supplementary Figure S1A and S1B) and the expression of HGS mRNA were measured, and the data demonstrated that knockdown or overexpression of circHGS did not affect HGS mRNA levels in BCa cells (Supplementary Figure S1C). Next, CCK8 assays (, Supplementary Figure S1D) and colony formation assays (), Supplementary Figure S1E and S1F) were performed to estimate the proliferation ability of BCa cells. The results showed that circHGS knockdown obviously suppressed the proliferation abilities of BCa cells. Conversely, overexpression of circHGS obviously enhanced the proliferative capacity of T24 and UM-UC-3 cells. Flow cytometry assay was performed to detect the effects of circHGS on BCa cell cycle. The results showed that knockdown of circHGS induced cell cycle arrest at G0/G1 phase and decreased the proportion of S phase in T24 and UM-UC-3 cells. However, circHGS overexpression induced cell cycle progression from G0/G1 phase to S phase (), Supplementary Figure S1G and S1H). In addition, western blot showed that circHGS overexpression increased the expression of Cyclin D1, CDK6 and CDK4, and knockdown of circHGS inhibited the expression of Cyclin D1, CDK6 and CDK4 (), suggesting that knockdown of circHGS may arrest cell cycle progression by downregulating the expression of cyclin D1, CDK6 and CDK4, thereby inhibiting BCa cell proliferation.
Figure 2. The effects of circHGS on BCa cell proliferation and cell cycle. (a–c) the proliferation ability of T24 cells treated with siRNA targeting circHGS or circHGS overexpression plasmid was measured by CCK8 assays and colony formation assays. (d–g) Cell cycle progression was detected by flow cytometry assay in T24 cells treated with siRNA targeting circHGS or circHGS overexpression plasmid. (h-k) Western blot assays were performed to detect the expression of Cyclin D1, CDK6 and CDK4 in BCa cells.
Knockdown of circHGS inhibited BCa cell migration and invasion, but circHGS overexpression showed the opposite results
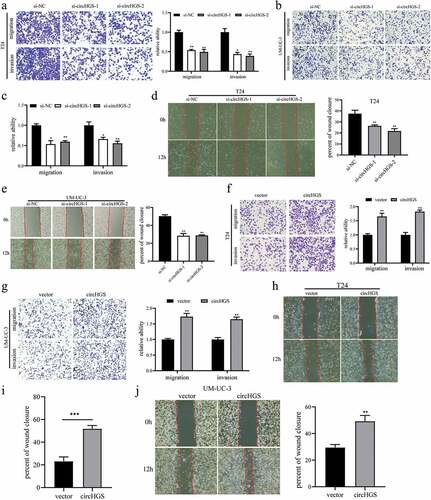
To further explore the roles of circHGS on BCa cell migration and invasion, Transwell assays and wound-healing assays were performed and the results showed that silencing circHGS markedly inhibited the migration and invasion of BCa cells ()). In contrast, circHGS overexpression showed a promoting effect in BCa cells ()).
Figure 3. CircHGS enhances the migration and invasion abilities of BCa cells. (a-c) the migration and invasion abilities of BCa cells transfected with siRNA targeting circHGS were detected by Transwell assay. (d-e) Wound-healing assays were applied to detect the migration ability of BCa cells treated with siRNA targeting circHGS. (f-g) the migration and invasion abilities of BCa cells transfected with circHGS overexpression plasmid were detected by Transwell assay. (h-j) Wound-healing assays were applied to detect the migration ability of BCa cells treated with circHGS overexpression plasmid. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01, ***P < 0.001.
CircHGS acts as a sponge for miR-513a-5p in BCa cells
Our data demonstrated that circHGS functions as an oncogene in BCa cells, however, the underlying mechanism is still unclear. Numerous studies have shown that circRNAs in the cytoplasm can serve as sponges for miRNAs [Citation9,Citation16,Citation19,Citation20]. Our previous results proved that circHGS mainly exists in the cytoplasm. Therefore, bioinformatics was performed to identify miRNAs that could bind to circHGS. Three miRNAs (miR-513a-5p, miR-1184 and miR-1205) were predicted as potential downstream genes of circHGS by using the CircInteractome, miRanda and RNAhybrid databases (). The binding sites of circHGS and these three miRNAs are shown in .
Figure 4. CircHGS served as a sponge for miR-513a-5p. (a) RNAhybrid, miRanda and CircInteractome databases were used to predict potential miRnas that could bind to circHGS. (b) Three potential miRnas were predicted by bioinformatics. (c) the binding sites of circHGS and miR-513a-5p, miR-1184, or miR-1205. (d) Dual-luciferase reporter gene assays were conducted to determine the interactions between circHGS and miR-513a-5p, miR-1184 or miR-1205 in T24 and UM-UC-3 cells. (e) the binding sites between circHGS-wt and miR-513a-5p. (f) Dual-luciferase reporter gene assays were used to detect the interactions between miR-513a-5p and circHGS-wt or circHGS-mut in BCa cells. (g) Stable knockdown efficiencies of circHGS in T24 and UM-UC-3 cells. (h-i) the relative expression levels of 3 miRnas in stably low-expressing circHGS T24 and UM-UC-3 cells were measured by qRT – PCR. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01.
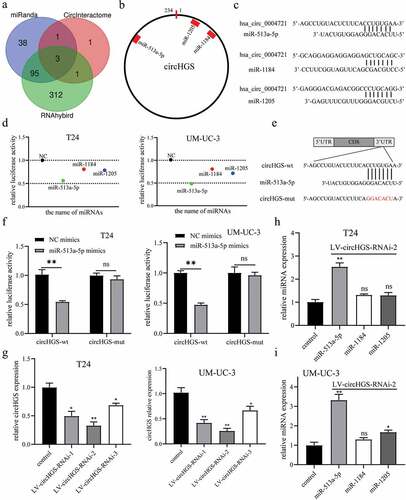
To determine which miRNA can interact with circHGS in BCa cells, GP-miRGLO plasmids containing the circHGS wild-type sequence (circHGS-wt) and the corresponding circHGS mutant sequence (circHGS-mut) were constructed for dual-luciferase reporter gene assays. Then, circHGS-wt plasmids were cotransfected with three candidate miRNAs into BCa cells. The results showed that the dual-luciferase activity decreased most significantly after miR-513a-5p mimics cotransfection with circHGS-wt plasmids in BCa cells (). Nevertheless, the luciferase activity was not changed significantly after NC mimics or miR-513a-5p mimics cotransfection with circHGS-mut plasmids in BCa cells (. To further identify downstream miRNAs regulated by circHGS, stable circHGS-knockdown T24 and UM-UC-3 cells were constructed by lentivirus, and the infection efficiencies were detected (). qRT – PCR results showed that only miR-513a-5p levels were obviously elevated when circHGS was effectively knocked down in BCa cells (). Therefore, we chose miR-513a-5p for subsequent study. In brief, circHGS inhibited miR-513a-5p expression and served as a sponge for miR-513a-5p in BCa.
MiR-513a-5p suppressed BCa cell proliferation, migration and invasion by targeting VEGFC
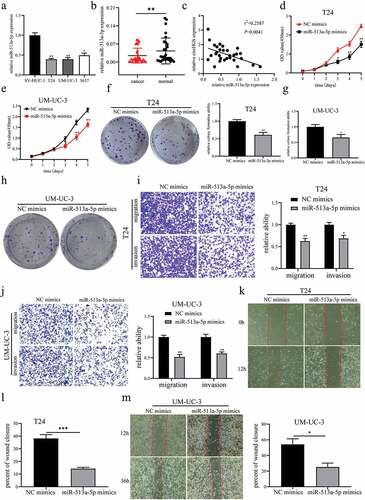
It has been reported that miR-513a-5p is an antitumor gene in BCa [Citation21]. Our results showed that the levels of miR-513a-5p were downregulated in BCa tissues and cells compared to paired normal tissues and SV-HUC-1 cells (). The correlation analysis showed that the expression of miR-513a-5p was negatively related with circHGS expression in BCa (). Moreover, compared to the control, the abilities of proliferation, migration and invasion were significantly suppressed when BCa cells were transfected with miR-513a-5p mimics ()).
Figure 5. MiR-513a-5p suppressed BCa cell proliferation, migration and invasion. (a-b) the expression of miR-513a-5p was downregulated in BCa tissues and cells. (c) Correlation of circHGS and miR-513a-5p relative expression in 30 BCa tissues. (d-h) CCK8 assays and colony formation assays were performed to detect the proliferation abilities of T24 and UM-UC-3 cells treated with miR-513a-5p mimics. (i-j) Transwell assays were used to detect the effects of miR-513a-5p overexpression on the migration and invasion of BCa cells. (k-m) Wound-healing assays were applied to detect the migration abilities of BCa cells treated with miR-513a-5p mimics. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01, ***P < 0.001.
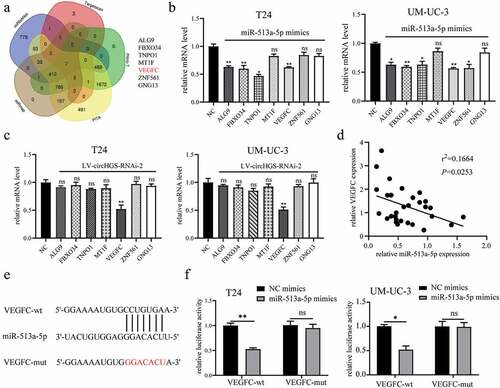
To identify the target gene regulated by circHGS/miR-513a-5p in BCa cells, the miRNANet, TargetScan, miRmap, micro-T and PITA databases were applied to predict potential target genes of miR-513a-5p, and 7 possible target genes were ultimately selected (). Numerous studies have shown that miRNAs can interact with the 3’-UTRs of target genes to inhibit their expression [Citation9]. Our data revealed that only VEGFC was regulated by both miR-513a-5p and circHGS at the mRNA level in BCa cells (). Additionally, overexpression of miR-513a-5p and circHGS knockdown markedly inhibited the expression of VEGFC at the protein level (). Therefore, VEGFC was selected as the downstream target gene of circHGS/miR-513a-5p. Then, qRT – PCR was performed to detect the expression of VEGFC in 30 BCa tissues and correlation analysis showed that the expression of VEGFC was negatively correlated with miR-513a-5p expression (). According to the TargetScan database, as shown in , miR-513a-5p could bind to the VEGFC 3’-UTR, and the binding sites between miR-513a-5p and VEGFC were consistent with those of miR-513a-5p and circHGS. Furthermore, the luciferase reporter gene assays showed that the luciferase activity of a reporter gene plasmid containing the VEGFC-wt sequence was significantly weakened by the presence of miR-513a-5p mimics in BCa cells (). The above results indicated that miR-513a-5p suppressed BCa cell proliferation, migration and invasion by targeting VEGFC.
Figure 6. VEGFC was a target gene of miR-513a-5p. (a) Bioinformatics analysis was performed to predict potential target genes of miR-513a-5p. (b) the effects of miR-513a-5p on predicted target gene expression levels in BCa cells. (c) the effects of circHGS knockdown on predicted target gene expression levels. (d) Correlation of VEGFC and miR-513a-5p relative expression in 30 BCa tissues. (e) the binding sites of miR-513a-5p and VEGFC-wt. (f) Dual-luciferase reporter gene assays were performed to determine the interactions between miR-513a-5p and VEGFC-wt or VEGFC-mut in T24 and UM-UC-3 cells. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01.
MiR-513a-5p reversed the oncogenic roles of circHGS in BCa cells
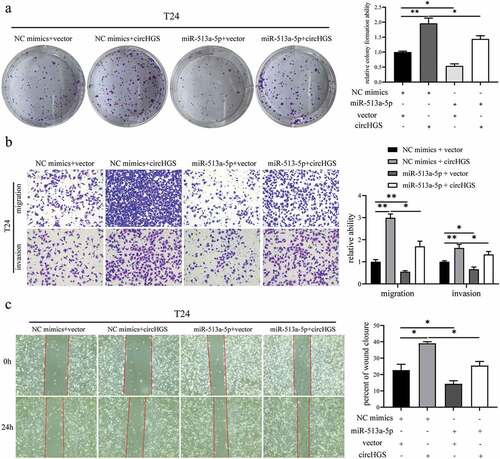
To determine whether circHGS plays an oncogenic role by acting as a ceRNA for miR-513a-5p, rescue experiments were applied in BCa cells. Then, NC mimics or miR-513a-5p mimics and vector or circHGS overexpression plasmids were cotransfected into BCa cells. Colony formation assays showed that the proliferative abilities induced by circHGS overexpression could be partly weakened by miR-513a-5p mimics in BCa cells (, Supplementary Figure S2A). The same rescue result was also obtained in BCa cell Transwell invasion assays (, Supplementary Figure S2B). Furthermore, as shown by Transwell migration assays and wound healing assays, miR-513a-5p mimics obviously eliminated the increased cell migration ability caused by circHGS overexpression in BCa cells (, Supplementary Figure S2C). In conclusion, miR-513a-5p can abrogate the tumor-promoting effects induced by circHGS overexpression in BCa cells.
Figure 7. MiR-513a-5p reversed the oncogenic roles of circHGS in vitro. (a) a colony formation assay indicated that cell proliferation ability of T24 cells transfected with circHGS overexpression plasmid was abolished after cotransfection with miR-513a-5p mimics. (b) Transwell migration and Matrigel invasion assays showed that cell migration and invasion capabilities of T24 cells transfected with circHGS overexpression plasmid were abrogated after cotransfection with miR-513a-5p mimics. (c) Wound-healing assays demonstrated that cell migration ability of T24 cells transfected with circHGS overexpression plasmid was counteracted after cotransfection with miR-513a-5p mimics. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01.
CircHGS upregulated VEGFC expression and activated the AKT/mTOR signaling pathway by sponging miR-513a-5p
To investigate the impacts of circHGS on the miR-513a-5p/VEGFC axis in BCa cells, western blot experiments and rescue assays were conducted. Western blot results indicated that both circHGS knockdown and miR-513a-5p overexpression inhibited the expression of VEGFC, whereas circHGS overexpression displayed a promoting effect in BCa cells ()). Overexpression of circHGS enhanced VEGFC expression, but miR-513a-5p mimics partly eliminated the upregulation, showing that circHGS served as a ceRNA for miR-513a-5p to enhance the expression of VEGFC in BCa cells ()).
Figure 8. The effects of circHGS and miR-513a-5p on VEGFC and the AKT/mTOR signaling pathway. (a) the effects of silencing circHGS on VEGFC and the AKT/mTOR signaling pathway in T24 and UM-UC-3 cells. (b) the effects of miR-513a-5p overexpression on the target gene VEGFC and the AKT/mTOR signaling pathway in BCa cells. (c) the effects of circHGS overexpression on VEGFC and the AKT/mTOR signaling pathway in T24 and UM-UC-3 cells. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01, ***P < 0.001.
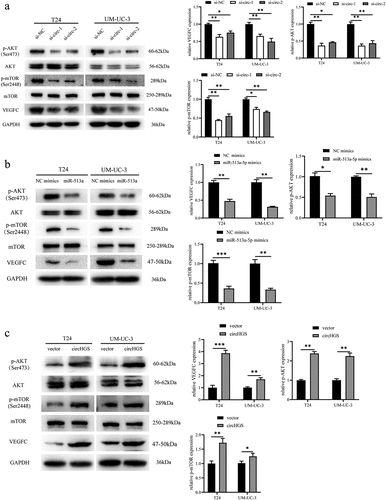
Figure 9. CircHGS upregulated VEGFC expression and activated the AKT/mTOR signaling pathway by sponging miR-513a-5p. (a-d) CircHGS overexpression upregulated VEGFC expression and activated the AKT/mTOR signaling pathway, effects that were abolished by miR-513a-5p overexpression in T24 and UM-UC-3 cells. Data are shown as the means ± SD of three independent experiments, * P < 0.05, **P < 0.01.
The protein kinase B and mammalian target of rapamycin (AKT/mTOR) signaling pathway is a critical signaling pathway in human tumors and is involved in cancer cell proliferation, invasion, and migration [Citation22,Citation23]. TCGA indicated that the AKT/mTOR pathway was altered in 72% of BCa [Citation24]. Multiple studies have also demonstrated that the AKT/mTOR pathway is frequently activated in muscle-invasive and metastatic BCa [Citation25,Citation26]. VEGFC is a positive regulator of the AKT/mTOR pathway in BCa cells [Citation27]. Given that circHGS and miR-513a-5p can regulate VEGFC expression at the mRNA and protein levels in BCa cells, western blot was performed to further explore whether they can also regulate the AKT/mTOR pathway. The results showed that both circHGS knockdown and miR-513a-5p overexpression suppressed the activation of the phosphorylated AKT/mTOR signaling pathway (). However, circHGS overexpression promoted the expression of phosphorylated AKT/mTOR protein, but the AKT/mTOR protein levels were not changed (). Moreover, rescue experiments showed that circHGS overexpression enhanced the expression of phosphorylated AKT/mTOR, while the promoting effects induced by circHGS could be attenuated by the presence of miR-513a-5p mimics in BCa cells ()). These results indicated that circHGS enhanced VEGFC expression and activated the AKT/mTOR signaling pathway by sponging miR-513a-5p.
Knockdown of circHGS inhibited BCa tumorigenesis
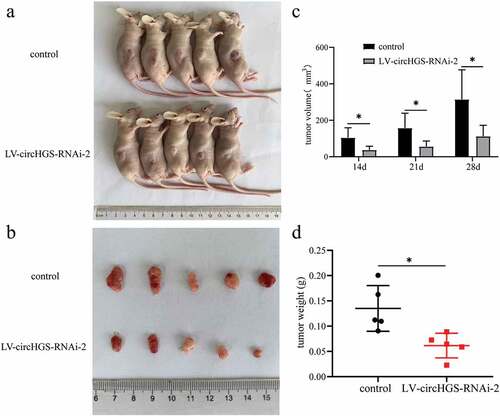
To study the biological roles of circHGS in vivo, we constructed stable low circHGS-expressing UM-UC-3 cells by using lentivirus (LV-circHGS-RNAi-2 group). Then, the control cells and LV-circHGS-RNAi-2 group cells were injected subcutaneously into the right backs of BALB/c nude mice (). Subcutaneous tumors were measured for the longest and widest diameters every week when the tumor was visible. Four weeks later, the nude mice were sacrificed. The results showed that the weights and volumes of tumors in the LV-circHGS-RNAi-2 group were markedly reduced compared to those in the control group (), indicating that knockdown of circHGS obviously suppressed tumor growth in vivo.
Figure 10. Knockdown of circHGS inhibited tumor growth in vivo. (a) Subcutaneous tumors in BALB/c nude mice. (b) Tumor tissues were excised from BALB/c nude mice in the control and LV-circHGS-RNAi-2 groups. (c) Tumor volumes of xenograft tumors at different time points. (d) Compared with the control group, the tumor weight was significantly reduced in the LV-circHGS-RNAi-2 group. Data are shown as the means ± SD of three independent experiments, * P < 0.05.
Discussion
With the development of high-throughput technology, abundant abnormally expressed circRNAs have been found to play crucial roles in the progression of human cancers. In contrast to previous linear mRNAs, circRNAs are special noncoding RNAs and exhibit closed circular structures [Citation28]. CircRNAs are conserved, abundant and stable [Citation29]. Due to the lack of 5’caps and 3’ poly (A) tail structures, circRNAs are more resistant to degradation by RNase R [Citation12]. Since circRNAs are stably present in the cytoplasm, they have great potential as disease biomarkers [Citation30,Citation31]. Mechanistically, circRNAs play biological roles by serving as miRNA sponges, interacting with RNA binding proteins, participating in protein coding and regulating transcription [Citation32]. As oncogenes or tumor suppressors, circRNAs were discovered to be associated with BCa proliferation, migration and invasion [Citation9]. For instance, hsa_circ_0014130 promotes BCa cell proliferation, migration and invasion by serving as a ceRNA for miR-132-3p [Citation15]. CircNUDT21 upregulates MDM2 expression by acting as a miR-16-1-3p sponge to promote BCa proliferation and invasion [Citation17].
In our study, we investigated the circRNA expression profiles of human BCa tissues and normal bladder tissues from the GEO database (GSE97239) and first identified a novel circRNA, circHGS. CircHGS is formed by back-splicing of exons 10, 11, and 12 of HGS. We found that circHGS was elevated in BCa tissues and cell lines. Moreover, the expression of circHGS was positively correlated with tumor grade and pathological T stage. In vitro experiments indicated that knockdown of circHGS might arrest cell cycle progression at G0/G1 phase by downregulating the expression of Cyclin D1, CDK6 and CDK4. In addition, silencing circHGS inhibited BCa cell proliferation, migration and invasion, whereas overexpression of circHGS showed an enhancing effect. Moreover, knockdown of circHGS markedly suppressed tumor growth in vivo. Given that circHGS mainly exists in the cytoplasm, we speculated that circHGS might play biological roles by serving as a miRNA sponge. Then, bioinformatics analysis was conducted and miR-513a-5p was predicted to be the potential downstream miRNA of circHGS. The dual-luciferase reporter gene assays indicated that the luciferase activity was obviously decreased after cotransfection of miR-513a-5p mimics with circHGS-wt plasmid, suggesting that circHGS binds to miR-513a-5p. Moreover, knockdown of circHGS significantly upregulated the level of miR-513a-5p in T24 and UM-UC-3 cells. Rescue experiments also showed that overexpression of miR-513a-5p partly eliminated the oncogenic roles of circHGS in BCa cells, demonstrating that circHGS serves as a sponge of miR-513a-5p in BCa cells.
MicroRNAs (miRNAs) are a class of endogenous small RNAs of approximately 20–23 nucleotides in length [Citation33] that play important roles in regulating tumor cell proliferation, migration and invasion [Citation34]. MiR-513a-5p was reported to play antitumor roles in gastric cancer, liver cancer, renal cell carcinoma and BCa [Citation21,Citation35–38]. Then, qRT – PCR was used to detect miR-513a-5p expression in BCa. The results showed that the expression of miR-513a-5p was decreased in BCa and was inversely correlated with circHGS expression. Subsequently, functional experiments indicated that overexpression of miR-513a-5p obviously inhibited BCa cell proliferation, migration and invasion.
It has been reported that miRNAs mainly bind to the 3’ UTRs of target genes, leading to translation inhibition or the induction of target gene degradation [Citation39,Citation40]. To investigate the target genes of the circHSG/miR-513a-5p axis, bioinformatics analysis was performed, and VEGFC was ultimately identified as the target gene. Western blot results indicated that both circHGS knockdown and overexpression of miR-513a-5p inhibited the expression of VEGFC at the protein level, while overexpression of circHGS promoted the expression of VEGFC in BCa cells. Moreover, circHGS overexpression partly eliminated the inhibitory effects of miR-513a-5p on VEGFC protein in BCa cells. Numerous studies have shown that VEGFC is an oncogene in BCa and can regulate the AKT/mTOR signaling pathway [Citation27,Citation41,Citation42]. As shown by western blot, both circHGS knockdown and miR-513a-5p overexpression inhibited the expression of phosphorylated AKT/mTOR proteins, and miR-513a-5p mimics partially reversed the promoting impacts of circHGS overexpression on phosphorylated AKT/mTOR proteins. In summary, these results showed that circHGS promotes BCa cell proliferation, migration and invasion via the miR-513a-5p/VEGFC/AKT/mTOR signaling pathway ().
Conclusion
In summary, we first identified the upregulated circular RNA HGS in BCa and determined that circHGS enhances BCa cell proliferation, migration and invasion. Mechanistically, circHGS promotes BCa progression by serving as a sponge for miR-513a-5p and activating the VEGFC/AKT/mTOR signaling pathway. Importantly, our study may provide a novel target for BCa treatment.
Author contributions
HWL designed the study. YZ, LZ and HX performed the in vitro experiments. YZ drafted the manuscript. YZ and ZL carried out the statistical analyses. HWL and ZL revised the manuscript. YZ, HX and RQC performed the animal experiments. SHL, HX and RQC collected clinical samples. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the EthicsCommittee of Affiliated Hospital of Guangdong Medical University.Informed consent was obtained from all patients involved in thestudy. All animal care and experiments were performed according to the guidelines of the Laboratory Animal Ethics Committee of GuangdongMedical University.
Supplemental Material
Download Zip (12.4 KB)Acknowledgment
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2164454
Data availability statement
The data analyzed in this study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Lenis AT, Lec PM, Chamie K, et al. Bladder cancer: a review. JAMA. 2020;324(19):1980–1991.
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108.
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249.
- Tran L, Xiao JF, Agarwal N, et al. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21(2):104–121.
- Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388(10061):2796–2810.
- Witjes JA, Bruins HM, Cathomas R, et al. European Association of urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur Urol. 2021;79(1):82–104.
- Wu Z, Liu J, Dai R, et al. Current status and future perspectives of immunotherapy in bladder cancer treatment. Sci China Life Sci. 2021;64(4):512–533.
- Cambier S, Sylvester RJ, Collette L, et al. EORTC nomograms and risk groups for predicting recurrence, progression, and disease-specific and overall survival in non–muscle-invasive stage Ta–T1 urothelial bladder cancer patients treated with 1–3 years of maintenance bacillus calmette-guérin. Eur Urol. 2016;69(1):60–69.
- Yang X, Ye T, Liu H, et al. Expression profiles, biological functions and clinical significance of circRnas in bladder cancer. Mol Cancer. 2021;20(1):4.
- Li M, Liu Y, Zhang X, et al. Transcriptomic analysis of high-throughput sequencing about circRNA, lncRNA and mRNA in bladder cancer. Gene. 2018;677:189–197.
- Li C, Fu X, He H, et al. The biogenesis, functions, and roles of circrnas in bladder cancer. Cancer Manag Res. 2020;12:3673–3689.
- Yang Q, Li F, He AT, et al. Circular RNAs: expression, localization, and therapeutic potentials. Mol Ther. 2021;29(5):1683–1702.
- Huang S, Yang B, Chen BJ, et al. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109(5–6):401–407.
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRnas. Molecular Cell. 2017;66(1):9–21.e7.
- Li G, Guo BY, Wang HD, et al. CircRNA hsa_circ_0014130 function as a miR-132-3p sponge for playing oncogenic roles in bladder cancer via upregulating KCNJ12 expression. Cell Biol Toxicol. 2021;9. DOI:10.2139/ssrn.3822281.
- Zhu J, Luo Y, Zhao Y, et al. circEHBP1 promotes lymphangiogenesis and lymphatic metastasis of bladder cancer via miR-130a-3p/TGFβR1/VEGF-D signaling. Mol Ther. 2021;29(5):1838–1852.
- Chen L, Li W, Li Z, et al. circNUDT21 promotes bladder cancer progression by modulating the miR-16-1-3p/mdm2/p53 axis. Mol Ther Nucleic Acids. 2021;26:625–636.
- Qiu F, Liu Q, Xia Y, et al. Circ_0000658 knockdown inhibits epithelial-mesenchymal transition in bladder cancer via miR-498-induced HMGA2 downregulation. J Exp Clin Cancer Res. 2022;41(1):22.
- Su Y, Feng W, Shi J, et al. circRIP2 accelerates bladder cancer progression via miR-1305/tgf-β2/smad3 pathway. Mol Cancer. 2020;19(1):23.
- Jin M, Lu S, Wu Y, et al. Hsa_circ_0001944 promotes the growth and metastasis in bladder cancer cells by acting as a competitive endogenous RNA for miR-548. J Exp Clin Cancer Res. 2020;39(1):186.
- Xia W, Chen C, Zhang MR, et al. LncRNA PCAT6 aggravates the progression of bladder cancer cells by targeting miR-513a-5p. Eur Rev Med Pharmacol Sci. 2020;24(19):9908–9914.
- Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–132.
- FT AM. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. 2017;407:153–189.
- NR SA. Targeting the PI3K/AKT/mTOR pathway in bladder cancer. Methods Mol Biol. 2018;1655:335–350.
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322.
- Iyer G, Al-Ahmadie H, Schultz N, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31(25):3133–3140.
- Wang Y, Xing QF, Liu XQ, et al. MiR-122 targets VEGFC in bladder cancer to inhibit tumor growth and angiogenesis. Am J Transl Res. 2016;8(7):3056–3066.
- Patop IL, Wust S, Kadener S. Past, present, and future of circRnas. EMBO J. 2019;38(16):e100836.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157.
- Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36(1):152.
- Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274.
- Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49–57.
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36.
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRnas: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465.
- Lu MD, Liu D, Li YX. LINC01436 promotes the progression of gastric cancer via regulating miR-513a-5p/ape1 axis. Onco Targets Ther. 2020;13:10607–10619.
- Xu W, Li K, Song C, et al. Knockdown of lncRNA LINC01234 suppresses the tumorigenesis of liver cancer via sponging miR-513a-5p. Front Oncol. 2020;10:571565.
- Li J, Huang C, Zou Y, et al. Circular RNA MYLK promotes tumour growth and metastasis via modulating miR-513a-5p/vegfc signalling in renal cell carcinoma. J Cell Mol Med. 2020;24(12):6609–6621.
- Zhu K, Miao C, Tian Y, et al. lncRNA MIR4435-2HG promoted clear cell renal cell carcinoma malignant progression via miR-513a-5p/klf6 axis. J Cell Mol Med. 2020;24(17):10013–10026.
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207.
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–433.
- Chen C, Luo Y, He W, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404–421.
- Ping Q, Shi Y, Yang M, et al. LncRNA DANCR regulates lymphatic metastasis of bladder cancer via the miR-335/VEGF-C axis. Transl Androl Urol. 2021;10(4):1743–1753.