ABSTRACT
In the recent decades, human genome engineering has been one of the major interesting research subjects, essentially because it raises new possibilities for personalized medicine and biotechnologies. With the development of engineered nucleases such as the Zinc Finger Nucleases (ZFNs), the Transcription activator-like effector nucleases (TALENs) and more recently the Clustered Regularly Interspaced short Palindromic Repeats (CRISPR), the field of human genome edition has evolved very rapidly. Every new genetic tool is broadening the scope of applications on human tissues, even before we can completely master each of these tools. In this review, we will present the recent advances regarding human genome edition tools, we will discuss the numerous implications they have in research and medicine, and we will mention the limits and concerns about such technologies
DISCOVERING THE SYSTEM
The concept of genetic engineering has been studied over the latest few decades. In 1974, scientists produced the first transgenic mouse from viral DNA,Citation1 followed by the production of the first knock-outs a few years later.Citation2,3 Before this, Friedmann et al. proposed the idea of modifying the human genome to treat diseases in 1972.Citation4
In 1990, this concept was proven on human when Ashanti DeSilva, a patient with a severe immunodeficiency (SCID), showed a temporary response to retroviral gene therapy.Citation5 At that time, gene therapy was drawing a lot of attention from industries, investors, medical doctors, and researchers. In 2000, a French team directed by Alain Fischer provided the first evidence that gene therapy could completely cure life-threatening genetic diseases,Citation6 raising attention for gene therapy to its peak. Unfortunately, one of the boy treated started to develop a leukemia-like condition, resulting in a suspension of gene therapy trials in France, USA, Germany, Japan and Italy.Citation7 The field had been severely affected by this event, urging scientists to create more sophisticated and secure tools to modify the genome.
In the 1990s, gene therapy utilized a "copy and paste" strategy (genome addition); in the beginning of the 21st century, 3 different tools emerged, allowing a precise "cut and paste" of the human genome (genome editing). Today, genome edition is a very active area of research, due to the advent of engineered nucleases. By combining engineered nucleases with the new generation of whole genome sequencing (WGS) technology, we believe that the future à la carte medicine is within reach, providing the ability to modify cells, tissues, and organs with high precision. This review will focus on the advancements of human genome editing, its potential applications in research and medicine, the problems it raises, and current limitations of this technology.
UPGRADING THE GENETIC TOOLBOX
Engineered nucleases have changed the approach we use to edit genetic information. It is now possible to target specific genetic changes in a selected locus, to either insert or edit DNA. It is the same principle for all engineered nucleases currently available. An engineered virus containing the custom designed nuclease and the edited nucleotide DNA sequence are transfected into human cells. Once inside the cell, the engineered nuclease cleaves a specific site of chromosomal DNA to induce a double stranded break (DSB). Using the cell's DNA repair machinery, the original nucleotide sequence will be substituted by the newly edited sequence of interest. This causes permanent genome modification.
Zinc-finger nucleases
The oldestCitation8 and the best described human genome edition tools are Zinc-finger nucleases (ZFNs). They are custom dimeric nucleases designed by the fusion ofCitation1 a highly specific DNA-binding domain andCitation2 a DNA-cleavage domain. Three to 6 Zinc Finger repeats compose this binding domain and each of them can recognize 3 nucleotides. Thus, the final DNA-binding domain is able to recognize sequences ranging from 9 to 18 nucleotides. In addition, the DNA-cleavage domain is constituted of a FokI endonuclease that will induce a double stranded break in the DNA once the custom Zinc Finger motif recognizes and binds to a target sequence. To work efficiently, Fok1 endonuclease needs to dimerize. Eventually, when both zinc finger nucleases bind to their recognition sites, DNA will be cut. This will trigger endogenous DNA repair systems and induce a desired genome modification.Citation9
Transcription activator-like effectors
Although each zinc finger recognizes 3 nucleotides, not every nucleotide triplet has a specified zinc finger. The possibility to cover any sequence of the genome is limited. Thus, it may be difficult to create custom zinc finger nucleases for some specific sequences. To overcome these limitations, Transcription activator-like effectors (TALENs) were createdCitation10 a few years later and were chosen as the Method of the Year 2011 by Nature.Citation11 The main mechanism remains the same as zinc finger nucleases, by taking advantage of a highly specific custom nuclease. However, as opposed to ZFNs, TALENs' DNA-binding domain is composed of repeats of about 34 TAL effectors. Each of these effectors is able to recognize a single nucleotide, theoretically covering any nucleotides sequences.Citation12 Once TALENs binds to the target sequence, the Fok1 endonuclease will induce a cut in the DNA, triggering repair mechanisms of the cell. The sequence of interest is then substituted to the original sequence.
Clustered regularly interspaced short palindromic repeats
ZFNs and TALENs have greatly helped scientists to shape the future of genome editing, but the main obstacle remains their fabrication. By definition, they are custom nucleases, and protein engineering is difficult and time consuming. Ultimately, the price of creating engineered nucleases is very high. Recently, researchers have put a lot of effort and attention into a new technology called Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs). CRISPR is initially an immune mechanism found in bacteria and archea, capable of destroying foreign DNA.Citation13 This system uses a 20-nucleotide guided-RNA in order to recognize one sequence of interest. Then, a Cas9 endonuclease induces a double stranded break in this precise location.Citation14 The essential difference is the use of RNA instead of protein to recognize a nucleotide sequence, resulting in much easier and faster manipulations. Moreover, as opposed to previous methods, CRIPSR can target many genes at once (multiplex), paving the way to more complicated and elegant genome editing.
CRISPR presents numerous advantages; it is very efficient, multiplex, easy to make, and generally inexpensive for bigger productions. Thus, CRISPR represents a brand new editing technology for loss-of-function screening of the whole genome. However, one of the major limitations is that it has a lower specificity than the previous tools. This results in higher chances of editing unintended DNA regions, leading to modifications of the genome at unknown locations (off targets effects). This is a critical aspect when researchers want to translate gene therapy research into the clinics.
PLAYING WITH THE HUMAN GENETIC SOFTWARE FOR RESEARCH
To Create human isogenic cell iines
Engineered molecular scissors have been used for decades in research. ZFNs,Citation15 TALENs,Citation16 and more recently, CRISPRsCitation17 have greatly facilitated the creation of mutant animals for research. They can also be used to directly study human tissues by creating human isogenic cell lines. In the past few years, the study of human diseases has greatly benefited from the input of induced pluripotent stem cells (iPSc), taken directly from patients. However, because people possess a unique genome, different genetic backgrounds may trigger variable phenotypes in vitro. This complicates the interpretation of disease mechanisms. Thus, correcting the genetic mutation of a patient, in order to provide an isogenic control human iPS cell is of great interest. ZFNsCitation18 have been used to correct mutations associated to Parkinson disease in a human iPS cell line and to accurately study the phenotype. Conversely, the same strategy can be applied to create mutant lines from healthy iPS cells. A group generated 15 mutant lines linked to various diseases (dyslipidemia, insulin resistance, hypoglycemia, lipodystrophy, motor neuron death, and hepatitis C infection) with their healthy isogenic controls.Citation19 The creation of such cell lines enables scientists to study pathogenesis after differentiation from iPS cells, to comprehend precise molecular biology mechanisms, such as the regulation of transcription, gene promoter, cell signaling, and eventually drug screening on different genetic backgrounds ().
FIGURE 1. The use of gene editing tools in research. The ability to edit the human genome can be used in research to manufacture human isogenic cell lines,Citation1 to model human cancerCitation2 to make large-scale genetic screeningCitation3 and to create a new generation of humanized animal models.Citation4 Human isogenic cell lines can represent a powerful tool to study with more accuracy human diseases (1a) human cell and molecular biology (1b) and perform drug screening on controlled genetic backgrounds (1c). Engineered nucleases can also be used to model human cancer, mimicking and correcting translocations induced by human cancer.Citation3 The new CRISPR technology can be applied for large-scale genetic screening to help answering unsolved questions linked to development and organogenesis (3a) and eventually be applied and used in tissue engineering technology (3b). Humanized mice models can facilitate the study of human pathology, human immune function, cancer, cell therapy, drug screening and infectious diseases.
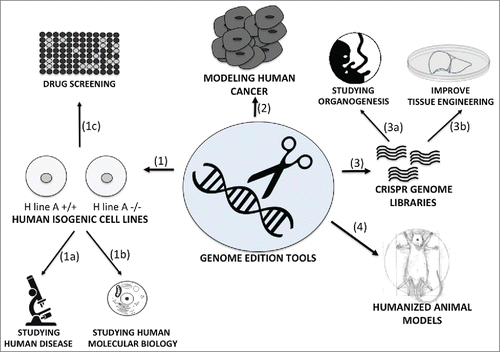
To model cancer
Genome-editing technologies can be powerful tools to characterize cancer mechanisms and provide potential treatments. They can easily cause mutations - frameshifts, deletions, and inversion - to all mechanisms linked to cancer. For example, they can be used to induce or correct chromosomal translocations that are present in numerous cancers, leading to gene fusion acting as oncogenes. Recently, a team has induced the same translocations found in Ewing sarcoma and anaplastic large cell lymphoma (ALCL) in human cell lines. Most importantly, this team was able to correct translocations of a patient cell line with ALCL, providing new concepts to cure certain types of cancersCitation20 ().
To study organogenesis
During the past few decades, RNAi-based technologies have been used for whole genome screening in order to identify the molecular network controlling development, the assembly and maturation of organs.Citation21-23 This has been a precious tool to identify important developmental genes but we still ignore many key players that drive organ development. This is illustrated by our imperfect differentiation protocols for many lineages in vitro, as well as the current hurdles in organogenesis. However, CRISPR is now been used for large-scale genetic screening in human cellsCitation24-27 and could help solve developmental questions. Indeed, as opposed to RNAi-based screening, CRISPR genome scale lentiviral single guide RNA (sgRNA) libraries can generate a complete loss-of-function, preserved in time through differentiation, but also target intergenic regions (non-transcribed sequences such as promoters, enhancers, etc.). This has the potential to revolutionize the approach to organogenesis, by identifying essential actors that could not be isolated with previous whole genome screening technologies ().
Moreover, combined with the very innovative field of tissue engineering, this has the potential to drastically transform the way we study diseases, by focusing more on human tissues. Recently, a group used iPS cells from a patient with polycystic kidney disease (PKD) to build kidney organoids, providing proof of concept that organogenesis could facilitate the study of kidney related diseases.Citation28 Another group used CRISPR to correct the cystic fibrosis transmembrane conductor receptor (CFTR) locus in stem cells from patients with cystic fibrosis.Citation29 The disease phenotype was corrected when these stem cells were expended in stable epithelial 3D-organoids, emphasizing the advantages of combining such genome editing technologies with engineered organogenesis (). Accordingly, introducing mutations in healthy human iPS cells through genome editing technologies to study pathogenesis in 3D would be very valuable. Human genome editing organ modeling could complement animal mutant models when they cannot mimic human disease or simply when researchers reach the limits of traditional animal studies.
To create animal models with human organ systems
The generation of chimeric (human/animal) models has become a valuable tool in research to study human diseases, in order to overcome limitations associated with traditional animal models. Two strategies currently prevail: injecting human cells in an animal model with impairments of specific molecular pathways (repopulated humanized model) or inserting human genes in the DNA of an animal model (genetically humanized model). Both cases involve genetic modifications of the original animal model ().
The best examples of repopulation humanized models involve mice with human haematopoietic cellsCitation30 or human liver.Citation31 For instance, the most common system to grow a humanized liver involves immunodeficient mice (Rag2–/–/Il2rg –/–) with a deficient fumarylacetoacetate hydrolase enzyme (Fah-/-). These mice are unable to metabolize 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC). The hepatocytes die when exposed to this drug and will eventually be replaced by injected human hepatocytes.Citation31 However, because these models require immunodeficiency, numerous diseases cannot be studied accurately unless they also have a human haematopoietic system.Citation32 Another repopulation example consisted of injecting human iPS cells in a mouse blastocyst with a knock out for Pdx1 gene to grow a human pancreas in an animal.Citation33 This technique called blastocyst complementation provided a proof of principle that it is possible to grow organs in vivo. However, growing complete human organs requires the impairment of several developmental pathways (of every cell type composing each organ). In general, all of these repopulation techniques are limited because they require very sophisticated genetic alterations. Repopulated humanized model approaches could greatly benefit from CRISPR multiplex technology in the years to come.
Numerous genetically humanized models have been described and are currently used in research to study the human immune function, cancer, cell therapy, drug screening and infectious diseases.Citation34 The possibility to target specific and multiple loci within DNA could help to manufacture and optimize next generation of genetically humanized animal models. Some studies already took advantage of CRISPR technology to easily create immunodeficient mice modelsCitation35 and it is now possible to envision sophisticated animal models carrying a humanized drug metabolism or immune system.
USE THE CUT AND PASTE TECHNOLOGY FOR GENOME THERAPY
Genomic editing also brings tremendous opportunities for gene therapies. This field is monitored very closely and is gaining a lot of interest again due to its recent advances and clinical trials. We now envision the possibility to cure genetic and non-genetic diseases by editing cells and tissues ex vivo and maybe one day to edit organs directly in vivo.
To cure genetic diseases
Curing monogenic diseases has already been the focus of gene therapy trials in the 1990s. Similarly, a group worked on immunodeficiency (X-SCID) and used ZFNs to correct the mutation on human primary T cells.Citation36 A few years later, an IL2RG transgene was inserted with ZFNs into CD34+ cells derived from human bone marrow of X-SCID patients. These cells were used for xenotransplantation inside SCID mice, resulting in long-term human haematopoietic chimerism by immune-competent human haematopoietic progenitor cells.Citation37
There are also several examples of iPS cell corrections from patients. For instance, using a combination of ZFNs and piggyBac technology, a European group was able to achieve a bi-allelic correction of a point mutation that causes α-1 antitrypsin deficiency (A1ATD). After differentiation in iPS-derived hepatocytes, these cells were able to express and restore the function of A1AT.Citation38 Another study utilized ZFNs for the correction of α-thalassemiaCitation39 and chronic granulomatous disease.Citation40 Similarly, TALENs and piggyBac were used to correct a point mutation in patient derived iPS cells with Sickle Cell disease.Citation41 Thus, numerous studies using iPS cells proved that we could successfully correct monogenic disease phenotypes in vitro. Certainly, iPS-derived gene editing cells will lead to interesting gene therapy trials in the near future.
Some researchers have tried to use gene-editing technologies directly in vivo by transducing a virus containing the gene of interest and the engineered nuclease. Researchers were able to restore homeostasis in a neonatal mouse model of hemophilia after in vivo genome editing using ZFNs, providing an evidence of the principle that ZFNs works in vivo.Citation42 However, one major hurdle in adult cells is that, unless the targeted cells have a growth advantage, the efficiency will be limited, thus resulting in an insufficient expression of protein. To avoid this, the same team has proposed to use a "safe harbor," a locus with a high transcriptional activity, providing an access to high expression of proteins. Using the Albumin locus in the liver, they achieved to rescue the phenotype of an adult mouse model of hemophilia by injecting the engineered adeno-associated viral vector (AAV) in the tail-vein.Citation43 This work provides a potential platform for secreted protein production by editing organs directly in vivo using engineered nuclease technologies.
To cure non-genetic diseases
It might not be the first application one considers when mentioning gene therapy, but genomic modifications can also be applied to non-genetic diseases. A clinical trial for HIV using genome edition has started and already completed several stepsCitation44 (NCT01044654; NCT00842634). The idea for this therapy arose a few years ago, when an HIV patient with an acute myeloid leukemia received a stem cell transplant from a donor with a CCR5 deletion, an essential co-receptor for HIV infection. This resulted in the clearance of HIV in this patient, referred as the "Berlin patient".Citation45 Here, the strategy is similar, targeting the genetic elimination of CCR5 in CD4+ T cell in HIV patients using ZFNs.Citation46 Another clinical trial focused on the treatment of glioblastoma. Upon removal of the glucocorticoid receptor gene with ZFNs and addition of glucocorticoids, CD8+ T cells have shown the ability to target and destroy glioblastoma tumor cells (NCT01082926).
There are also promising example of in vivo genome editing to cure non-genetic diseases. A South African team has disrupted the HBV viral genome at 4 different locations using TALENs. They tested the efficiency of their system in vitro on HepG2 cells and in vivo in a murine injection model of HBV replication.Citation47 Despite remaining problems, the ability to disrupt viral DNA using genome-editing technology would greatly improve viral treatments in the future. Another group created several VEGF isoforms expressed from the endogenous gene by zinc finger transcription factors. They injected these adeno-ZFP vectors into the quadriceps muscle of CD-1 mice. It induced a substantially greater VEGF-A stimulated improved angiogenesis and wound healing.Citation48 Such strategies have been considered to treat diabetic neuropathy and amyotrophic lateral sclerosis. With a continuous improvement of these techniques, we begin to foresee the use of genetic edition to modulate diseased organs directly in vivo.
WAITING FOR THE NEXT RELEASE
The recent advances in gene editing technology have the potential to revolutionize the field of biological research and medicine. The immediate impact of CRISPR technology in research is enormous, introducing an easy way to amend the human genome. This will bring elegant modifications of human cell lines (knockins and knockouts) to study molecular biology, organogenesis, and pathogenesis and to create a new generation of humanized animal models. It will also provide a new method for whole genome screening on human cells. Despite the amount of sequenced data that we have gathered throughout the years, there are still many mysteries to unveil, such as how certain DNA sequences function inside the cell. Thus, such screening tools should unravel numerous well-kept mysteries of the human genome. Surely, this could also be applied for high throughput gene disruptions in drug discovery.
From a clinical perspective, genome-editing technologies will serve as a base for personalized medicine in the future. However, there are still several hurdles to overcome before using these technologies on human cells, tissues, and organs. The off target effect mentioned earlier remain one of the major obstacles of this technology. Due to the necessity to edit the genome ex vivo to facilitate the screening and monitor off target effects, haematopoietic stem cells are easily targeted cells for therapy. In the future, we will need to improve our genetic tools in order to eliminate any off target effects and to improve the gene-edition efficiency to modify an entire organ directly in vivo.
Despite these attractive applications, the possibilities that such technology offer is unlimited. Providing turnkey solutions for genome editing may lead to uncontrolled creations of modified organisms, plants, fungi, and pathogens. Researchers have reported the creation of a modified virus that mice would inhale, allowing CRISPR system to directly induce mutations to create a model for lung cancer.Citation49 The potential of creating such tools on human is already a concern for scientists.Citation50 Others already see a risk for gene doping in athletics.Citation51 Moreover, ethical concerns are still a major issue. After a study published in April 2015 by a Chinese teamCitation52 that used CRISPR to modify human embryos, the fear of "playing god" surfaced once more. Thus, a debate concerning the proper and ethical use of such technologies was brought up in and beyond the scientific community.Citation53 Though great caution must be taken with this technology, it certainly has a tremendous potential to bring a new renaissance in numerous fields, from genetic research to biotechnologies and medicine.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- Jaenisch RM, B. Simian Virus 40 DNA Sequences in DNA of Healthy Adult Mice Derived from Preimplantation Blastocysts Injected with Viral DNA. Proc Natl Acad Sci 1974; 71(4):1250-4; PMID:4364530; http://dx.doi.org/10.1073/pnas.71.4.1250
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 1985; 317:230-4; PMID:2995814; http://dx.doi.org/10.1038/317230a0
- Robertson E, Bradley A, Kuehn M, Martin Evans. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature 1986; 323:445-8; PMID:3762693; http://dx.doi.org/10.1038/323445a0
- Friedmann T, Roblin R. Gene therapy for human genetic disease? Science 1972; 174(4025):949-55; http://dx.doi.org/10.1126/science.175.4025.949
- Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, Clerici M, Shearer G, Chang L, Chiang Y, Tolstoshev P, et al. T lymphocyte-directed gene therapy for ADA- SCID: initial trial results after 4 years. Science 1995; 20(270):475-80; http://dx.doi.org/10.1126/science.270.5235.475
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, et al. Gene Therapy of Human Severe Combined Immunodeficiency (SCID)–X1 Disease. Science 2000; 288:699-72; http://dx.doi.org/10.1126/science.288.5466.669
- Check E. A tragic setback. Nature 2002; 420:116-8
- Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 1985; 4(6):1609-14; PMID:4040853
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 2010; 11(9):636-46; PMID:20717154; http://dx.doi.org/10.1038/nrg2842
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009; 326:1509-12; PMID:19933107; http://dx.doi.org/10.1126/science.1178811
- Baker M. Method of the Year 2012. Nature Methods 2012; 9(1):1-1; http://dx.doi.org/10.1038/nmeth.1852
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 2013; 14(1):49-55; PMID:23169466; http://dx.doi.org/10.1038/nrm3486
- Ishino Y S H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 1987; 169(12):5429-33; PMID:3316184
- Barrangou R. Cas9 Targeting and the CRISPR Revolution. Science 2014; 344:707-8; PMID:24833384; http://dx.doi.org/10.1126/science.1252964
- Meyer M, Ortiz O, Hrabe de Angelis M, Wurst W, Kuhn R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc Natl Acad Sci U S A 2012; 109(24):9354-9; PMID:22660928; http://dx.doi.org/10.1073/pnas.1121203109
- Wefers B, Meyer M, Ortiz O, Hrabé de Angelis M, Hansen J, Wurst W, Kühn R. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. PNAS 2013; 110(10):3782-7; PMID:23426636; http://dx.doi.org/10.1073/pnas.1218721110
- Fujii W, Kawasaki K, Sugiura K, Naito K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res 2013; 41(20):e187; PMID:23997119; http://dx.doi.org/10.1093/nar/gkt772
- Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 2011; 146(2):318-31; PMID:21757228; http://dx.doi.org/10.1016/j.cell.2011.06.019
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 2013; 12(2):238-51; PMID:23246482; http://dx.doi.org/10.1016/j.stem.2012.11.011
- Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, René O, Katibah GE, Zhang L, Holmes MC, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res 2013; 23(7):1182-93; PMID:23568838; http://dx.doi.org/10.1101/gr.147314.112
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 2004; 428:431-7; PMID:15042092; http://dx.doi.org/10.1038/nature02371
- Jiang D, Zhao L, Clapham DE. Genome-Wide RNAi Screen Identifies Letm1 as a Mitochondrial Ca2+/H+ Antiporter. Science 2009; 326(5949):144-7; PMID:19797662; http://dx.doi.org/10.1126/science.1175145
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. Haploid Genetic Screens in Human Cells Identify Host Factors Used by Pathogens. Science 2009; 326:1231-5; PMID:19965467; http://dx.doi.org/10.1126/science.1178955
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 2014; 509(7501):487-91; PMID:24717434; http://dx.doi.org/10.1038/nature13166
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 2014; 343:80-4; PMID:24336569; http://dx.doi.org/10.1126/science.1246981
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014; 343:84-7; PMID:24336571; http://dx.doi.org/10.1126/science.1247005
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 2014; 32(3):267-73; PMID:24535568; http://dx.doi.org/10.1038/nbt.2800
- Xia Y, Nivet E, Sancho-Martinez I, Gallegos T, Suzuki K, Okamura D, Wu MZ, Dubova I, Esteban CR, Montserrat N, et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat Cell Biol 2013; 15(12):1507-15; PMID:24240476; http://dx.doi.org/10.1038/ncb2872
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013; 13(6):653-8; PMID:24315439; http://dx.doi.org/10.1016/j.stem.2013.11.002
- Ito R, Takahashi T, Katano I, Ito M. Current advances in humanized mouse models. Cell Mol Immunol 2012; 9(3):208-14; PMID:22327211; http://dx.doi.org/10.1038/cmi.2012.2
- Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, Kay MA, Finegold M, Grompe M. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol 2007; 25(8):903-10; PMID:17664939; http://dx.doi.org/10.1038/nbt1326
- Bility MT, Cheng L, Zhang Z, Luan Y, Li F, Chi L et al. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog 2014; 10(3):e1004032; PMID:24651854; http://dx.doi.org/10.1371/journal.ppat.1004032
- Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee YS, Usui J, Knisely AS, et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 2010; 142(5):787-99; PMID:20813264; http://dx.doi.org/10.1016/j.cell.2010.07.039
- Brehm MA, Shultz LD, Greiner DL. Humanized mouse models to study human diseases. Curr Opin Endocrinol Diabetes Obes 2010; 17(2):120-5; PMID:20150806; http://dx.doi.org/10.1097/MED.0b013e328337282f
- Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, Zhang N, Zhu K, Xu J, Hu B, et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 2014; 46:49-55; PMID:24269190; http://dx.doi.org/10.1016/j.biocel.2013.10.010
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005; 435:646-51; PMID:15806097; http://dx.doi.org/10.1038/nature03556
- Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, Holmes MC, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature; 510:235-40; PMID:24870228; http://dx.doi.org/10.1038/nature13420
- Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordóñez A, Hannan NR, Rouhani FJ, et al. Targeted gene correction of α 1 -antitrypsin deficiency in induced pluripotent stem cells. Nature 2011; 478:391-4; PMID:21993621; http://dx.doi.org/10.1038/nature10424
- Chang CJ, Bouhassira EE. Zinc-finger nuclease-mediated correction of α-thalassemia in iPS cells. Gene therapy 2012; 120:3906-14
- Zou J S C, Chou BK, Choi U, Pan J, Wang H, Dowey SN, Cheng L, Malech HL. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells : functional correction by zinc finger nuclease – mediated safe harbor targeting. Blood 2011; 117(21):5561-72; PMID:21411759; http://dx.doi.org/10.1182/blood-2010-12-328161
- Sun N, Zhao H. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng 2014; 111(5):1048-53; PMID:23928856; http://dx.doi.org/10.1002/bit.25018
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. In vivo genome editing restores hemostasis in a mouse model of hemophilia. Nature 2010; 475:217-21; http://dx.doi.org/10.1038/nature10177
- Sharma R, Anguela XM, Doyon Y, Wechsler T, DeKelver RC, Sproul S, Paschon DE, Miller JC, Davidson RJ, Shivak D, et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 2015; 126(15):1777-84; PMID:26297739; http://dx.doi.org/10.1182/blood-2014-12-615492
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 2014; 370(10):901-10; PMID:24597865; http://dx.doi.org/10.1056/NEJMoa1300662
- Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, Schneider T, Hofmann J, Kücherer C, Blau O, et al. Long-Term Control of HIV by CCR5 Delta32/ Delta32 Stem-Cell Transplantation. N Engl J Med 2009; 370(7):392-8
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 2008; 26:808-16; PMID:18587387; http://dx.doi.org/10.1038/nbt1410
- Bloom K, Ely A, Mussolino C, Cathomen T, Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol Ther 2013; 21(10):1889-97; PMID:23883864; http://dx.doi.org/10.1038/mt.2013.170
- Rebar EJ, Huang Y, Hickey R, Nath AK, Meoli D, Nath S, Chen B, Xu L, Liang Y, Jamieson AC, et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med 2002; 8(12):1427-32; PMID:12415262; http://dx.doi.org/10.1038/nm1202-795
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014; 516(7531):423-7; PMID:25337876; http://dx.doi.org/10.1038/nature13902
- Ledford H. CRISPR, the disruptor. Nature 2015; 522(7554):20-4; PMID:26040877; http://dx.doi.org/10.1038/522020a
- Gullans JEaS. Genetically enhanced Olympics are coming. Nature 2012; 487:297; PMID:22810679; http://dx.doi.org/10.1038/487297a
- Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015; 6(5):363-72; PMID:25894090; http://dx.doi.org/10.1007/s13238-015-0153-5
- Cyranoski D, Reardon S. Embryo editing sparks epic debate. Nature 2014; 520:593-4; http://dx.doi.org/10.1038/520593a
