ABSTRACT
Upregulation of utrophin, the autosomal homologue of dystrophin, can compensate dystrophin deficiency in Duchenne Muscular Dystrophy (DMD) although the therapeutic success is yet to be achieved. The present study has identified Poly (C) binding protein 2 (PCBP2) as a post-transcriptional suppresser for the expression of utrophin-A, the muscle-specific utrophin isoform. This study confirms nuclear retention of utrophin-A mRNA in C2C12 cells, which is mediated by PCBP2. Further investigation demonstrates PCBP2-dependent nuclear retention of follistatin mRNA as well. Its involvement in nuclear retention of mRNA sheds light on a novel function of PCBP2 that makes utrophin-A mRNA less available in cytosol. PCBP2, therefore, may be a target to de-repress utrophin-A expression in DMD.
Introduction
Utrophin, the autosomal homologue of dystrophin, could compensate dystrophin deficiency in mdx mice, the Duchenne Muscular Dystrophy (DMD) model [Citation1]. Although many isoforms exist, utrophin-A, the muscle-specific isoform, is of utmost interest and is found to be localized mostly at neuromuscular and myotendinous junctions in adults [Citation2]. However, in the embryo and regenerating muscles, utrophin-A is expressed throughout Gthe sarcolemma [Citation2]. Efforts have been made to upregulate utrophin to eliminate the DMD phenotype in mdx mice using its reported modulators [Citation3–5].
Discordance in utrophin-A protein and mRNA level in regenerating mouse muscle suggested post-transcriptional regulation of utrophin-A expression [Citation6]. K-homology splicing regulator protein (KSRP) and miRNAs bind 3ʹUTR of utrophin-A mRNA and regulate its expression [Citation7,Citation8]. Mouse utrophin-A 5ʹUTR confers the internal ribosome entry site (IRES), which is regulated by eEF1A2 [Citation9,Citation10]. Elements in utrophin-A 5ʹUTR repress cap-dependent translation [Citation11]. The post-transcriptional regulation of utrophin-A is complex as its transcription initiates from multiple start sites, resulting in variations in its 5ʹUTR [Citation12,Citation13].
Identification of trans-acting regulators of utrophin-A expression is of therapeutic interest. Since the 5ʹUTR of mouse utrophin-A has a poly C stretch, a potential target for Poly (C) binding protein 2 (PCBP2), we have investigated the PCBP2-mediated regulation of utrophin-A expression in mouse myoblast C2C12 cells. PCBP2 is a sequence-specific RNA binding protein consisting of three KH domains [Citation14]. PCBP2 plays an important role in mRNA stabilization and regulates IRES-mediated translation of many viruses like hepatitis C virus, cadicivirus and Type-I picornavirus [Citation15–17]. Translational silencing of Lox and human papillomavirus type 16 L2 mRNAs is associated with PCBP2 [Citation18].
In the present study, we have found that PCBP2 binds the 5ʹUTR of utrophin-A mRNA in C2C12 cells. Binding of PCBP2 to the 5ʹUTR downregulates utrophin-A expression at the protein level. We present evidence of PCBP2 and 5ʹUTR interaction-mediated nuclear retention of utrophin-A mRNA in C2C12 cells. Also, according to our observation, nuclear retention of utrophin mRNA is evident in RAW264.7 and B16F10 cells. Analysis of recently published RNA-seq data of whole muscle and its nuclei indicates nuclear retention of utrophin transcripts in wild-type (WT) and DMD (dystrophin exon 51 deleted) mice [Citation19]. Our results suggest that PCBP2 binding is sufficient to retain an mRNA within the nucleus. Follistatin mRNA is also retained within the nucleus in a PCBP2-dependent manner. The study provides insight into a novel function of PCBP2, which plays a key role in post-transcriptional repression of utrophin-A expression.
Materials and methods
Constructs
Amplified with Fw-5ʹ-AAAACATATGGACGCCGGTGTGACT-3′and Rev-5ʹ-AAGGGAATTCCTAGCTGCAC
CCCATCCC-3ʹ, mouse PCBP2 ORF was cloned in pET28a+at the EcoRI–NdeI site, under the T7 promoter to overexpress His-tagged PCBP2. Amplified with Fw-5ʹ-AATAAGCTTGATAATGGACACGGTGTGA-3ʹ and Rev 5ʹ-AAAAGAATTCCTAGCTGCTCCCCATG-3ʹ, PCBP2
ORF was cloned in pcDNA3.1+ at the HindIII-EcoRI site to develop pcDNA-pcbp2. The PCR product (Fw-5ʹ-GCCAGGGTTTTCCCAGTCACGACG-3ʹ/Rev-5ʹ-GCAGCAAGCTTGAATTAGCCTGATGGTGA
GAGAGACAAACAAGGCTTTTCTCCAAGGGATATTT-3ʹ) using shRNA vector pBS/U6 and the annealing
product of 5ʹ-AGCTTGTAGCCTGATGGTGAGAGTGACTTTTTG-3′and 5′-AATTCAAAAAGTCACTCTCACCA
TCAGGCTACA -3ʹ were inserted in pBS/U6 at KpnI-EcoRI as described [Citation20] to develop pBS/U6-psi2 for PCBP2 silencing. 18nt-long poly C stretch of mouse utrophin-A 5ʹUTR, prepared by annealing of5ʹ-CATGCCCCCCCCCCCACCCCCC-3ʹ and 5ʹ-CATGGGGGGGTGGGGGGGGGGG-3ʹ oligos, was inser-ted upstream of firefly ORF at the NcoI site of pGL3-Control to form PolyC-NcoI. The 5ʹ and 3ʹ regions flanking the polyC stretch were amplified with primer pairs 5ʹ-ACTGCCATGGGTTGTGGAGTC
GCCCTTCC-3ʹ and 5ʹ-CGACAAGATCCAAAAGATCCAAAATTGACACCCGA-3ʹand 5ʹ-CAATTTTGGATCTTT
TGGATCTTGTCGGGCTT-3ʹ& 5ʹ-TCGTCCATGGCTTGAATGAGTTTCAGTATAATCCAAAGG-3′, respectively.
The products were then mixed, melted, annealed and elongated to generate CDEL, the poly C deleted 5′UTR. Upon sequence verification, it was then cloned at the NcoI site of pGL3-Control to obtain pCDEL.
RNA-IP
RNA-IP was performed as described [Citation21]. We used an 80% confluent 100 mm plate of C2C12 for RNA-IP. Formaldehyde-treated cells, harvested in vanadyl ribonucleoside complexes (400 µM) (NEB) supplemented with RIPA buffer [Citation21], were sonicated and added to protein-A agarose beads, coated with either rabbit anti-PCBP2 antibody (1:60) (Abcam EPR14858) or mouse Lsm1 antibody (Santacruz, sc-398,552) and supplemented with BSA (2.5%), vanadyl ribonucleoside complexes (400 µM) and RNaseOUT (200 U/ml). After washing with RIPA-HS buffer [Citation21], the protein-RNA complex was disrupted in elution buffer [Citation21] for 45 mins at 70°C. cDNA was synthesized by MMLV reverse transcriptase (Epicentre) and random hexamer using DNaseI-treated RNA isolated with TriZol (Invitrogen). RT-PCR was performed using primers for utrophin-A (Fw-5ʹ-GCCAACATTTCCCGACCTT-3ʹ/Rev-5ʹ-ATAGTCAGTGTTTGGTTCCCTCAGT-3ʹ) and follistatin (Fw-5ʹ-TCACCTGATTCACACTGAAC-3ʹ/Rev-5ʹ-CGCGATTCAATGGACGTC-3ʹ) and luciferase (Fw-5ʹ-AAAGTTGC
CGAGGAGTT-3ʹ/Rev-5ʹ-CCCTTCTTTGGCCTTTTGAGG-3ʹ).
Purification of His-tagged PCBP2
His-tagged PCBP2 was purified using Ni-NTA agarose (Qiagen) according to the manufacturer’s protocol followed by dialysis with a Tub-O-Dialyzer (G-Biosciences) against RNA-Protein binding buffer (15 mM NaCl, 1 mM MgCl2 and 50 mM Tris pH 7.4).
In vitro transcription
In vitro transcription was performed using T7 RNA polymerase (NEB) at 37°C for 2 hours. The ratio of GTP to m7G5ʹppp5ʹG capped analogue was maintained at 1:4 for preparation of capped mRNA. For capped, poly A-tailed mRNA reporter preparation, amplified products from 5′Luc [Citation7] (Fw-5ʹ-GAGTACTTAATACGACTCACTATAGGGTTGTGGAGTCGCCCTTCCC-3ʹ & Rev-5ʹ-T50TACACGGCGATCTTTC
CGCCCTTCTTGGC-3ʹ), pGL3-Control(Fw-5ʹ-GAGTACTTAATACGACTCACTATAGGATGGAAGACGCCAAA
AACAT-3ʹ &Rev-5ʹ-T50TACACGGCGATCTTTCCGCCCTTCTTGGC-3ʹ) and pRL-TK (Fw-5ʹ-GAGTACTTAATACGACTCACTATAGGGCCCTCTCCCTCCCCCCCCCCTAA-3ʹ& Rev-5ʹ-T50TTATTGTTCATTTT
TGAGAACTCGCTC-3ʹ) were used as templates. 5ʹUTR and CDEL, amplified from 5ʹLuc and pCDEL, respectively, with primer pair (Fw-5ʹ-ACTGCTAATACGACTCACTATAGGGGTTGTGGAGTCGCCCTTC-3ʹ & Rev-5ʹ-CTTGAATGAGTTTCAGTATAATCCAAAGG-3ʹ) were used as templates to prepare RNA for ITC.
Isothermal titration calorimetry
For the ITC, 0.2 mM His-tagged PCBP2 was taken into the cell and 0.5 mM in vitro transcribed 507nt 5ʹUTR or CDEL was taken into the syringe. The experiment was performed at 25°C in the NANO ITC (TA Instruments).
Cell culture and transfection
Cells were cultured in high glucose DMEM media (HiMedia) supplemented with 10% FBS (Invitrogen) with or without streptomycin (0.1 mg/Lit, HiMedia) and neomycin (0.05 mg/Lit, HiMedia). DNA and RNA transfections were carried out with Lipofectamine2000® (Invitrogen) according to the manufacturer’s protocol. For analysis of mRNA stability, cells were treated with actinomycin D (10 µg/ml) and RNA was isolated at different time points.
Immunoblotting
For whole cell lysate, 106–108 C2C12 cells were harvested and lysed in NP-40 buffer [150 mM NaCl, 50 mM Tris pH 8.0, 1% NP-40 (G-Biosciences)]. Nuclear pellets were prepared as described [Citation22]. Pellets were then resuspended and lysed in RIPA buffer [Citation21]. All buffers were supplemented with 1X cOmpleteTM Mini, EDTA-free Protease Inhibitor Cocktail (Roche). Electrophoresis was performed in 3–15% Tris-Acetate gel [Citation23]. After transfer [Citation23], the membrane was blocked in 2.5% BSA and incubated with rabbit anti-PCBP2 antibody (Abcam; ab184962) (1:1000) or rabbit α-Tubulin antibody (CST; #2144) (1:1000) or mouse utrophin antibody [MANCHO3(8A4), deposited to the DSHB by Morris, G.E.] (1:50) or mouse Lamin A/C antibody [MANLAC1(4A7), deposited to the DSHB by Morris, G.E.] (1:50) [Citation24–26]. The membrane was incubated with either anti-rabbit-HRP antibody (Millipore) or anti-mouse-HRP antibody (Millipore). Bands were detected with ECL solution in Bio-Rad XRS+.
Luciferase assay
50,000 C2C12 cells in each well of the 24-well plate were seeded the day before transfection. 1 µg of 5ʹLuc or an equimolar amount of pGL3-Control or pCDEL and 1 ng of pRL-TK plasmids per well were used. For RNA transfection, 1 µg of in vitro transcribed m7G5ʹppp5ʹG-capped poly A-tailed 5ʹLuc or an equimolar amount of pGL3-Control reporter and 1 ng of renilla luciferase mRNAs were used in each well. We performed reporter assay 6 h and 3 h after DNA and RNA transfection, respectively, using a Dual-Glo® Luciferase Assay System (Promega) in the GloMax® 20/20 Luminometer (Promega). Luciferase activity from in vitro transcribed reporter mRNAs was measured after 90 minutes of in vitro translation in Rabbit Reticulocyte Lysate (Promega) according to the manufacturer’s protocol.
RNA isolation from the nuclear and cytosolic fraction
For isolation of nuclear and cytosolic RNA separately, we followed the protocol as described [Citation22]. The swelled cell pellet resuspended in ice-cold RSBG40 [Citation22] was centrifuged to collect the cytoplasmic fraction. The pellet was resuspended in RSBG40 supplemented with sodium deoxycholate [3.3% (w/w)] and Tween 40 [6.6% (v/v)]. The supernatant was added to the cytosolic fraction. The pellet was again resuspended in RSBG40 and centrifuged at 10,000 rpm for 5 mins at 4°C. Finally, the nuclear pellet was resuspended in RSBG40. RNA isolation was performed by TriZol (Invitrogen) followed by DNaseI (NEB) treatment.
qPCR
cDNA was prepared from 2 µg of RNA by MMLV reverse transcriptase (Epicentre), oligo-dT (Invitrogen) or random hexamer (Invitrogen). qPCR was performed using iTaq™ Universal SYBR® Green Supermix (Bio-Rad) with primer pairs specific for utrophin-A (Fw-5ʹ-GCCAACATTTCCCGACCTT-3ʹ/Rev-5ʹ-ATAGTCAGTGTTTGGTTCCCTCAGT-3ʹ), GAPDH (Fw-5ʹ-ACTGAGC
ATCTCCCTCACAATTTC-3ʹ/Rev-5ʹ-TCCCTAGGCCCCTCCTGTT-3ʹ), β- actin (Fw-5ʹ-CGTGCGTGACATCAA
AGAGAAGC-3ʹ/Rev-5ʹ-CCCAAGAAGGAAGGCTGGAAAAG-3ʹ), firefly luciferase (Fw-5ʹ-AAAGTTGCCGA
GAGTT-3ʹ/Rev-5ʹ-CCCTTCTTTGGCCTTTSTGAGG-3ʹ) and follistatin (Fw-5′-TCACCTGATTCACACTGAAC
-3′/Rev-5′-CGCGATTCAATGGACGTC-3′) in StepOne™ (Applied Biosystems). We used the 2−ΔΔCT method for the determination of the relative abundance of transcripts [Citation27].
RNA FISH
Mouse utrophin-A-specific 48 Stellaris RNA FISH probes (Suppl Table ST1) were designed with Stellaris Probe Designer. RNA FISH was performed with Quasar 570-labelled Stellaris RNA FISH probes (Biosearch) using wash buffer A, hybridization buffer and wash buffer B as per the protocol of Biosearch.
Microscopy and image analysis
Images were captured with a Leica SP8 confocal platform using an oil immersion 63X objective and deconvoluted using Leica Lighting Software. Nuclear and cytosolic puncta (red) was counted manually by two individuals separately without the knowledge of the sample and data collated. The nuclear vs cytosolic ratio of puncta was represented as the bar graph. Each cell was accounted for sampling for every experimental set.
RNA-seq data analysis
Normalized gene RPKM (Reads Per Kilobase per Million mapped reads) values were downloaded from NCBI GEO database (GSE156496), which were then used for clustering, plotting and determining the gene expression value. The box plot of RPKM values shows the even distribution of data across all the samples. The hierarchical clustering with the heatmap of RPKM values shows the separation between the samples belonging to different groups. The PCA plot of RPKM values also shows the clustering of data based on variance between the two groups. To determine the expression values of GAPDH, utrophin and PCBP2 genes, we used log transformed RPKM values of three samples in each group and performed the student’s t-test.
SnRNA-seq data analysis
The gene count data were downloaded from the NCBI GEO database (GSE156497) as three files, where barcodes.tsv saves barcode information, genes.tsv saves gene information and matrix.mtx saves the count data in MatrixMarket format. The data set was loaded into the R by function read_MM from the R package Matrix [Citation28] and further downstream analysis was performed using R package Seurat (v3.1.5) [Citation29]. For each sample, single-nucleus transcriptomes with fewer than 200 or more than 4,000 genes or with more than 15,000 UMIs were further filtered out from the analysis. Subsequently, data were log-normalized and scaled and principal component analysis was performed using the top 2,000 genes that showed highly variable expression in the integrated data set. Cell clusters were called using the first 15 principal components under a clustering resolution of 0.6. Dimensional reduction was performed by Uniform manifold approximation and projection (UMAP) using the first 15 principal components. Three-dimensional UMAP was generated by using the RunUMAP function from R package Seurat, which is a much faster visualization and preserves more of the global structure in data. To check the expression of genes in the data, we used violin plots using the VlnPlot function from R package Seurat. For estimating the percent of nuclei expressing the gene of interest (GOI), we subset the nuclei expressing GOI and divided it with the total number of nuclei left after normalization and scaling of data.
Statistical analysis
All statistical analyses were performed using GraphPad Prism (ver.5) software.
Results
PCBP2 interacts with utrophin-A 5ʹUTR
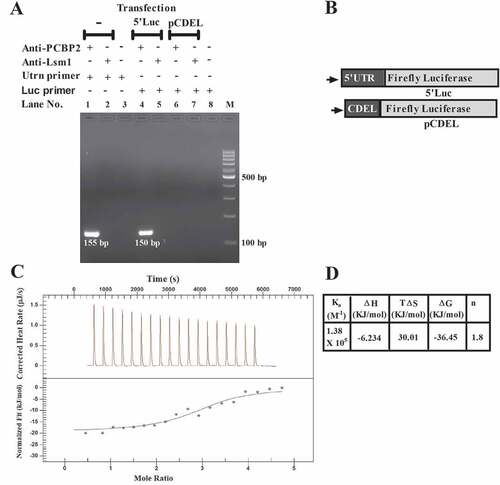
The mouse utrophin-A 5ʹUTR has poly C stretch (Suppl Fig. S1), target for Poly (C) binding protein 2 (PCBP2), which is a known post-transcriptional modulator of many viral and cellular transcripts. In RNA-IP using C2C12 lysate with the anti-PCBP2 antibody, we have detected the utrophin-A 3ʹUTR-specific 155bp RT-PCR product, indicating PCBP2 binding with full-length mRNA (). Upon transfection with the reporter construct 5ʹLuc, which contains 507nt 5ʹUTR of the mouse utrophin-A fragment at the upstream of the firefly luciferase ORF [Citation7], in RNA-IP with the anti-PCBP2 antibody, we have detected the luciferase-specific 150bp RT-PCR product, whereas with pCDEL, which contains utrophin-A 5′UTR devoid of poly C stretch (), no product is detected in RNA-IP with the same antibody (). In all three experiments, antibody against Lsm1, a non-specific RNA-binding protein, was used as control. In order to confirm the in vitro interaction between PCBP2 and 507nt-long utrophin-A 5ʹUTR fragment, the Isothermal Titration Calorimetric (ITC) study was carried out. The interaction has been confirmed from ITC thermogram () analysis. The calculated binding constant (Ka) value is 1.38 x 105M−1. Thermodynamically, the interaction is exothermic (ΔH = −6.234 kJ/mol) and stoichiometry (n) of the interaction is found to be ≈ 2, indicating that one molecule of protein forms complex with two molecules of RNA. PCBP2 is known to have three KH domains. As KH domains interact with RNA, binding of two RNA molecules with one molecule of PCBP2 suggests that out of three, two KH domains are involved whilst interacting with utrophin-A 5ʹUTR in vitro. This is in agreement with the previous report, which demonstrated that only first and third KH domains of PCBP2 have high affinity for poly rC [Citation30]. Here, it is important to note that when ITC experiment was performed with RNA corresponding to CDEL in the same condition, no interaction was detected (Suppl Fig. S2).
Figure 1. Interaction between PCBP2 and mouse utrophin-A 5ʹUTR.
PCBP2 as a post-transcriptional modulator of utrophin-A expression
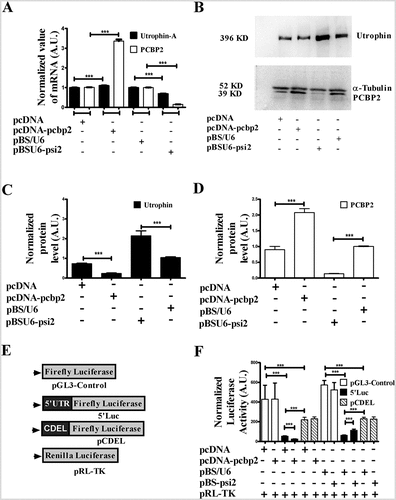
To investigate how PCBP2 might influence utrophin-A expression, we overexpressed PCBP2 with the construct pcDNA-pcbp2 in mouse myoblast C2C12 cells. PCBP2 was silenced with pBSU6-psi2. The expression level of utrophin-A and PCBP2 in transfected cells was determined through qPCR and Western blot. The cells transfected with empty vectors, pcDNA and pBS/U6 were used as corresponding controls. Upon PCBP2 overexpression, abundance of utrophin-A mRNA is slightly increased and there is a minute decrease in the utrophin-A mRNA level with silencing of PCBP2 (). Interestingly, in western blot (), utrophin is found to be decreased by more than 80% upon PCBP2 overexpression and upregulated 2-fold upon PCBP2 silencing (). The results, thus, suggest PCBP2-mediated post-transcriptional repression of utrophin-A expression. The reporter activity of 5ʹLuc is far less than that from pGL3-Control () because of the translational repressors present in the utrophin-A 5ʹUTR fragment [Citation11]. In PCBP2 overexpressed cells, we observe a decrease in luciferase activity from 5ʹLuc (). Upon PCBP2 silencing, luciferase activity from 5ʹLuc is increased compared to control (). In contrast, luciferase activity from pGL3-Control and pCDEL remains unchanged upon PCBP2 overexpression and silencing (). These data do indicate that 5ʹUTR of utrophin-A regulates protein expression in a PCBP2-dependent manner. In order to investigate the mechanistic basis of PCBP2-mediated utrophin-A regulation, we determined the utrophin-A mRNA level at different time points upon actinomycin-D treatment in PCBP2 overexpressed and silenced C2C12 cells. As there is no significant change in utrophin-A mRNA decay (Suppl Fig. S3), we conclude that the regulation is not mediated through mRNA stability. In order to check if PCBP2 and 5′UTR interaction exerts any inhibition of translation, in vitro transcribed m7G5ʹppp5ʹG-capped, poly-A-tailed 5′Luc reporter mRNA was translated in Rabbit Reticulocyte Lysate. The slightly reduced luciferase activity in the presence of purified His-tagged PCBP2 (Suppl Fig. S4) suggests that PCBP2 and utrophin-A 5′UTR-mediated suppression of utrophin expression cannot be explained exclusively with the translational inhibition given the high magnitude of repression as evidenced in . We then aimed to check PCBP2-utrophin-A 5′UTR-mediated translation regulation, if any, with mRNA transfection in C2C12 cells. A previous report has suggested that in contrast to its DNA counterpart, after lipofection, reporter mRNA is mostly present in discrete particles outside the nucleus [Citation31] and therefore, translation regulation can be addressed using this approach. In vitro transcribed m7G5ʹppp5ʹG-capped, poly-A-tailed reporter 5ʹLuc mRNA although has lower luciferase expression compared to pGL3-Control in C2C12 cells, no significant change in reporter activity is found with the 5ʹLuc mRNA reporter upon PCBP2 overexpression and silencing (Suppl Fig. S5). We, therefore, conclude that the utrophin-A 5ʹUTR-PCBP2-mediated post-transcriptional repression does not primarily target translation.
Figure 2. PCBP2 is a modulator of utrophin-A expression.
Nuclear retention of utrophin-A mRNA is mediated through PCBP2
Having confirmed that the PCBP2-utrophin-A 5ʹUTR interaction-mediated post-transcriptional repression of utrophin-A expression is mainly pre-translational, we looked into other probable mechanisms. Although mRNAs are usually exported to cytosol very efficiently, nuclear retention of mRNA happens sometimes [Citation32–34]. Since PCBP2 is found in the nuclear fraction of C2C12 cells (Suppl Fig. S6), we asked if this mechanism operates in PCBP2-mediated repression of utrophin expression. We isolated RNA from nuclear and cytosolic fractions of C2C12 cells. The nuclear and cytosolic fractionation was confirmed with Western blot using nuclear marker Lamin A/C and cytosolic marker α-Tubulin (). The abundance of utrophin-A, β-actin and GAPDH in two fractions was analysed with qPCR. Using GAPDH as endogenous control, the relative nuclear to cytosolic abundance of both utrophin-A and β-actin was calculated as 2−ΔΔCT, where ΔΔCT = [(CT Nuclear (Target) – CT Nuclear (GAPDH)) – (CT Cytosolic (Target) – CT Cytosolic (GAPDH))] (). Nuclear to cytosolic abundance (henceforth nuclear retention) of utrophin-A is more than 7-fold than that of β-actin in control cells. This suggests that compared to β-actin, utrophin-A transcript mostly resides within the nucleus. We determined the nuclear retention of utrophin-A and β-actin upon overexpression and silencing of PCBP2 (). Upon PCBP2 overexpression, nuclear retention of utrophin-A is increased 4-fold, whereas it is reduced by 8-fold upon PCBP2 silencing. In contrast, nuclear retention of β-actin remains unchanged upon variation of PCBP2 level. The nuclear retention of utrophin-A mRNA is also observed in mouse macrophage like RAW264.7 and melanoma B16F10 cell lines (Suppl Fig. S7)
Figure 3. Interaction between PCBP2 and utrophin-A 5ʹUTR helps in the retention of utrophin-A transcript within the nucleus.
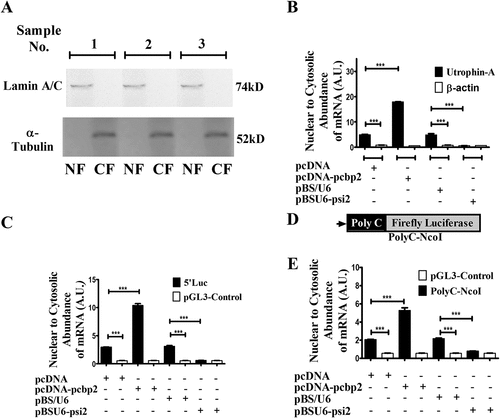
In pcDNA-transfected cells, nuclear retention of mRNA from the 5ʹLuc reporter is much higher than that produced from pGL3-Control (), indicating a pivotal role of utrophin-A 5ʹUTR in nuclear retention of the reporter transcript. The nuclear retention of 5ʹLuc mRNA is much more pronounced when PCBP2 was overexpressed. In contrast, nuclear retention of transcript from pGL3-Control remains insensitive to PCBP2 overexpression (). Upon PCBP2 silencing, nuclear retention of 5ʹLuc mRNA is decreased (). Hence, the observation confirms that utrophin-A 5ʹUTR and PCBP2 interaction assist reporter transcript to remain within the nucleus.
We introduced the poly C stretch (5ʹCCCCCCCCCCCACCCCCC3ʹ) of utrophin-A 5ʹUTR at the upstream of reporter ORF in pGL3-Control to construct PolyC-NcoI (). Nuclear retention of the mRNA from the PolyC-NcoI reporter is increased upon PCBP2 overexpression (). It is remarkable that silencing of PCBP2 reduces its nuclear retention at the level similar to that of mRNA from pGL3-Control.
These observations confirm that transcripts having utrophin-A 5ʹUTR or poly C stretch are mostly retained within the nucleus in a PCBP2-dependent manner. However, the extent of nuclear retention is found to be much higher in the case of transcript with the 507nt utrophin-A 5ʹUTR fragment compared to mRNA carrying only poly C stretch. This suggests the possible role of other cis and/or trans-acting factors in nuclear retention of utrophin-A mRNA.
The localization of utrophin-A mRNA in C2C12 cells was further checked using RNA FISH. Since mouse utrophin-A has many transcript variants differing at their 5′ terminus [Citation12,Citation13], the probes were designed over the coding region in order to rule out any effect due to the altered isoform ratio. In the empty vector transfected cells, utrophin-A mRNAs, represented as puncta, are observed in the cytosol and in the nucleus (, C & E). The distribution pattern of utrophin-A transcript is very similar to that found in the non-transfected C2C12 cells (Suppl Fig. S8A). Upon overexpression of PCBP2, the nuclear to cytosolic abundance of utrophin-A mRNA is increased and upon silencing, a sharp decrease in the same has been observed in RNA FISH experiment (, D & E), which conclusively demonstrates PCBP2-mediated nuclear retention of utrophin-A mRNA in C2C12 cells.
Figure 4. Localization of utrophin-A transcript in C2C12 cells and transcriptomic analysis of TA muscle from WT and DMD (ΔEx51) mice.
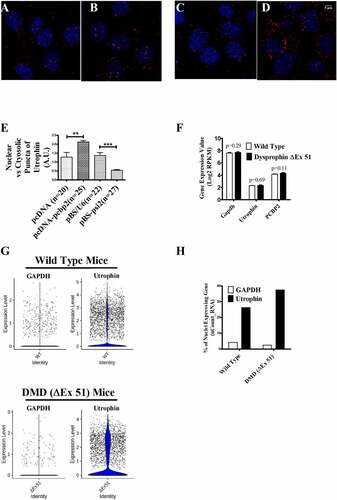
To further investigate the nuclear retention of utrophin transcript in muscle myofiber in an in vivo system, we compared the transcriptomic data sets of tibialis anterior (TA) muscle isolated from 4-wk-old DMD mice (dystrophin ΔExon 51) with that of WT mice [Citation19]. From the normalized RNA-seq data (Suppl. Fig S9A,B and C) of intact TA muscle from WT and DMD mice, we have found the expression of utrophin as well as the control housekeeping gene GAPDH (). Although the expression level of each of these genes is comparable in WT and DMD TA muscles, the abundance of GAPDH is much higher than that of utrophin (). We also observe the expression of PCBP2 in the muscle of both WT and DMD mice (). Then, we normalized and analysed the single nuclear RNA sequencing (snRNA-seq) data of nuclei isolated from TA muscle of WT (suppl. Fig. S9D & E) and DMD (Suppl. Fig. S9G & H) mice at 4 wk of age, which captured specifically the count of nuclear transcripts. Nuclear transcriptomes were visualized using a UMAP plot of all the nuclei from WT (7080 nuclei) and DMD (4455 nuclei) TA muscle and we have identified 15 different clusters of nuclei, coloured according to their transcriptional signatures (suppl. Fig. S9F & I). Considering all the clusters of nuclei in data sets, we have found an increase in the transcript count of utrophin compared to GAPDH in both the WT and the DMD TA muscles ( & H). As utrophin transcripts are less abundant in the whole muscle and more abundant in the isolated muscle nuclei than GAPDH, the in vivo transcriptome data further support our experimental findings in cultured cells and suggest nuclear retention of utrophin transcript in muscle.
PCBP2-mediated nuclear retention of follistatin mRNA
If interaction with PCBP2 retains a transcript within the nucleus, the phenomenon should not be limited to utrophin-A only. We, therefore, searched for transcripts having potential PCBP2-binding sites. The follistatin 5ʹUTR (Suppl Fig. S10) has poly C stretch and could be a potential PCBP2 interactor. The in vivo interaction between PCBP2 and follistatin mRNA was confirmed with RNA-IP (Suppl Fig. S11A) using the anti-PCBP2 antibody. We checked whether PCBP2 could confine the follistatin mRNA within the nucleus. Compared to control, in PCBP2-overexpressing cells, the nuclear retention of follistatin mRNA is ~3.3-fold (Suppl Fig. S11B). The result was reversed upon PCBP2 silencing (Suppl Fig. 11B). These observations further confirm that interaction with PCBP2 helps a transcript to be preferentially retained within the nucleus.
Discussion
The key to success in achieving utrophin upregulation in DMD is to identify underlying molecular events that regulate its expression. Hence, trans-acting repressors operating in its post-transcriptional regulation could be effective therapeutic targets. The present study reports PCBP2 as a post-transcriptional repressor of utrophin-A expression. Although PCBP2 overexpression downregulates utrophin protein, we have noted a slight increase in utrophin-A mRNA (). It is to be noted that the effects of two phenomena, the nuclear retention and low-level rise of utrophin-A mRNA, are conflicting. Evidently, nuclear retention overweighs the effect of the other and as a result, PCBP2 upregulation eventually downregulates the utrophin-A protein level.
As a sorting bin with many ribonucleases and RNA binding proteins, the nucleus plays a pivotal role in determining the fate of a transcript. Ribonucleoproteins (RNPs) like ALYREF and Exon Junction Complex (EJC) help an mRNA to be efficiently exported to the cytosol, whereas ARS2 and ZC3H18 are examples of nuclear proteins connected to the nuclear ribonucleolytic machinery. In general, it is believed that the mRNAs not properly processed are failed to be efficiently exported to the cytosol and are subjected to rapid nuclear decay [Citation32]. However, many long non-coding RNAs (lncRNAs), like XIST, are known to be retained within the nucleus, but escape nuclear degradation. The protein known to provide nuclear retention of XIST is hnRNP U [Citation35]. Although many lncRNAs are retained within the nucleus, only a handful of mRNAs is to date known to behave the same. A recent study has identified the Alu repeated derived 42-nt long sequence having consensus RCCTCCC (R = A/G) and hnRNP K as its interacting partner for nuclear accumulation of RNAs [Citation33]. In another study, a cytosine-rich motif has also been found in nuclear enrichment sequences of RNA [Citation34]. In the present study, we demonstrate that a stretch of C interrupted with single A derived from utrophin-A 5ʹUTR is sufficient for nuclear retention of a transcript in a PCBP2-dependent manner. RNA-seq analysis of whole TA muscle and its nuclei also suggests nuclear retention of utrophin transcript in the WT and DMD muscles. Moreover, the PCBP2-mediated nuclear retention is not restricted to utrophin-A mRNA only. We have demonstrated that nuclear accumulation of follistatin transcript is also PCBP2-dependent (Suppl Fig. S11). In different cell types, PCBP2 has been reported to be present in the nucleus and cytosol [Citation36]. In cytosol, it is found in the P-body [Citation37], whereas its localization in any nuclear substructure has not been reported. Therefore, understanding mechanistic details underlying its role in nuclear retention of mRNA requires further investigation.
In conclusion, the present study has identified PCBP2-mediated nuclear retention of the transcript, a novel mechanism that makes utrophin-A mRNA less abundant in the cytosol. Although many transcriptional modulators of utrophin-A have been identified, its upregulation, sufficient to ameliorate the DMD phenotype, is yet to be accomplished. Since the present study has demonstrated that the silencing of PCBP2 upregulates utrophin protein in mouse myoblast C2C12 cells, it provides proof of principle that PCBP2 inhibition could be a novel therapeutic strategy to upregulate utrophin expression. The pharmacological inhibitor of PCBP2 although may not be very specific for utrophin upregulation, CRISPR-mediated deletion of the PCBP2 binding site or use of Site Blocking Oligonucleotide may be more utrophin specific. Recently, ~2-fold utrophin upregulation in C2C12 cells by pharmacological activation of eEF1A2 and Site Blocking Oligonucleotide targeting the miRNA let-7 c binding site at utrophin-A 3′UTR have been reported [Citation10,Citation38]. Strategies targeting PCBP2-mediated repression could, therefore, be developed to further upregulate utrophin expression in combination with other strategies, which targets different pathways.
Authors contributions
UB and GG designed the study. GG performed most of the experiments. SS performed the biophysical study. RNA FISH was performed by GG, DP and SDa. Microscopy and imaging were performed by SDc. Image analysis was performed by GG and SDc. VS and SSR analysed RNA-seq and SnRNA-seq data. GG, UB, SS, SDc, VS, JN and SSR analysed the data and wrote the paper.
Supplemental Material
Download Zip (2.2 MB)Acknowledgments
This work was supported by the Science Engineering Research Board (EMR/2016/003040) and Department of Biotechnology (BT/PR15082/GBD/27/305/2011), Govt. of India to UB. GG and SS received their fellowships from the University of Kalyani. The authors acknowledge the infrastructural support developed by the DST-PURSE programme in the University of Kalyani. The support of Dr. Arnab Gupta, Indian Institute of Science, Education and Research, Kolkata, in confocal imaging is gratefully acknowledged.
Disclosure statement
No potential conflict of interest was reported by the author(s)
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Tinsley JM, Potter AC, Phelps SR, et al. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 1996;384(6607): 349–353.
- Weir AP, Burton EA, Harrod G, et al. A- and B-utrophin have different expression patterns and are differentially upregulated in mdx muscle. J Biol Chem. 2002;277: 45285–45290.
- Krag TOB, Bogdanovich S, Jensen CJ, et al. Heregulin ameliorates the dystrophic phenotype in mdx mice. Proc Natl Acad Sci USA. 2004;101(38): 13856–13860.
- Tinsley JM, Fairclough RJ, Storer R, et al. Daily treatment with SMTC1100, a novel small molecule utrophin upregulator, dramatically reduces the dystrophic symptoms in the mdx mouse. PLoS One. 2011;6(5): e19189.
- Péladeau C, Ahmed A, Amirouche A, et al. Combinatorial therapeutic activation with heparin and AICAR stimulates additive effects on utrophin A expression in dystrophic muscles. Hum Mol Genet. 2016;25(1): 24–43.
- Gramolini AO, Karpati G, Jasmin BJ. Discordant expression of utrophin and its transcript in human and mouse skeletal muscles. J Neuropathol Exp Neurol. 1999;58(3): 235–244.
- Basu U, Lozynska O, Moorwood C, et al. Translational regulation of utrophin by miRNAs. Plos One. 2011;6(12): e29376.
- Amirouche A, Tadesse H, Miura P, et al. Converging pathways involving microRNA-206 and the RNA-binding protein KSRP control post-transcriptionally utrophin A expression in skeletal muscle. Nucleic Acids Res. 2014;42: 3982–3997.
- Miura P, Thompsona J, Chakkalakala J, et al. The utrophin A 5ʹUTR confers IRES-mediated translational control during regeneration of skeletal muscle fibers. J Biol Chem. 2005;280: 32997–33005.
- Péladeau C, Adam N, Bronicki LM, et al. Identification of therapeutics that target eEF1A2 and upregulate utrophin A translation in dystrophic muscles. Nat Commun. 2020;11(1): 1990.
- Ghosh T, Basu U. Cis-acting sequence elements and upstream open reading frame in mouse utrophin-A 5′-UTR repress cap-dependent translation. PLoS One. 2015;10: e0134809.
- Perkins KJ, Davies KE. Alternative utrophin mRNAs contribute to phenotypic differences between dystrophin-deficient mice and Duchenne muscular dystrophy. FEBS Lett. 2018;592(11): 1856–1869.
- Malik D, Basu U. Repression-free utrophin-A 5ʹUTR variants. Mol Biol Res Commun. 2019;8: 129–133.
- Leffers H, Dejgaard K, Celis JE. Characterisation of two major cellular Poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230(2): 447–453.
- Spangberg K, Schwartz S. Poly(C)-bindging protein interacts with the hepatitis C virus 5ʹ untranslated region. J Gen Virol. 1999;80: 1371–1376.
- Sweeney TR, Abaeva IS, Pestova TV, et al. The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J. 2014;33(1): 76–92.
- Asnani M, Pestova TV, Hellen CUT. PCBP2 enables the cadicivirus IRES to exploit the function of a conserved GRNA tetraloop to enhance ribosomal initiation complex formation. Nucleic Acids Res. 2016;44: 9902–9917.
- Collier B, Goobar-Larsson L, Sokolowski M, et al. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J Biol Chem. 1998;273(35): 22648–22656.
- Chemello F, Wang Z, Li H, et al. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc Nat Acad Sci. 2020;117(47): 29691–29701.
- Sui G, Shi Y. Gene silencing by a DNA vector-based RNAi technology. Methods Mol Biol. 2005;309: 205–218.
- Selth LA, Gilbert C, Svejstrup JQ. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb Protoc. 2009;2009(6): pdb prot5234.
- Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39(4): 356–362.
- Cubillos-Rojas M, Amair-Pinedo F, Tato I, et al. Simultaneous electrophoretic analysis of proteins of very high and low molecular mass using Tris-acetate polyacrylamide gels. Electrophoresis. 2010;31(8): 1318–1321.
- Manilal S, Randles KN, Aunac C, et al. A lamin A/C beta-strand containing the site of lipodystrophy mutations is a major surface epitope for a new panel of monoclonal antibodies. Biochim Biophys Acta. 2004;1671(1–3): 87–92.
- Shi G, Abbott KN, Wu W, et al. Dnase1L3 regulates inflammasome-dependent cytokine secretion. Front Immunol. 2017;8: 1–16.
- Trajanovska S, Ban J, Huang J, et al. Muscle specific kinase protects dystrophic mdx mouse muscles from eccentric contraction-induced loss of force-producing capacity. J Physiol. 2019;597(18): 4831–4850.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25: 402–408.
- Bates D, Maechler M. 2010. Matrix: sparse and dense matrix classes and methods. R package version 0.999375-43, http://cran.r-project.org/package=Matrix
- Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5): 411–420.
- Dejgaard K, Leffers H. Characterisation of the nucleic-acid-binding activity of KH domains different properties of different domains. Eur J Biochem. 1996;241(2): 425–431.
- Barreau C, Dutertre S, Paillard L, et al. Liposome-mediated RNA transfection should be used with caution. RNA. 2006;12(10): 1790–1793.
- Garland W, Jensen TH. Nuclear sorting of RNA. Wiley Interdiscip Rev RNA. 2020;11: e1572.
- Lubelsky Y, Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555(7694): 107–111.
- Shukla CJ, McCorkindale AL, Gerhardinger C, et al. High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 2018;37(6): e98452.
- Sakaguchi T, Hasegawa Y, Brockdorff N, et al. Control of chromosomal localization of Xist by hnRNP U family molecules. Dev Cell. 2016;39(1): 11–12.
- Ghanem LR, Chatterji P, Liebhaber SA. Specific enrichment of the RNA-binding proteins PCBP1 and PCBP2 in chief cells of the murine gastric mucosa. Gene Expr Patterns. 2014;14(2): 78–87.
- Yoshimura M, Honda H, Sasagasako N. PCBP2 is downregulated in degenerating neurons and rarely observed in TDP-43-positive inclusions in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2021;80(3): 220–228.
- Sengupta K, Loro L, Khurana TS. PMO based let 7c site blocking oligonucleotide (SBO) mediated utrophin upregulation in mdx mice, a therapeutic approach for Duchenne muscular dystrophy (DMD). Sci Rep. 2020;10: 21492.
