ABSTRACT
As one of the most common forms of RNA modification, N6-methyladenosine (m6A) RNA modification has attracted increasing research interest in recent years. This reversible RNA modification added a new dimension to the post-transcriptional regulation of gene expression. In colorectal cancer (CRC), the role of m6A modification has been extensively studied, not only on mRNAs but also on non-coding RNAs (ncRNAs). In the present review, we depicted the role of m6A modification in CRC, systematically elaborate the interaction between m6A modification and regulatory ncRNAs in function and mechanism. Moreover, we discussed the potential applications in clinical.
1. Introduction
As the most abundant RNA modification in eukaryotic cells [Citation1], N6-methyadenosine (m6A) RNA modification played a vital role in cancer onset and development [Citation2], which has been well studied [Citation3]. m6A modification was first discovered in the 1970s [Citation4], related research has become a new hot spot in the epigenetic study since the new understanding of the reversibility and dynamic process of m6A modification in 2011 [Citation5]. With the rapid advancement of sequencing technology, m6 A modification was found not only on messenger RNAs (mRNAs) but also on non-coding RNAs (ncRNAs), especially regulatory ncRNAs [Citation6–10]. Until now, it has been shown that more than 7000 coding RNAs and 300 ncRNAs contain m6A sites [Citation3,Citation11]. ncRNAs were a group of endogenous RNA molecules that were not translated into proteins but played an important role in regulating gene expression and disease progress [Citation12,Citation13]. ncRNAs could be divided into two categories, housekeeping ncRNAs and regulatory ncRNAs. Regulatory ncRNAs were usually considered as key regulatory RNA molecules in the cancer process, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) [Citation13,Citation14].
Colorectal cancer (CRC) was one of the most common malignant tumours, caused a vast medical burden with its increasing mortality (the second leading cause of cancer-related death) [Citation15–17]. The occurrence and metastasis of CRC were considered a multi-factor, multi-step process, in which aberrant regulation of m6A modification was widely involved [Citation18–20]. While previous studies have reported dysregulated m6A modification in CRC tumorigenesis [Citation18–20], few reviews focused on the association between m6A modification and regulatory ncRNAs in CRC. To better understand the roles of m6A modification in CRC, the present study systematically reviewed the up-to-date research progress of m6A modification and its interaction with regulatory ncRNAs in CRC.
2. Regulation of m6A modification
m6A sites had a typical consensus sequence ‘RRACH’ (R = G or A; H = A, C, or U), and were enriched in 3ʹ untranslated regions (UTRs) and around stop codons in mRNAs or near the last exon in ncRNAs [Citation3,Citation11]. The effect of m6A modification was determined by m6A regulatory proteins, which consisted of m6A methyltransferases ‘writers’, m6A demethylases ‘erasers’, and m6A-binding proteins ‘readers’ [Citation21].
m6A modification was installed by a methyltransferase complex (MTC), consisting of Methyltransferase-like 3 (METTL3), Methyltransferase-like 14 (METTL14), and Vir-like m6A methyltransferase-associated (KIAA1429/VIRMA), RNA-binding motif protein 15/15B (RBM15/15B), zinc finger CCCH domain-containing protein 13 (ZC3H13) and other wirters [Citation22]. Among them, METTL3 acted as the primary catalytic subunit bound to the methyl donor S-adenosylmethionine (SAM) and catalysed methyl group transfer, and METTL14 was required for stabilizing METTL3 conformation and substrate RNA binding [Citation23–25]. In addition, it was reported that Methyltransferase-like 16 (METTL16) was a novel m6A methyltransferase to control cellular SAM level and installed m6A marks onto the U6 small nuclear RNA [Citation7,Citation26].
As shown in (), RNA m6A modification could be reversibly removed, and the demethylation process required erasers, a-ketoglutaric acid (a-KG) and molecular oxygen (O2) as cosubstrates and ferrous iron (Fe2+) as a cofactor [Citation27]. Alpha-ketoglutarate dependent dioxygenase (FTO) was the first demethylase of m6A modification [Citation5], catalysed the oxidative demethylation of m6A on mRNA. alkB homolog 5 (ALKBH5) one of the two identified m6A demethylases, showed a preference for target transcripts that had a consensus sequence [Citation28]. And alkB homolog 3 (ALKBH3) was a recently discovered demethylase that preferred mitochondrial transfer RNAs (tRNAs) [Citation29].
Figure 1. Reversible m6A modification on Eukaryotic RNAs. a. reversible m6A modification; b. the function of m6A modification
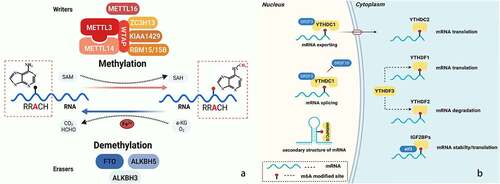
m6A modifications exerted different biological functions by recruiting specific reader proteins, including the YTH domain family (YTHDF1–3, YTHDC1, and YTHDC2), IGF2BPs (IGF2BP1/2/3), Heterogeneous nuclear ribonucleoprotein C/G (HNRNPC/G), Heterogeneous nuclear ribonucleoprotein A2B1 (HNRNPA2B1), and other proteins. These readers played different roles in the nucleus and cytoplasm (). In addition, it was reported that METTL3 could also serve as an m6A reader to promote the translation of target RNAs [Citation30].
3. m6A modification on mRNAs in CRC
Maintaining proper m6A modification levels on mRNAs was essential for normal bioprocesses and development, dysregulation of m6A modification was usually associated with cancers [Citation2,Citation3,Citation31–33]. Herein, we sorted out related articles in CRC and classified them into different parts by m6A regulators.
3.1. Aberrant m6A Writers
As the earliest identified m6A methyltransferase [Citation34], METTL3 had been deeply studied in recent years. In CRC, previous articles reported that METTL3 was highly expressed [Citation35–37] and acted as an oncogene. Li et al. [Citation31] found that METTL3 was highly expressed in metastatic CRC and was associated with a poor prognosis. Mechanistically, METTL3 led to methylation of SRY-box transcription factor 2 (SOX2) transcripts, which were subsequently recognized by IGF2BP2 to prevent the degradation of SOX2 mRNA. Xiang et al. [Citation38] demonstrated that METTL3 was upregulated in CRC and promoted cancer proliferation, with MYC being identified as a target downstream that could be enhanced by IGF2BP1. Zhou et al [Citation39] indicated that METTL3 epigenetically repressed Yippee-like 5 (YPEL5) in an m6A-YTHDF2-dependent manner by targeting the m6A site in the coding sequence region of the YPEL5 transcript. Liu and Sun et al. [Citation40] found that Sec62 homolog, preprotein translocation factor (Sec62) upregulated by the METTL3-mediated m6A modification promotes the stemness and chemoresistance of CRC by binding to β-catenin and enhancing Wnt signalling. Furthermore, the oncogenic function of METTL3-mediated m6A modification was reported in other related articles [Citation41–47], as shown in (). However, Deng et al. [Citation48] concluded that METTL3 might play a tumour-suppressive role in CRC cell proliferation, migration, and invasion. They demonstrated that METTL3 inhibited CRC progression by modulating the p38/ extracellular-signal-regulated kinase (ERK) pathway.
Table 1. The roles of m6A modification on coding RNAs in CRC
As another critical component of MTC, recent studies suggested that METTL14 played an inhibitory role in CRC progression. Chen and Xu et al. [Citation32] unveiled that decreased METTL14 could enhance the expression of SRY-related high-mobility-group box 4 (SOX4) through an m6A YTHDF2-dependent way, then promoted SOX4-mediated Epithelial-mesenchymal transition (EMT) process and PI3K /AKT signalling pathway. And Wang et al. [Citation49] demonstrated that METTL14 enhanced tumour suppressor Kruppel-like factor 4 (KLF4) mRNA stability in an m6A IGF2BP2-dependent way. In addition, other writers like WTAP, METTL16 were up-regulated in CRC [Citation50,Citation51].
3.2. Aberrant m6A Erasers
In CRC, the expression of FTO was increased [Citation51,Citation52]. Yue et al. [Citation53] indicated that FTO activated MYC by reducing the m6A modification of MYC. Zhang and Gao et al. [Citation52] raised that FTO activated the MZF1/c- Myc axis to promote CRC cell proliferation. FTO increased MZF1 expression by demethylating MZF1 mRNA to play its oncogenic role, which was mentioned in lung cancer [Citation54]. However, Liu et al [Citation55] found that reduced FTO protein expression was correlated with a high recurrence rate and poor prognosis in resectable CRC patients. Mechanistically, FTO exerted a tumour-suppressive role by inhibiting the expression of metastasis-associated protein 1 (MTA1) in an m6A-IGF2BP2 dependent way, Demethylation decreased its mRNA stability. In addition, ALKBH5 was down-expressed in CRC [Citation56,Citation57].
3.3. Aberrant m6A Readers
The YTH domain family were overexpressed and extensively involved in CRC progress. Bai and Yang et al. [Citation58] found that overexpressed YTHDF1 played a vital oncogenic role in CRC, and they proposed that YTHDF1 recognized and promoted the translation of m6A-modified FZD9 and Wnt6 mRNA. Nishizawa et al. [Citation59] discovered that YTHDF1 was overexpressed in CRC, which was transcriptionally regulated by MYC. YTHDF2, YTHDF3, and YTHDC2 et al. have also been mentioned [Citation37,Citation51,Citation57,Citation60–62] in CRC-related studies ().
IGF2BPs were newly reported m6A readers. In CRC, Xiang et al. [Citation38] indicated that MYC mRNA expression might be regulated in an m6A-IGF2BP1 dependent manner. IGF2BP2 and IGF2BP3 were reported to regulate target mRNAs stability in CRC cells [Citation31,Citation41,Citation45,Citation63].
4. m6A modification on regulatory ncRNAs in CRC
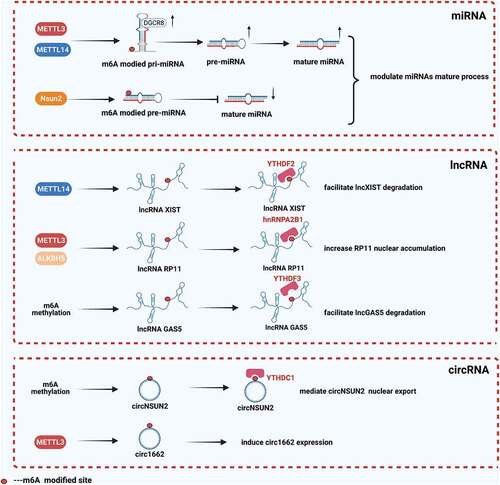
In addition to mRNAs, ncRNAs could also be regulated by m6A modification [Citation6–10]. An increasing number of studies have reported that m6A modification on ncRNAs was closely related to the occurrence and development of CRC (), the results were followed.
Table 2. The role of m6A modification on regulatory ncRNAs in CRC
4.1. m6A on miRNAs
miRNAs were short non-coding RNAs around 22 nucleotides, they could endogenously express and regulate mRNA post-transcriptional gene expression [Citation64]. Perious studies have verified that either METTL3 or METTL14 was required for the engagement of primiRNAs by DiGeorge Syndrome Crisis Area Gene 8 (DGCR8) [Citation65,Citation66]. In CRC, Peng et al. [Citation67] indicated that overexpression of METTL3 increased the level of miR-1246 by promoting the maturation of miR-1246 from pri-miR-1246, which further enhanced the metastatic capacity of CRC. The result of the m6A RNA immunoprecipitation (MeRIP) assay showed that m6A modification was enriched with pri-miR-1246 sequence, and the expression level of pri-miR-1246 was upregulated while miR-1246 level was decreased in METTL3-knockdown CRC cells. Similarly, Chen and Xu et al. [Citation68] found METTL14 was downregulated in CRC, which inhibited the pri-miR-126 processes in an m6A-dependent manner. They immunoprecipitated DGCR8 and detected pri-miR-375 bound to DGCR8, the result shown that the expression levels of pri-miR-375 bound to DGCR8 were significantly increased in METTL14-overexpressing CRC cells. Yang et al. [Citation69] revealed that NOP2/Sun RNA methyltransferase 2 (Nsun2) contributed to the down-regulation of miR-125b via the m6A-dependent way in CRC, interfering with the mature of miR-125b. Mature miR-125b came from pri-miR-125b1 and pri-miR-125b2, they revealed that the RNA methyltransferase NSun2 interfered with the mature processing of miR-125b by activating m6A modificaition on pre-miR-125b2, but not pri-miR-125b2.
4.2. m6A on lncRNAs
LncRNAs referred to those ncRNAs that are longer than 200 nucleotides [Citation70], previous studies have shown that m6A modification on lncRNAs was involved in the CRC progression [Citation70]. Zuo and Su et al. [Citation71] found that m6A levels of lncRNAs in CRC tissues were significantly up-regulated. Wu et al. [Citation72] revealed that m6A modification was involved in the upregulation of lncRNA RP11 via increasing its nuclear accumulation in CRC cells. The m6A RNA-immunoprecipitation (RIP) qPCR showed the m6A sites were enriched in RP11, and they found overexpression of METTL3 increased RP11 expression while ALKBH5 decreased RP11 expression. Moreover, they indicated that overexpressed METTL3 promoted the binding between HNRNPA2B1 and RP11. Yang et al. [Citation73] identified that METTL14 inhibited oncogenic lncRNA X inactivate-specific transcript (XIST) through an m6A dependent pathway. They found that m6A modification on XIST promoting YTHDF2- induced RNA degradation in CRC.
4.3. m6A on circRNAs
CircRNAs belonged to a new class of ncRNAs formed by covalently closed loops through back splicing, played important roles in various biological functions [Citation74]. The existence of m6A modification in circRNAs was confirmed by the interaction between circRNAs and YTHDF1/YTHDF2. Meanwhile, knockdown of METTL3 significantly affected m6A level on circRNAs [Citation75,Citation76]. Chen and Yuan et al. [Citation77] reported that METTL3-induced circ1662 promoted CRC cell invasion and migration. Mechanistically, METTL3 induced circ1662 expression by marking m6A sites in flanking reverse complementary sequences. Meanwihle, Chen et al. [Citation63] found that YTHDC1 could bind to circNSUN2 to facilitate its export from the nucleus to the cytoplasm in an m6A dependent manner, the latter was upregulated in CRC patients with liver metastasis (LM) and predicted poorer patient survival. In addition, Guo et al. [Citation78] found that m6A modification was involved in regulating the degradation of circ3823, which was highly expressed in CRC. While the mechanism of m6A modification on circRNAs was still vague, these studies proved that m6A modification existed in the ring structure of circRNAs, suggesting that m6A modification could be a regulator to affect circRNA.
5. Regulation of m6A modification by ncRNAs
5.1 ncRNAs regulated the process of m6A modification
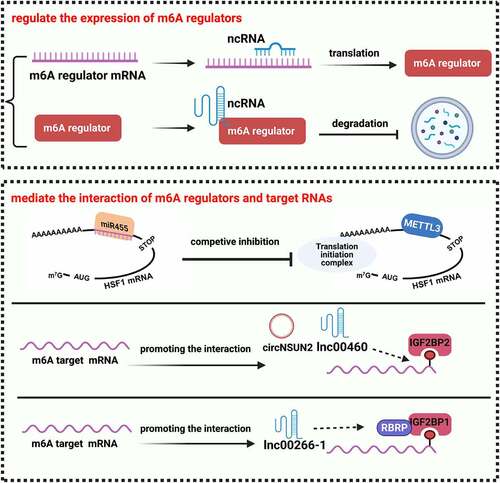
As explained above, m6A modification had regulatory effects on ncRNAs, including their mature, accumulation, transport, and degradation. Interestingly, regulatory ncRNAs mediated the process of m6A modification conversely ().
A previous study reported that m6A peaks were enriched at miRNA target sites on mRNAs, revealing a solid link between m6A modification and miRNAs. In CRC, Song and Feng et al. [Citation46] reported that miR455-3p inhibited m6A modification on heat shock transcription factor 1 (HSF1) mRNA, thus repressing HSF1 translation. The m6A site on the 3ʹUTR region of HSF1 was completely complementary to the seed sequence of miR455-3p, thus formed a competitive inhibition between miR455-3p and METTL3. Besides, lncRNAs could also regulate the interaction between m6A regulators and their target mRNAs. Zhu et al. [Citation79] discovered the lncRNA LINC00266-1 encoded a 71-amino acid peptide, which mainly interacted with the RNA-binding proteins (RBP) including IGF2BP1, thus named RNA binding regulatory peptide (RBRP). RBRP bound to IGF2BP1 and strengthened m6A recognition by IGF2BP1 to increase the mRNA stability and expression of c-Myc [Citation79]. Hou et al. [Citation45] demonstrated that LINC00460 enhanced interaction between IGF2BP2 and HMGA1 (High Mobility Group A1), thereby regulating mRNA stability and expression of HMGA1. Moreover, Chen et al. [Citation63] reported that YTHDF1-induced circNSUN2 promoted the interactions between IGF2BP2 and HMGA2, and enhanced the mRNA stability of HMGA2 through the formation of a circNSUN2/IGF2BP2/HMGA2 RNA−protein ternary complex.
5.2 ncRNAs regulated the expression of m6A regulators
m6A regulators themselves as protein-coding genes could be modified by methylation or acetylation or other transcriptional or post-transcription regulations, including the regulation of ncRNAs.
ncRNAs could regulate m6A modification by targeting the mRNAs of m6A regulators. In CRC, Yue et al. [Citation53] found that miR-96 mediated m6A modification by regulating AMP-activated alpha 2 (AMPKα2), which led to overexpression of FTO. And low expressed miR-1266 was reported to promote the occurrence and progression of CRC by directly targeting FTO [Citation80]. Wen et al. [Citation81] uncovered a negative feedback loop between lncRNA GAS5 and m6A reader YTHDF3: In normal, lncRNA GAS5 bound with Yes1 associated transcriptional regulator (YAP) to attenuate YAP-mediated transcription of YTHDF3; In tumour process, YTHDF3 could conversely bind with m6A-modified GAS5 to trigger its decay (Supplementary Figure S1). Moreover, Wang et al [Citation82] also found that lncRNA LINRIS bound to a site of IGF2BP2 and blocked its degradation through the ubiquitination-autophagic pathway, thereby maintaining the MYC-mediated glycolysis and the proliferation of CRC cells.
6. Therapeutic Implications of m6A modification
6.1. m6A modification as biomarkers and therapeutic targets
Given that dysregulation of m6A level and aberrant expression of m6A regulators were widely participated in the cancer progression, m6A modification and its regulatory proteins seemed to be potential biomarkers and therapeutic targets in CRC.
Most m6A related-genes were upregulated in CRC, while METTL14, YTHDF3, and ALKBH5 were downregulated [Citation37,Citation83]. Wang et al. [Citation50] discovered that the differentiation of CRC was closely relevant to m6A modification: high total m6A and high expression of METTL3, METTL16, WTAP were relevant to poor prognosis. Liu et al. [Citation83] found that the expression pattern of m6A regulators including METTL3, METTL14, METTL16, FTO and, ALKBH5 were associated with the clinical outcomes of CRC patients. Li et al. [Citation31] revealed that METTL3 was highly expressed in metastatic CRC tissues and associated with a poor prognosis. Low expression of METTL14 in CRC tissues associated with poor overall survival was revealed in Chen’s study [Citation32], and the discovery of the METTL14/SOX4 axis on CRC metastasis would aid in exploring efficient therapeutic target. And Bai et al. [Citation58] revealed that overexpressed YTHDF1 was associated with tumour depth and tumour size in CRC, silencing YTHDF1 could significantly inhibit the Wnt/β-catenin pathway activity in CRC cells, which provided a potential therapeutic target for CRC.
Drug resistance was responsible for treatment failure and/or cancer recurrence. Lan et al. [Citation84] discovered that the total m6A level and the METTL3 expression were increased in CRC tissues from Oxaliplatin (OX) -resistant patients, targeting METTL3-mediated m6A modification might be a promising adjuvant therapeutic strategy for OX-resistant CRC patients. Mohammad B. Uddin et al. [Citation36] found that silencing METTL3 with neplanocin A to suppressing m6A formation in p53 pre-mRNA could re-sensitize CRC cells to anticancer drugs. Moreover, YTHDF1 mediated cisplatin through the GLS1-glutamine metabolism axis was validated by an in vivo xenograft mouse model [Citation85], which revealed a novel sight contributing to overcoming chemo-resistant CRC.
Li et al. suggested [Citation56] that m6A erasers contributed to the efficacy of immunotherapy and identified that ALKBH5 regulated anti-PD-1 therapy response. Wang et al. [Citation35] uncovered that METTL3/14 regulated immune responses to anti-PD-1 therapy in CRC.
6.2. Potential application of the interaction between m6A modification and ncRNAs
The discovery of potent and selective inhibitors for m6A modification was urgent important in cancer treatment. Niu et al. [Citation86] found inhibitors targeting 2-oxoglutarate (2OG) and iron-dependent oxygenases, and DAA (3-deazaadenosine) was shown to block the introduction of m6A into mRNA substrates by inhibiting the hydrolysis of SAH [Citation87]. In addition, inhibitors of FTO including rhein, meclofenamic acid (MA), and MA2 have been mentioned in Acute myeloid leukaemia (AML) treatment [Citation88,Citation89].
However, the effect of these m6A inhibitors was not specific, and not targeted for particular m6A regulators or processes. It was speculated that the ncRNA regulatory mechanisms mentioned above could be used as breakthrough points in targeted therapy. Based on upstream regulation of m6A regulators by ncRNAs, we proposed a novel strategy for the CRC patients’ treatments: targeting unique ncRNAs to regulate their downstream m6A regulators and m6A processes (Supplementary Figure S2). For example, using small molecule that specifically acted on the consensus sequences of ncRNAs, the levels of m6A modification were changed, which affected the expression of downstream genes and thereby regulating the biological functions on CRC cells. And the clinical application of ncRNAs in cancer therapy has been widely studied, such as delivering ncRNA mimics or antisense oligonucleotide of ncRNAs and small molecular compounds [Citation90], which provided the possibility for the novel strategy.
However, effective therapeutic clinical application of ncRNAs and m6A modification has been faced lots of challenges, the specific mechanism of ncRNAs for use in targeted therapy needed to be further warranted.
7. Discussion
In the past few years, serval reviews about m6A modification in CRC have been published [Citation18–20]. In contrast, the present review especially focused on the interaction of m6A modification with regulatory ncRNAs in CRC, which included m6A modification on regulatory ncRNAs and the regulation of m6A modification by ncRNAs.
In CRC, the m6A modification on regulatory ncRNA regulated the maturation, transportation, stability, and degradation of the target ncRNAs (). While m6A modification on ncRNAs had the same reversible process, the function of m6A modification on ncRNAs mainly depended on types of its target ncRNAs: m6A modification on miRNAs mainly regulated the mature process of miRNAs, m6A modification on lncRNAs regulated their translation and degradation. Meanwhile, ncRNAs also regulated the m6A modification in the process of post-transcriptional regulation (). We summarized that ncRNAs could regulate m6A modification in two ways: mediating the expression of m6A regulators; participating in the interaction of m6A regulators with their target RNAs (). For example, miR455-3p competitively inhibited the binding of METTL3 to the target site on HSF1 mRNA; The expression of FTO was regulated by miR-96 and miR-1266 [Citation53,Citation80], IGF2BPs were habitually regulated by lncRNAs [Citation45,Citation79,Citation82]. Furthermore, m6A-modified circNSUN2 enhanced HMGA2 mRNA stability through the m6A-IGF2BP2 dependent way [Citation63], and there was a negative feedback loop between lncRNA GAS5 and m6A reader YTHDF3 [Citation81]. The above researches all proved the interplay between m6A modification and regulatory ncRNAs, which was helpful to explain lots of paradoxes in cancer progression.
Table 3. m6A regulators modulated by regulatory ncRNAs in CRC
In CRC, METTL3 was the most studied m6A regulator. Highly expressed METTL3 served as an oncogene in cancer progression through targeting different downstream, such as SOX2, MYC, HMGA1, and YPEL5. However, it was controversial that Deng et al. [Citation48] pointed out METTL3 was low expressed and played a tumour-suppressive role through p38/ERK pathways in CRC. Similarly, the inconsistent conclusions of METTL3 in different cancer types have been mentioned in other articles. We supposed that the dual role of METTL3 might be attributed to background differences of target genes. He et al. [Citation91] revealed that the functional effects of m6A on downstream processes might be heterogeneous greatly and depended on binding target sites of RBP. Besides, the heterogeneity of the tissue samples might partly explain the different expressions of METTL3, Liu et al. [Citation92] indicated that the expression level of METTL3 was related to tumour stage and grade. And noteworthily, METTL3 was also mentioned as a reader in the cytoplasm, promoting the mRNA translation independent of its methyltransferase activity [Citation30].
Contrary to METTL3, METTL14 acted as a tumour suppressor in CRC, which was contradictory for two core components of MTC demonstrated opposite effects on cancer progression. It seemed that METTL3 or METTL14 possessed methylation activity alone under certain conditions, while the METTL3-METTL14 complex displayed much higher catalytic activity [Citation24,Citation93], and with targets specificity. Moreover, Wang et al. [Citation49] found that overexpressed methyl CpG binding protein 2 (MeCP2) occupied reciprocity between METTL3 and METTL14, which indicated that there was a competitive relationship between MeCP2 and METTL3 with METTL14. We speculated that the overexpression of METTL3 and the depletion of METTL14 had some feedback in the MTC. However, the exact mechanism of the MTC functions awaited structural investigation.
As one of the most vital proto-oncogenic transcription factors in tumorigenesis, MYC controlled the transcription of at least 15% of the human genes [Citation94]. Though lack of knowledge the accurate m6A sites on MYC mRNA, MYC was reported as one of the primiary target genes of m6A modification in CRC [Citation38,Citation52,Citation53,Citation82], directly or indirectly. Xiang and Liang et al. [Citation38] pointed out that METTL3 methylated MYC mRNA to enhance its expression, while Yue et al. [Citation53] found that FTO might demethylate MYC mRNA to enhance expression. We speculated that FTO regulated MYC expression in an indirected way as we noticed that FTO modulated MYC expression through the FTO-m6A-MZF1 axis [Citation52]. Besides, Nishizawa et al. [Citation59] discovered that YTHDF1 was overexpressed in CRC, which was transcriptionally regulated by MYC.
Given that m6A modification played significant roles in the regulation of metabolism, stemness, metastasis and, drug resistance in CRC, key m6A modification regulators might be identified as the potential targets in the diagnosis and treatment of CRC. Moreover, we believed that upstream regulation of m6A regulators by ncRNAs provided a new insight for the treatment. Application of unique ncRNAs to regulate downstream m6A regulators might be more accurate and personalized for CRC patients.
8. Conclusions
In this review, we systematically summarized the biological functions of m6A regulators, investigated the interaction between m6A modification and regulatory ncRNAs in CRC. As two critical parts of post-transcriptional regulation, it was vital to figuring out the m6A-mediated post-transcriptional regulation on regulatory ncRNAs and the way regulatory ncRNAs modulated m6A regulators. In the present review, we first deeply investigated the interaction between m6A modification and regulatory ncRNAs in CRC, while other published reviews only mentioned about. Mechanistically, we summarized in detail the way ncRNAs regulated the process of m6A modification. In function, we concluded that the m6A modification on miRNA mainly regulates its maturation process, and the m6A modification on lncRNA regulates its translation and degradation. More importantly, uniquely proposed a novel strategy for CRC patients’ treatments based on the regulation of m6A modification by ncRNAs.
Abbreviations
Notes on contributor
Yiling Ge: Conceptualization, Methodology, Writing - Original Draft, Writing - Review & Editing. Tong Liu: Methodology, Writing - Review & Editing. Chuntao Wang: Visualization, Investigation. Yanqiu Zhang: Methodology, Funding acquisition.
Siyi Xu: Software, Validation. Yiyi Ren: Visualization.Yanlu Fen: Visualization. Lihong Yin: Supervision, Validation. Yuepu Pu: Supervision, Validation. Geyu Liang: Conceptualization, Project administration. All authors approved the final version of the manuscript.
Supplemental Material
Download MS Word (224.6 KB)Disclosure statement
The authors have declared that no competing interest exists.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev (in eng). 2015 Jul 1;29(13):1343–1355.
- He L, Li H, Wu A, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer (in eng). 2019 Dec 4;18(1):176.
- Ping XL, Sun B-F, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res (in eng). 2014 Feb;24(2):177–189.
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A (in eng). 1974 Oct;71(10):3971–3975.
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol (in eng). 2011 Oct 16;7(12):885–887.
- Linder B, Grozhik AV, Olarerin-George AO, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods (in eng). 2015 Aug;12(8):767–772.
- Warda AS, Kretschmer J, Hackert P, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep (in eng). 2017 Nov;18(11):2004–2014.
- Wen S, Wei Y, Zen C, et al. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer (in eng). 2020 Dec 12;19(1):171.
- Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science (in eng). 2016 Jun 17;352(6292):1408–1412.
- Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: roles and Therapeutic Implications in Cancer. Cancer Cell (in eng). 2020 Mar 16;37(3):270–288.
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3ʹ UTRs and near stop codons. Cell (in eng). 2012 Jun 22;149(7):1635–1646.
- Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J Pathol (in eng). 2010 Jan;220(2):126–139.
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell (in eng). 2014 Mar 27;157(1):77–94.
- Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics (in eng). 2014 Jan;9(1):3–12.
- Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin (in eng). 2020 May;70(3):145–164.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (in eng). 2020 Jan;70(1):7–30.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (in eng). 2018 Nov;68(6):394–424.
- Li J, Liang L, Yang Y, et al. N(6)-methyladenosine as a biological and clinical determinant in colorectal cancer: progression and future direction. Theranostics in eng). 2021;11(6):2581–2593.
- Li Q, He W, Wan G. Methyladenosine Modification in RNAs: classification and Roles in Gastrointestinal Cancers. Front Oncol in eng). 2020;10:586789.
- Fang Z, Hu Y, Hu J, et al. The crucial roles of N(6)-methyladenosine (m(6)A) modification in the carcinogenesis and progression of colorectal cancer. Cell Biosci (in eng). 2021 Apr 9;11(1):72.
- Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res (in eng). 2018 Jun;28(6):616–624.
- Deng X, Su R, Weng H, et al. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res (in eng). 2018 May;28(5):507–517.
- Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell (in eng). 2016 Jul 21;63(2):306–317.
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol (in eng). 2014 Feb;10(2):93–95.
- Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell (in eng). 2018 Feb 1;22(2):191–205.e9.
- Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell (in eng). 2017 May 18;169(5):824–835.e14.
- Jia G, Yang C-G, Yang S, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett (in eng). 2008 Oct 15;582(23–24):3313–3319.
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell (in eng). 2013 Jan 10;49(1):18–29.
- Ueda Y, Ooshio I, Fusamae Y, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep (in eng). 2017 Feb 13;7(1):42271.
- Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell (in eng). 2016 May 5;62(3):335–345.
- Li T, Hu PS, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6) A-IGF2BP2-dependentmechanism in colorectal carcinoma. Mol Cancer (in eng). 2019 Jun 24;18(1):112.
- Chen X, Xu M, Xu X, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer (in eng). 2020 Jun 17;19(1):106.
- Wang S, Sun C, Li J, et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett (in eng). 2017 Nov 1;(408):112–120.
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, et al. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem (in eng). 1994 Jul 1;269(26):17697–17704.
- Wang L, Hui H, Agrawal K, et al. m6A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J (in eng). 2020 Oct 15;39(20):e104514.
- Uddin MB, Roy KR, Hosain SB, et al. An N(6)-methyladenosine at the transited codon 273 of p53 pre-mRNA promotes the expression of R273H mutant protein and drug resistance of cancer cells. Biochem Pharmacol (in eng). 2019 Feb;160:134–145.
- Liu T, Li C, Jin L, et al. The prognostic value of m6A RNA methylation regulators in colon adenocarcinoma. Med Sci Monit (in eng). 2019 Dec 11;25:9435–9445.
- Xiang S, Liang X, Yin S, et al. N6-methyladenosine methyltransferase METTL3 promotes colorectal cancer cell proliferation through enhancing MYC expression. Am J Transl Res (in eng). 2020;12(5:1789–1806. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7270026/pdf/ajtr0012-1789.pdf
- Zhou D, Tang W, Xu Y, et al. METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Mol Oncol (in eng). 2021 Jan 7;15(8):2172–2184.
- Liu X, Su K, Sun X, et al. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/β-catenin pathway. J Exp Clin Cancer Res (in eng). 2021 Apr 15;40(1):132.
- Shen C, Xuan B, Yan T, et al. m(6) A-dependentglycolysis enhances colorectal cancer progression. Mol Cancer (in eng). 2020 Apr 3;19(1):72.
- Xu J, Chen Q, Tian K, et al. m6A methyltransferase METTL3 maintains colon cancer tumorigenicity by suppressing SOCS2 to promote cell proliferation. Oncol Rep (in eng). 2020 Sep;44(3):973–986.
- Zhu W, Si Y, Xu J, et al. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med (in eng). 2020 Mar;24(6):3521–3533.
- Zhang Y, Kang M, Zhang B, et al. m(6)A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer (in eng). 2019 Dec 18;18(1):185.
- Hou P, Meng S, Li M, et al. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J Exp Clin Cancer Res (in eng). 2021 Feb 1;40(1):52.
- Song P, Feng L, Li J, et al. β-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer (in eng). 2020 Aug 24;19(1):129.
- Chen H, Gao S, Liu W, et al. RNA N(6)-methyladenosine methyltransferase METTL3 facilitates colorectal cancer by activating the m(6) A-GLUT1-mTORC1Axis and is a therapeutic target. Gastroenterology (in eng). 2021 Mar;160(4):1284–1300.e16.
- Deng R, Cheng Y, Ye S, et al. m(6)A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther (in eng). 2019;12:4391–4402.
- Wang S, Gan M, Chen C, et al. Methyl CpG binding protein 2 promotes colorectal cancer metastasis by regulating N6-methyladenosine methylation through methyltransferase-like 14. Cancer Sci (in eng). 2021 Jun 7;112(8):3243–3254.
- Wang S, Fan X, Zhu J, et al. The differentiation of colorectal cancer is closely relevant to m6A modification. Biochem Biophys Res Commun (in eng). 2021 Feb 8;546:65–73.
- Zhuang J, Lin C, Ye J. m6A RNA methylation regulators contribute to malignant progression in rectal cancer. J Cell Physiol (in eng). 2020 Sep;235(9):6300–6306.
- Zhang Z, Gao Q, Wang S. Kinase GSK3β functions as a suppressor in colorectal carcinoma through the FTO-mediated MZF1/c-Myc axis. J Cell Mol Med (in eng). 2021 Feb 2. DOI:10.1111/jcmm.16291.
- Yue C, Chen J, Li Z, et al. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J Exp Clin Cancer Res (in eng). 2020 Nov 12;39(1):240.
- Liu J, Ren D, Du Z, et al. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun (in eng). 2018 Aug 25;502(4):456–464.
- Ruan DY, Li T, Wang Y-N, et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene (in eng). 2021 Jul 3;40(33):5168–5181.
- Li N, Kang Y, Wang L, et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U S A (in eng). 2020 Aug 18;117(33):20159–20170.
- Ji L, Chen S, Gu L, et al. Exploration of potential roles of m6A regulators in colorectal cancer prognosis. Front Oncol (in eng). 2020;10:768.
- Bai Y, Yang C, Wu R. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol (in eng). et al. 2019;9:332.
- Nishizawa Y, Konno M, Asai A, et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget (in eng). 2018 Jan 26;9(7):7476–7486.
- Xu D, Shao J, Song H, et al. The YTH domain family of N6-Methyladenosine “readers” in the diagnosis and prognosis of colonic adenocarcinoma. Biomed Res Int (in eng). 2020;2020:9502560.
- Zhang J, Cheng X, Wang J, et al. Gene signature and prognostic merit of M6a regulators in colorectal cancer. Exp Biol Med (Maywood) (in eng). 2020 Sep;245(15):1344–1354.
- Tanabe A, Tanikawa K, Tsunetomi M, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett (in eng). 2016 Jun 28;376(1):34–42.
- Chen RX, Chen X, Xia L-P, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun (in eng). 2019 Oct 16;10(1):4695.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (in eng). 2004 Jan 23;116(2):281–297.
- Alarcón CR, Lee H, Goodarzi H, et al. N6-methyladenosine marks primary microRNAs for processing. Nature (in eng). 2015 Mar 26;519(7544):482–485.
- Ma J-Z, Yang F, Zhou -C-C, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543.
- Peng W, Li J, Chen R, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res (in eng). 2019 Sep 6;38(1):393.
- Chen X, Xu M, Xu X, et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol Ther (in eng). 2020 Feb 5;28(2):599–612.
- Yang L, Ma Y, Han W, et al. Proteinase-activated receptor 2 promotes cancer cell migration through RNA methylation-mediated repression of miR-125b. J Biol Chem (in eng). 2015 Oct 30;290(44):26627–26637.
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (in eng). 2007 Jun 8;316(5830):1484–1488.
- Zuo L, Su H, Zhang Q, et al. “Comprehensive analysis of lncRNAs N(6)-methyladenosine modification in colorectal cancer. Aging (Albany NY) (in eng). 2021 Jan 20;12. DOI:10.18632/aging.202383.
- Wu Y, Yang X, Chen Z, et al. m(6) A-inducedlncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer (in eng). 2019 Apr 13;18(1):87.
- Yang X, Zhang S, He C, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer (in eng). 2020 Feb 28;19(1):46.
- Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer (in eng). 2020 Feb 15;19(1):30.
- Zhou C, Molinie B, Daneshvar K, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep (in eng). 2017 Aug 29;20(9):2262–2276.
- Zhang L, Hou C, Chen C, et al. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol Cancer (in eng). 2020 Jun 10;19(1):105.
- Chen C, Yuan W, Zhou Q, et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics (in eng). 2021;11(9):4298–4315.
- Guo Y, Guo Y, Chen C, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer (in eng). 2021 Jun 25;20(1):93.
- Zhu S, Wang J-Z, Chen D, et al. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun (in eng). 2020 Apr 3;11(1):1685.
- Shen XP, Ling X, Lu H, et al. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur Rev Med Pharmacol Sci (in eng). 2018 Dec;22(23):8220–8226.
- Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer (in eng). 2019 Oct 16;18(1):143.
- Wang Y, Lu JH, Wu QN, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer (in eng). 2019 Dec 2;18(1):174.
- Liu X, Liu L, Dong Z, et al. Expression patterns and prognostic value of m(6) A-relatedgenes in colorectal cancer. Am J Transl Res (in eng). 2019;11(7):3972–3991. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6684930/pdf/ajtr0011-3972.pdf
- Lan H, Liu Y, Liu J, et al. Tumor-associated macrophages promote oxaliplatin resistance via METTL3-mediated m6A of TRAF5 and necroptosis in colorectal cancer. Mol Pharm (in eng). 2021 Feb 8;18(3):1026–1037.
- Chen P, Liu XQ, Lin X, et al. Targeting YTHDF1 effectively re-sensitizes cisplatin-resistant colon cancer cells by modulating GLS-mediated glutamine metabolism. Mol Ther Oncolytics (in eng). 2021 Mar 26;20:228–239.
- Yi N, Arabella W, Ziyou L, et al. N6-methyladenosine modification: a novel pharmacological target for anti-cancer drug development. Acta Pharm Sin B. 2018;8(6):833–843.
- Bader JP, Brown NR, Chiang PK, et al. 3-Deazaadenosine, an inhibitor of adenosylhomocysteine hydrolase, inhibits reproduction of Rous sarcoma virus and transformation of chick embryo cells. Virology (in eng). 1978 Sep;89(2):494–505.
- Chen B, Ye F, Yu L, et al. Development of Cell-Active N6-methyladenosine RNA demethylase FTO Inhibitor. J Am Chem Soc (in eng). 2012 Oct 31;134(43):17963–17971.
- Huang Y, Yan J, Li Q, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res (in eng). 2015 Jan;43(1):373–384.
- Shusuke T, Timothy JZ, Ajay G. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(1):188491.
- Zhang Z, Luo K, Zou Z, et al. Genetic analyses support the contribution of mRNA N(6)-methyladenosine (m(6)A) modification to human disease heritability. Nat Genet (in eng). 2020 Sep;52(9):939–949.
- Liu T, Yang S, Sui J, et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol (in eng). 2020 Jan;235(1):548–562.
- Wang X, Feng J, Xue Y, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature (in eng). 2016 Jun 23;534(7608):575–578.
- Carroll PA, Freie BW, Mathsyaraja H, et al. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front Med (in eng). 2018 Aug;12(4):412–425.