 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The RNA world hypothesis confers a central role to RNA molecules in information encoding and catalysis. Even though evidence in support of this hypothesis has accumulated from both experiments and computational modelling, the transition from an RNA world to a world where heritable genetic information is encoded in DNA remains an open question. Recent experiments show that both RNA and DNA templates can extend complementary primers using free RNA/DNA nucleotides, either non-enzymatically or in the presence of a replicase ribozyme. Guided by these experiments, we analyse protocellular evolution with an expanded set of reaction pathways made possible through the presence of DNA nucleotides. By encapsulating these reactions inside three different types of protocellular compartments, each subject to distinct modes of selection, we show how protocells containing DNA-encoded replicases in low copy numbers and replicases in high copy numbers can dominate the population. This is facilitated by a reaction that leads to auto-catalytic synthesis of replicase ribozymes from DNA templates encoding the replicase after the chance emergence of a replicase through non-enzymatic reactions. Our work unveils a pathway for the transition from an RNA world to a mixed RNA-DNA world characterized by Darwinian evolution, where DNA sequences encode heritable phenotypes.
1. Introduction
The RNA world hypothesis states that a living system composed entirely of RNA may have emerged prior to the origin of the current DNA-protein world. The ability of RNA to store information [Citation1,Citation2] and catalyse reactions [Citation3–5] suggest that they may indeed have played a key role in the emergence of a self-sustaining chemical system capable of undergoing Darwinian evolution. However, the plausibility of such an RNA world relies on explaining how self-replicating protocells encapsulating RNA molecules first emerged from a primordial soup containing basic chemical-building blocks. Several experiments have shown that both ribonucleotides [Citation6–8] and deoxy-ribonucleotides [Citation9,Citation10] can be spontaneously created in prebiotic environments. Non-enzymatic processes of concatenation and template-directed primer extension, albeit with [Citation11,Citation12] or without [Citation13] activated RNA nucleotides, along with environmental cycling [Citation14,Citation15] give rise to long RNA polymers that can fold into complex secondary structures [Citation16]. It seems plausible that the first ribozymes emerged by chance through such processes, as a result of extensive sampling of the sequence space through error-prone replication [Citation17] in a prebiotic environment. Recent work has provided evidence of phenotype bias, indicating that certain secondary structures which appear most frequently, while sampling sequences of fixed length happen to be the ones that are found in nature [Citation18]. Remarkably, those structures can be generated by sampling a relatively small region of the sequence space, implying that the chance of emergence of a functional bio-molecule may not be too small. However, the random creation of a few functional bio-molecules is not sufficient to guarantee their proliferation, especially in a spatially open environment dominated by selfish parasitic sequences. It is therefore imperative for template-directed replication processes to be encapsulated in compartments since such segregation can help in preventing parasites from overwhelming the system [Citation19]. If distinct ribozymes are created with small probabilities inside a protocell during replication by the rolling circle mechanism [Citation20–24], they can act to not only synergistically enhance each others’ formation but also increase the likelihood of creation of other functionally distinct ribozymes. This provides a pathway towards increasing the functional complexity of the protocell and can allow for the proliferation of protocells containing a functionally diverse set of ribozymes [Citation25]. However, such an evolution is not Darwinian in nature, and the ribozymes generated cannot be considered as heritable traits since they appear by chance during the error-prone replication process and are also prone to degradation. How then is it possible to create a self-sustained chemical system that does not rely on random ribozyme production for its survival? A possible resolution to this problem in the RNA world was proposed by Takeuchi et al. [Citation26] who showed that genome-like strands can appear inside vesicles due to symmetry-breaking between two complementary strands of a self-replicating ribozyme. One strand would then act as a genome and its complementary strand as an enzyme. Since there is very little free-energy difference between a sequence and its complement, it seems unlikely that the reverse complement of a template strand would fold into a complex secondary structure, that is characteristic of a ribozyme.
We believe the answer to the riddle of creating a self-sustained chemical replicator lies in the appearance of information-encoding DNA templates that act like primitive pseudo-genes. A collection of such pseudo-genes, each encoding a different functional phenotype, would then constitute a primitive albeit fragmented genome. However, unlike ribonucleotides [Citation27–29], spontaneous polymerization of free DNA nucleotides into long DNA strands has not yet been demonstrated by experiments. Therefore, the transition to a DNA world must have been facilitated by RNA. Indeed, experiments on mixed RNA-DNA template-primer systems have shown that RNA is capable of extending DNA primers both non-enzymatically [Citation30] and under the action of polymerase ribozymes [Citation31–33]. Non-enzymatic and enzymatic reverse transcription of RNA strands might have been the primary mechanism of DNA strand creation in a primordial world. These results [Citation31–33] are particularly significant because they suggest that the transition from an RNA world to a DNA-protein world passed through an intermediate epoch where life may have been based on DNA genomes and ribozymes that took on the role of proteins.
In this work, we show how the presence of DNA nucleotides increases the space of possible reactions and opens up the possibility of creating single-stranded DNA sequences that can act as information-encoding templates. Even though the error-prone non-enzymatic template-directed replication process produces mostly useless sequences (parasites), we assume that bio-molecules like replicase ribozymes can be created with small probabilities. The replication of RNA templates using DNA nucleotides can also create, with a small probability, the DNA sequences encoding the replicase ribozymes. Those DNA sequences can act as templates, whose accurate, auto-catalytic replication with RNA nucleotides can create more copies of the replicase. Such templates, along with the replicase, can speed up the replication process inside a protocell which in-turn ensures rapid growth in the number of strands and enhanced likelihood of creation of new replicases and templates. A population of protocells can eventually evolve to increase the number of protocells containing both replicase-encoding templates and replicases, thereby marking the onset of Darwinian evolution characterized by the encoding of a heritable trait like the replicase ribozyme in DNA sequences. The nature of the protocellular compartment determines how likely it is for such protocells to dominate the population under reasonable conditions. Proliferation of such protocells is observed for coacervate droplets, water-in-oil droplets as well as vesicular compartments. However, coacervate droplets and water-in-oil droplets allow for the dominance of such protocells under diverse conditions that are even robust to decrease in the error threshold for accurate replication. Intriguingly, in our model, evolution leads to the spontaneous emergence of protocells that contain the replicase-encoding DNA templates in low copy numbers and replicases in high copy numbers. Our work shows how the creation of DNA-encoded heritable phenotypes through RNA-templated replication can lead to the emergence of protocells capable of undergoing Darwinian evolution.
2. Methods
The reactions inside each protocell depend on the nature of the components present in it. The presence of DNA nucleotides allows for the non-enzymatic replication of RNA templates using deoxy-ribonucleotides [Citation30,Citation34] leading to the creation of DNA strands. The number of strands inside a protocell initially grows through non-enzymatic template-directed replication based on the reactions described below (see section 2 of the electronic supplementary material for details of estimation of the replication rates). Enzymatic reactions are activated once a replicase emerges as a result of a chance non-enzymatic replication event. gives both the non-enzymatic and enzymatic replication rates for the various reactions and Supplementary Fig-S1 gives a pictorial representation of the different processes considered. The evolution of a population of such protocells occurs due to protocellular growth followed by division and selection in a manner that depends on the nature of the encapsulating compartment.
Table 1. Non-enzymatic and enzymatic replication rates for a template of length L = 200 nt inside a protocell of radius 57 nm along with the corresponding percentage errors ().
.
,
represent non-enzymatic and enzymatic replication rates from RNA to DNA etc.
2.1. Reactions
Initially, we consider protocells containing RNA templates only. Non-enzymatic replication of
by RNA nucleotides (with a rate
) will mostly create RNA parasites (P) (which are non-functional sequences that are neither templates nor ribozymes), due to the high error-rates of non-enzymatic reactions. However, following the argument given in the introduction, we assume that replicase ribozymes (R) can be created with a small probability
during the error-prone, non-enzymatic replication process (see reaction (1)). The chance creation of a replicase eventually leads to the emergence of a more complex reaction network as indicated by the blue arrows in . We also assume unlimited monomer supply by the environment to keep the rates independent of monomer concentrations.
on the RHS of reactions 1 indicates that both parasites (
) and replicases (
) can be produced albeit with different probabilities that are indicated in brackets.
Figure 1. Pictorial representation of the reaction network inside a protocell. Red (blue) arrows indicate non-enzymatic (enzymatic) reactions. Parasites are indicated by square filled boxes whereas all other reactants and products are indicated by filled circles. Light blue filled circles denote RNA templates, peach colour filled circles denote replicases, and yellow filled circles denote DNA templates. The type of monomer used in each reaction is indicated in brackets with rNTP and dNTP indicating RNA and DNA nucleotides respectively. The left panel shows the reactions possible initially in presence of RNA templates only. The middle panel shows the non-enzymatic reactions possible after the emergence of DNA templates. Ch-1, Ch-2 and Ch-3 denote three different channels for non-enzymatic replication of the three different types of DNA templates. The right panel (blue arrows) shows the enzymatic reactions possible upon emergence of a replicase ribozyme. The numbers 1–9 correspond to the reaction numbers specified in the main text. Arrows with multiple arrowheads denote the possible products of a reaction.
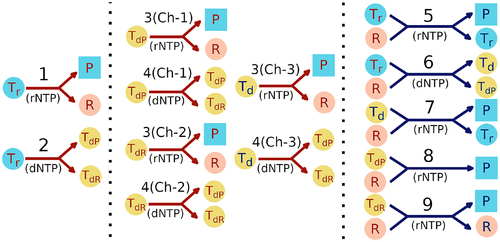
For the RNA world to be viable, sustained creation of replicases that can catalyse replication of template strands is essential. Non-enzymatic or enzymatic replication of a replicase, whose catalytic ability arises from its complex folded structure, is extremely difficult to achieve because folded segments can block replication of such RNA sequences. So far, the only way this can be achieved is by using trimer building blocks instead of monomers [Citation36]. However, that process requires an abundance of trimers that are an exact complement of triplet nucleotides that make up the structured template. The abundance of such trimers, created via spontaneous concatenation of free monomers in a primordial RNA world, is likely to be much smaller than the abundance of monomers. Therefore, the feasibility of replicating ribozyme sequences from structured templates in prebiotic scenarios remains questionable. This problem can be avoided if ribozymes are encoded in DNA sequences. We calculated the folding free energies of different RNA sequences and their DNA counterparts and observed that free energies of the DNA strands are times larger than the free energies of their RNA counterparts [Citation37–39]. Therefore, DNA strands are less likely to fold into complex secondary structures and more likely to act as templates.
The enzymatic replication of such ribozyme-encoding DNA templates using RNA nucleotides [Citation33] can then provide a higher fidelity pathway for creation of those ribozymes. Recent experiments showing RNA templated DNA synthesis [Citation32] and DNA templated RNA synthesis [Citation33] seem to suggest the plausibility of such a scenario of sustained ribozyme creation.
Non-enzymatic replication of by DNA nucleotides will create DNA templates, the majority of which will be non-functional since they will encode RNA parasites (
). However, such replication processes can also create, with a small probability
, DNA templates (
) that encode replicase ribozymes. The probabilities
and
for creation of
and
respectively are indicated in brackets in the reaction below.
Non-enzymatic replication of DNA templates (which is the DNA analog of
),
and
by RNA nucleotides will create parasites and replicases (rate
). Similarly, non-enzymatic replication of DNA templates
,
and
by DNA nucleotides can create
and
(rate
).
on the LHS of reactions 3–9 indicate that any one of them is used as a template and the corresponding template is therefore also present on the RHS of the reactions.
We assume the probability of and
creation to be same for all DNA templates
,
and
, because non-enzymatic replication processes have very high nucleotide misincorporation rates (see ). Therefore, such replication processes effectively lead to the creation of randomly sampled 200-mer sequences that are likely to be uncorrelated with the sequence of the underlying templates. It then seems reasonable to expect that the chances of non-enzymatic creation of
and
would be independent of the nature of the template.
Initially, the processes inside a protocell are driven only by non-enzymatic reactions. However, upon the chance emergence of replicase ribozymes, enzymatic replication of [Citation31] by RNA nucleotides (see reaction 5) can create new copies of the template as the accuracy of the enzymatic reactions is much higher compared to the non-enzymatic reactions. Nevertheless, parasites will continue to be created, since even enzymatic replication in a primordial world is error-prone () in the absence of proof-reading mechanisms. We define a function
, where
is the error-threshold, to quantify the likelihood of accurate replication under the action of a replicase ribozyme. This depends on the percentage error (
) during such enzymatic replications (see ). Fig S2 shows the variation of this probability of accurate replication for the three types of enzymatic replication processes with different values of the error threshold
. Enzymatic replication of
by DNA nucleotides [Citation32] will similarly create (see reaction 6) a DNA version
of the template
or DNA template
encoding a parasitic sequence, with different likelihoods modulated by the function
.
The three types of DNA templates in our model allow for three different enzymatic replications of DNA templates (with a rate ). Enzymatic replication of
by RNA nucleotides will create mainly parasites but can also produce an RNA template
with probability
(see reaction 7). In the case of
, enzymatic replication by RNA nucleotides will create only parasites (see reaction 8). Finally, auto-catalytic replication of
by RNA nucleotides can recreate a replicase depending on the error-threshold.
Reaction 9 marks the onset of one-way information flow from the DNA template sequence to the replicase it encodes and can be considered to be a manifestation of a primitive central dogma. provides a pictorial representation of the nine types of replication reactions that can occur inside a protocell. Additionally, we also consider degradation of both RNA and DNA molecules. As DNA is more stable than RNA, the degradation rate of DNA () is taken to be lower than RNA (
). We use
for all simulations.
The differential equations determining the time evolution of the abundances of the six different types of molecules, as a result of reactions (1)–(10), are given in the electronic supplementary material.
2.2. Protocellular compartments and population evolution
We considered N = 400 protocells each initially containing 10 RNA templates . To account for variation between different protocells, we assumed that the templates inside different protocells have different replicase creation probabilities, with the probability taken randomly from the range (
). The two key parameters in our model are the creation probability (
) of
via error-prone, non-enzymatic replication and the error-threshold (
) in case of enzymatic reactions. We varied these two parameters and carried out stochastic simulations of the evolving population. The dynamics of the evolving population can be best understood by tracking the average fraction of replicase per protocell, fraction of protocells containing both replicase
, the corresponding DNA strand (
) encoding it and the relative propensity of reaction 9 (see electronic supplementary material for details on how these quantities were estimated).
The three different types of protocellular compartments in our model are distinguished by the distinct selection mechanisms they undergo during the evolution of the protocellular population. gives a schematic representation of the different types of selection mechanisms.
Figure 2. Schematic representation of three different modes of protocellular competition: ,
corresponds to the
and
protocell.
denotes the total number of strands inside the
protocell,
is the upper limit of
and
is the fitness of the protocell. Vesicles: if
exceeds the upper limit
, while
,
will divide into two daughter vesicles and
is eliminated. Water-in-oil droplets:
and
are equally likely to eliminate each other through a random selection process. The surviving droplet divides into two daughter droplets. Coacervate droplets: if
contains larger number (or fraction) of ribozymes (
) compared to
;
and
is more likely to eliminate
and divide into two daughter protocells.
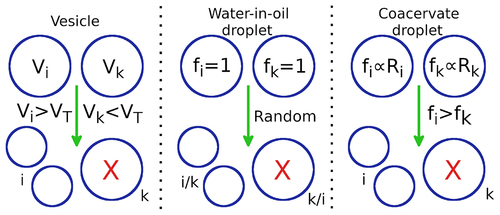
In the first scenario, our choice of protocellular compartments made out of water-in-oil droplets was inspired by recent RNA host-parasite experiments [Citation40–44]. In the experiments, the protocellular compartments are subjected to periodic washout-mixing cycles where a fraction of droplets is randomly removed from the population followed by supply of empty compartments which are then mixed with existing compartments leading to random redistribution of components from the filled to empty compartments. We model this system by considering pairwise competition between protocells that is initiated whenever the number of strands inside all droplets lies in the range (). During such competition, each droplet competes with another droplet chosen at random from the population; one of those two is then randomly selected to be eliminated, while the other divides into two daughter protocells with components of the surviving protocell being randomly distributed between the two daughter droplets.
As another alternative, we consider the competition between protocells whose fitness is determined by functional molecules like ribozymes encapsulated in it. Such genotype–phenotype coupling is an essential feature of Darwinian evolution since internal functional components can provide a fitness advantage thereby ensuring preferential selection of such protocells. Coacervate droplets formed by liquid–liquid phase separation [Citation45–48] can be considered to be an ideal candidate for those types of protocells. Such membraneless droplets can selectively partition biomolecules, sequester long RNA molecules [Citation49], undergo growth and division [Citation50] under environmental cycling [Citation51], support key prebiotic processes like catalysis [Citation45], template-directed primer extension [Citation52] and ligation [Citation53] and show enhanced catalytic activity of encapsulated ribozymes [Citation45,Citation53,Citation54]. Moreover, the presence of active ribozymes inside coacervate droplets has been shown to modulate droplet properties [Citation53,Citation55] thereby establishing a genotype–phenotype linkage and conferring on such droplets a potential fitness advantage under certain environmental conditions.
Therefore, in our coacervate droplet model, we consider a competition similar to the water-in-oil droplet model but with selection dependent not just on the number of strands but on the fraction of replicase ribozymes inside the droplet. This amounts to defining a fitness function for each droplet involved in pairwise competition with
. The droplet selected on the basis of its fitness divides into two daughter droplets, one of which replaces the droplet eliminated during pairwise competition.
Finally, vesicular compartments with lipid bilayer membranes were also considered. The volume of the vesicle is determined by the number but not the nature of strands inside it and vesicle division occurs when the number of strands reach an upper bound set to . During the division process, the components of the vesicle are randomly distributed among the two daughter vesicles, while an existing vesicle is eliminated with a probability proportional to the difference in the number of strands between itself and the dividing vesicle. This results in the preferential elimination of smaller vesicles [Citation56] while keeping the population size fixed.
These three different types of competition between protocells lead to distinct conditions for the dominance of protocells containing a replicase ribozyme whose synthesis is brought about primarily through template-directed, replicase-catalysed replication of a DNA sequence encoding the enzyme.
3. Results
The presence of DNA nucleotides inside a single protocell can enhance the process of replicase creation and ensure sustained growth of strands. When enzymatic reactions are activated due to the chance emergence of a replicase, the presence of even a single DNA template ensures rapid growth of both DNA and RNA strands that further facilitate replicase production (see section 5 of electronic supplementary material for details). In the sub-sections below, we describe how the encapsulation of strands in three different types of protocellular compartments affects the outcome of competition between protocells in an evolving population.
3.1. Water-in-oil droplet model
shows the heatmaps at equilibrium for the average fraction of replicase per droplet (panel A), fraction of droplets containing both replicase & the DNA template (
) encoding it (panel B); and the relative propensity of reaction 9 (panel C) which provides a measure of the efficacy of replicase creation from its encoded DNA template. A very low non-enzymatic probability of replicase creation (
) is not favourable for ensuring the presence of both
and
in majority of droplets (see lower part of . A low value of
leads to low copy numbers of both
and
through non-enzymatic processes which primarily lead to the creation of parasites (
) or parasite encoding DNA (
). The enzymatic creation of replicases through reaction 9 is suppressed since that pathway depends on the easy availability of both
and
. Even when an increase in the error threshold increases the probability of creation of
and
through reactions 5–7, it does not have an impact because of the suppression in creation of
and
ensures that the reactant concentrations for 5–7 remain quite low. Even if the probability of non-enzymatic creation of replicases is relatively high (upper left region of ), low enzymatic replication fidelity due to very low error thresholds makes parasite creation through enzymatic reaction channels 5–9 more likely, leading to lower average replicase fraction per droplet (see upper left region of .
Figure 3. Stochastic simulation of a population of water-in-oil droplets containing strands. Heatmaps for A: average fraction of replicase ribozymes per droplet; B: fraction of droplets containing both replicase () and replicase encoding DNA template (
); C: average reaction propensity of reaction 9 per droplet; with different non-enzymatic ribozyme creation probabilities and error thresholds of enzymatic replications. The heatmaps are generated by taking both ensemble average and time average of the quantities at equilibrium.
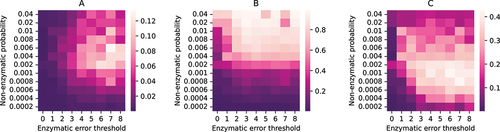
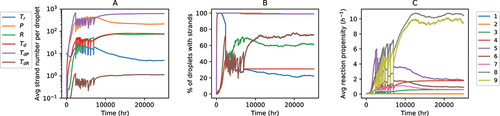
When the non-enzymatic replicase creation probability () as well as the error threshold (
) are high, replicases are produced with a high likelihood both non-enzymatically and enzymatically since the replicase phenotype is more robust to replication errors. However, the resultant replicases are predominantly used to catalyze reactions 5–7 as evident from the higher reaction propensities of these reactions in the upper right region of Fig S5(B). Replicases available for catalyzing their own formation are relatively fewer, and reaction 9 is sub-dominant (see upper right region of ). Moreover, since
and
are created with higher fidelity in this region of parameter space, the relative fraction of R is comparatively low as seen in . In contrast, reaction 9 is dominant for moderately low non-enzymatic replicase creation probability and moderate-to-high error-threshold (see centre-right region of leading to a comparatively larger average fraction of replicases per protocell. shows the time evolution of the average number of different strands () for a point lying in this region. The percentage of droplets containing both
and
(DNA template encoding
) is
() and the average propensity of reaction 9, responsible for replicase creation using a DNA template encoding the replicase, is large (). Even though parasites continue to be formed, primarily through reaction 8, efficient creation of replicases both non-enzymatically and enzymatically is sufficient to ensure that close to a majority of protocells contain both
and
. The continuous regeneration of both
and
can be sustained in the population even if the non-enzymatic reactions are eventually switched off after the onset of enzymatic replication of
(see Fig S6). Intriguingly, we also find symmetry breaking between the number of
and
with the average number of former and latter being
per droplet and
per droplet respectively (see ). The crucial role played by reaction 9 in sustaining replicase formation and aiding the proliferation of droplets containing both
and
is evident from Fig S7 which shows the outcome of switching off reaction 9. Doing so prevents the asymmetry in copy numbers from emerging drastically reduces the average number of both
and
to below one per droplet and the percentage of droplets containing both
and
to
% of the population. This indicates that replicase creation through non-enzymatic processes alone cannot drive the evolution of the protocellular population towards increasing functional complexity.
Figure 4. Stochastic simulation of a population of water-in-oil droplets (). Time evolution of the A: Average number of different types of strands per droplet; B: Percentage of droplets containing different types of strands; C: Average reaction propensities of the nine types of reactions per droplet. Panels A and B have the same legends.
3.2. Coacervate droplet model
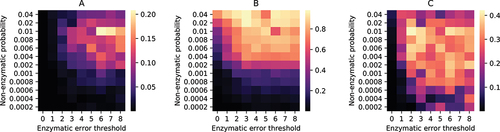
In the case of coacervate droplets, selection pressure acts in favour of droplets containing more ribozymes. The region of parameter space where the fraction of droplets containing both &
exceeds
50% also overlaps with the region where the reaction propensity of 9 is large () and is characterized by moderately low values (
) of non-enzymatic creation probability of a replicase.
creation inside coacervate droplets can be amplified by harnessing the products (
and
) of the replicase catalysed reaction 6 and utilizing them to drive the non-enzymatic reactions 3–4 occurring through reaction channels Ch-1 and Ch-3. Hence, replicase creation through reaction 9 leads to a positive feedback that enhances the propensity of non-enzymatic reactions and induces the creation of even more replicases. Droplets which exploit such positive feedback loops resulting from creation of
&
will be preferentially selected (see Fig S8, Fig-S9(B) and the text below it in the electronic supplementary material) over droplets containing only parasites and can flourish even after non-enzymatic reactions are turned off (Fig S10). The symmetry breaking between
&
is also observed in this region (Fig S9(A)), with the asymmetry between DNA templates and replicases vanishing in the absence of reaction 9 (see Fig S11).
Figure 5. Stochastic simulation of a population of coacervate droplets containing strands. Heatmaps for A: average fraction of replicase ribozymes per droplet; B: fraction of droplets containing both replicase () and replicase encoding DNA template (
); C: average reaction propensity of reaction 9 per droplet; with different non-enzymatic ribozyme creation probabilities and error thresholds of enzymatic replications. The heatmaps are generated by taking both ensemble average and time average of the quantities at equilibrium.
3.3. Vesicle model
For such compartments, the selection of a protocell depends only on the number but not on the type of strands in it, unlike the previous case. Since replicase formation is favoured both non-enzymatically (through reactions 1 and 3) and enzymatically (through reaction 9) for high and high
respectively, the fraction of replicase per protocell increases with the non-enzymatic replicase creation probability (
) and error threshold (
). Surprisingly, the parameter region of highest replicase fraction per vesicle is distinct from the region with the highest propensity of reaction 9, which occurs for low
(see Fig S12). For high
, reactions 5–8 collectively dominate over reaction 9 and the non-enzymatic reactions (compare Fig S12 and S13). Moreover, they use up replicases as catalysts producing parasites, or templates that feed the non-enzymatic reactions (see ); thereby reducing the propensity of reaction 9 which depends on the presence of both
and
. For low error-thresholds, the fidelity of replicase creation through reaction 9 is reduced thereby creating a negative feedback loop that reduces the propensity of enzymatic reactions 5–8 and leading to the decrease in number of both RNA and DNA templates. In this regime, the contribution of non-enzymatic reactions to the creation of replicase-encoding DNA template can be significant at high
(see Fig-S14 in the electronic supplementary material) and when those reactions are switched off, the average number of
per vesicle drops below 1 and the percentage of vesicles containing
drops below 50% (see Fig S15 in the electronic supplementary material). Even though reaction 9 is sub-dominant compared to other enzymatic reactions (see Fig S14(C)), it is essential for ensuring that the average number of replicases inside a vesicle exceeds the number of DNA templates encoding the replicase (see Fig S16 and section 8 of electronic supplementary material).
4. Discussion
Evolution of a protocellular population, aided by the coupling between growth in the number of internal components and division, can occur through physical processes only [Citation25,Citation56]. However, the onset of Darwinian evolution requires heritable and selectively advantageous phenotypes encoded in sequences to be accurately replicated so that they can spread through the population. In this work, we show how the transition to an epoch characterized by Darwinian evolution can be facilitated by the presence of DNA nucleotides. The RNA-templated creation of a DNA sequences using DNA nucleotides leads to new reaction pathways that increase the likelihood of replicase formation through both non-enzymatic and enzymatic channels. Once enzymatic pathways are established, they can help in sustainably creating replicases by increasing both the speed and accuracy of replicase formation. Such enzymatic replicase creation induces symmetry breaking in the number of replicases and replicase-encoding DNA templates, increasing the copy number of the former relative to the latter.
Changing the nature of the protocellular compartment affects the mode of selection and consequently the ability of protocells containing replicases to spread through the population. It is most desirable to obtain sustained replication and proliferation of a protocell containing both the encoded DNA template () and corresponding replicase (
) for low error thresholds and low values of non-enzymatic replicase creation probability. These conditions are satisfied for coacervate and water-in-oil droplet models, and we therefore conclude that those compartments will be more effective in ensuring the dominance of protocells containing
and
in high and low copy numbers, respectively. Although we used coacervates to demonstrate the importance of preferential selection of ribozyme encapsulating protocells on the evolution of the population, those results are applicable for any protocellular compartment that allows for genotype–phenotype coupling. Examples of such coupling have been previously demonstrated for fatty acid vesicles [Citation25,Citation57].
Our model does not incorporate enzymatic reactions that can create new replicase-encoding DNA templates () with high fidelity; such templates can be created through non-enzymatic reactions only. That is, most likely, the reason for the observed symmetry-breaking between the number of DNA-encoded replicases (
) and replicases (
) seen in and Fig S9(A), Fig S14(A) in the Supplementary Information file. Although we observed sustenance of protocells that contain both
and
in the population, such sustenance is possible due to the low degradation rates of
molecules. Hypothetically, the two alternate pathways for creating
enzymatically involves replication of
and
with DNA monomers. However, as discussed earlier, it is extremely difficult to replicate complex-folded RNA molecules like the replicase
. The alternative pathway involving DNA-templated DNA replication requires the presence of a DNA-dependent DNA-polymerase ribozyme. The 38–6 polymerase ribozyme generated in RNA evolution experiments is far less efficient in catalysing DNA-templated polymerization of DNA sequence using DNA monomers, capable of extending only very short, C-rich primers [Citation33]. Nevertheless, if we assume that the polymerase is also capable of catalysing the creation of new replicase-encoding DNA templates of adequate length, that would necessitate the addition of a new reaction to our model of the form
. Simulations including this reaction in the water-in-oil droplet model indicate that the fraction of protocells containing both
and
increases significantly compared to the case where the above reaction is absent (
). Moreover, in such a situation, the symmetry breaking between the number of replicases and DNA templates encoding replicases also disappears. Whether or not enzymatic synthesis of ribozyme encoding DNA templates would have been possible during the early stages of origin of life remains an open question for future experiments to address. Hence, a detailed investigation of this new model (with
) is beyond the scope of this work.
Even though several aspects of our model can be subject to experimental validation, we believe the most compelling test would be to demonstrate the efficacy of auto-catalytic replicase creation through reaction 9 inside a protocell. Although ribozyme-catalysed replication of sequences longer than 100 nucleotides has not yet been experimentally observed, a possible alternative approach might involve ribozyme-catalysed replication of different fragments of a complete DNA template encoding the ribozyme. The RNA fragments generated can then self-assemble to form the whole ribozyme [Citation58–60], perhaps with the help of auto-catalytic networks of the smaller ribozyme fragments [Citation61]. The versatility of the replicase thus created and its role in catalysing several other reactions, especially in the presence of both RNA and DNA nucleotides, can create a self-sustained chemical system inside a protocell. Although we focused on replicase creation only in this work, other functional ribozymes can also be produced in a similar fashion. We envisage a scenario where multiple functional ribozymes, each encoded by their specific DNA templates, gradually emerge, providing an increasing selective advantage to the protocell encapsulating them and consequently facilitating their proliferation in the population.
The epoch of evolution discussed here predates the Darwinian transition and is also likely to have been of a communal nature [Citation62] characterized by widespread horizontal transfer of sequence elements across protocells. It would be interesting to explore how such horizontal transfer could have driven the evolution towards increasing complexity characterized by the presence of functionally diverse components inside such progenotes. We hope that our work, which establishes a proof of concept for the emergence and proliferation of encoded heritable phenotypes, will motivate the design of novel in vitro evolution experiments that will eventually help in further unravelling the mystery of the origin of life.
Authors’ contributions
S.R.: conceptualization, data curation, formal analysis, investigation, methodology, software, visualization, writing-original draft, writing-review and editing; S.S.: conceptualization, formal analysis, methodology, resources, supervision, writing-original draft, writing-review and editing.
Supplementary_RNA_Biology.pdf
Download PDF (1.2 MB)Acknowledgments
SR was supported by an INSPIRE graduate fellowship given by SERB, India.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All codes used to generate the results in the manuscript and in the electronic supplementary material are available at this https://github.com/suvamroy/Codes/tree/master/RNA_DNA_WorldGithub link.
Supplemental data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2024.2355391.
Additional information
Funding
References
- Brenner S, Jacob F, Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961 May;190(4776):576–581. doi: 10.1038/190576a0
- Gros F, Hiatt H, Gilbert W, et al. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature. 1961 May;190(4776):581–585. doi: 10.1038/190581a0
- Kruger K, Grabowski PJ, Zaug AJ, et al. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982 nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7
- Stark BC, Kole R, Bowman EJ, et al. Ribonuclease p: an enzyme with an essential RNA component. Proc Natl Acad Sci. 1978 Aug;75(8):3717–3721. doi: 10.1073/pnas.75.8.3717
- Guerrier-Takada C, Gardiner K, Marsh T, et al. The RNA moiety of ribonuclease p is the catalytic subunit of the enzyme. Cell. 1983 dec;35(3):849–857. doi: 10.1016/0092-8674(83)90117-4
- Matthew WP, Gerland B, John DS. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009 May;459(7244):239–242. doi: 10.1038/nature08013
- Cafferty BJ, Fialho DM, Khanam J, et al. Spontaneous formation and base pairing of plausible prebiotic nucleotides in water. Nat Commun. 2016 Apr;7(1):11328. doi: 10.1038/ncomms11328
- Becker S, Feldmann J, Wiedemann S, et al. Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides. Science. 2019 Oct;366(6461):76–82. doi: 10.1126/science.aax2747
- Xu J, Chmela V, Green NJ, et al. Selective prebiotic formation of RNA pyrimidine and DNA purine nucleosides. Nature. 2020 June;582(7810):60–66. doi: 10.1038/s41586-020-2330-9
- Xu J, Green NJ, Russell DA, et al. Prebiotic photochemical coproduction of purine ribo- and deoxyribonucleosides. J Am Chem Soc. 2021 Sep;143(36):14482–14486. doi: 10.1021/jacs.1c07403
- Adamala K, Szostak JW. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2013 Nov;342(6162):1098–1100. doi: 10.1126/science.1241888
- Yeonwha Song E, Ivanhoe Jiménez E, Lin H, et al. Prebiotically plausible RNA activation compatible with ribozyme-catalyzed ligation. Angewandte Chemie. 2020 Dec;60(6):2952–2957. doi: 10.1002/anie.202010918
- Jauker M, Griesser H, Richert C. Copying of RNA sequences without pre-activation. Angewandte Chemie. 2015 Oct;54(48):14559–14563. doi: 10.1002/anie.201506592
- Damer B, Deamer D. The hot spring hypothesis for an origin of life. Astrobiology. 2020 Apr;20(4):429–452. doi: 10.1089/ast.2019.2045
- Roy S, Sengupta S. The effect of environment on the evolution and proliferation of protocells of increasing complexity. Life. 2022 Aug;12(8):1227. doi: 10.3390/life12081227
- Roy S, Bapat NV, Derr J, et al. Emergence of ribozyme and tRNA-like structures from mineral-rich muddy pools on prebiotic earth. J Theor Biol. 2020 Dec;506:110446. doi: 10.1016/j.jtbi.2020.110446
- Leu K, Kervio E, Obermayer B, et al. Cascade of reduced speed and accuracy after errors in enzyme-free copying of nucleic acid sequences. J Am Chem Soc. 2012 Dec;135(1):354–366. doi: 10.1021/ja3095558
- Dingle K, Ghaddar F, Šulc P, et al. Phenotype bias determines how natural RNA structures occupy the morphospace of all possible shapes. Mol Biol Evol. 2021 Sep;39(1). doi: 10.1093/molbev/msab280
- Shah V, de Bouter J, Quinn P, et al. Survival of RNA replicators is much easier in protocells than in surface-based, spatial systems. Life. 2019 Aug;9(3):65. doi: 10.3390/life9030065
- Kusumoto-Matsuo R, Kanda T, Kukimoto I. Rolling circle replication of human papillomavirus type 16 DNA in epithelial cell extracts. Genes Cells. 2010 Nov;16(1):23–33. doi: 10.1111/j.1365-2443.2010.01458.x
- Daròs J-A, Elena SF, Flores R. Viroids: an Ariadne's thread into the RNA labyrinth. EMBO Rep. 2006 Jun;7(6):593–598. doi: 10.1038/sj.embor.7400706
- Flores R, Gas M-E, Molina-Serrano D, et al. Viroid replication: Rolling-circles, enzymes and ribozymes. Viruses. 2009 Sep;1(2):317–334. doi: 10.3390/v1020317
- Tupper AS, Higgs PG. Rolling-circle and strand-displacement mechanisms for non-enzymatic RNA replication at the time of the origin of life. J Theor Biol. 2021 oct;527:110822. doi: 10.1016/j.jtbi.2021.110822
- Rivera-Madrinan F, Di Iorio K, Higgs PG. Rolling circles as a means of encoding genes in the RNA world. Life. 2022 Sep;12(9):1373. doi: 10.3390/life12091373
- Roy S, Sengupta S. Evolution towards increasing complexity through functional diversification in a protocell model of the RNA world. Proc R Soc B. 2021 Nov;288(1963). doi: 10.1098/rspb.2021.2098
- Takeuchi N, Hogeweg P, Kaneko K. The origin of a primordial genome through spontaneous symmetry breaking. Nat Commun. 2017 Aug;8(1). doi: 10.1038/s41467-017-00243-x
- Rajamani S, Vlassov A, Benner S, et al. Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Origins Life Evol Biosphere. 2007 Nov;38(1):57–74. doi: 10.1007/s11084-007-9113-2
- Huang W, James PF. One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J Am Chem Soc. 2006 Jul;128(27):8914–8919. doi: 10.1021/ja061782k
- Hassenkam T, Damer B, Mednick G, et al. AFM images of viroid-sized rings that self-assemble from mononucleotides through wet–dry cycling: Implications for the origin of life. Life. 2020 Nov;10(12):321. doi: 10.3390/life10120321
- Leu K, Obermayer B, Rajamani S, et al. The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucleic Acids Res. 2011 June;39(18):8135–8147. doi: 10.1093/nar/gkr525
- Horning DP, Joyce GF. Amplification of RNA by an RNA polymerase ribozyme. Proc Natl Acad Sci. 2016 Aug;113(35):9786–9791. doi: 10.1073/pnas.1610103113
- Samanta B, Joyce GF. A reverse transcriptase ribozyme. Elife. 2017 Sep;6. doi: 10.7554/eLife.31153
- Horning DP, Bala S, Chaput JC, et al. RNA-catalyzed polymerization of deoxyribose, threose, and arabinose nucleic acids. ACS Synth Biol. 2019 May;8(5):955–961. doi: 10.1021/acssynbio.9b00044
- Kervio E, Claasen B, Steiner UE, et al. The strength of the template effect attracting nucleotides to naked DNA. Nucleic Acids Res. 2014 May;42(11):7409–7420. doi: 10.1093/nar/gku314
- Bapat NV, Rajamani S. Effect of co-solutes on template-directed nonenzymatic replication of nucleic acids. J Mol Evol. 2015 Oct;81(3–4):72–80. doi: 10.1007/s00239-015-9700-1
- Attwater J, Raguram A, Morgunov AS, et al. Ribozyme-catalysed RNA synthesis using triplet building blocks. Elife. 2018 May;7. doi: 10.7554/eLife.35255
- Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706
- SantaLucia J, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct. 2004 June;33(1):415–440. doi: 10.1146/annurev.biophys.32.110601.141800
- Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2009 Oct;38(suppl_1):D280–D282. doi: 10.1093/nar/gkp892
- Ichihashi N, Usui K, Kazuta Y, et al. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat Commun. 2013 Oct;4(1). doi: 10.1038/ncomms3494
- Bansho Y, Furubayashi T, Ichihashi N, et al. Host–parasite oscillation dynamics and evolution in a compartmentalized RNA replication system. Proc Natl Acad Sci. 2016 Mar;113(15):4045–4050. doi: 10.1073/pnas.1524404113
- Yoshiyama T, Ichii T, Yomo T, et al. Automated in vitro evolution of a translation-coupled RNA replication system in a droplet flow reactor. Sci Rep. 2018 Aug;8(1). doi: 10.1038/s41598-018-30374-0
- Furubayashi T, Ueda K, Bansho Y, et al. Emergence and diversification of a host-parasite RNA ecosystem through Darwinian evolution. Elife. 2020 July;9. doi: 10.7554/eLife.56038
- Mizuuchi R, Furubayashi T, Ichihashi N. Evolutionary transition from a single RNA replicator to a multiple replicator network. Nat Commun. 2022 Mar;13(1). doi: 10.1038/s41467-022-29113-x
- Drobot B, Iglesias-Artola JM, Le Vay K, et al. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat Commun. 2018 Sep;9(1). doi: 10.1038/s41467-018-06072-w
- Christine DK. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc Chem Res. 2012 Feb;45(12):2114–2124. doi: 10.1021/ar200294y
- Poudyal RR, Pir Cakmak F, Keating CD, et al. Physical principles and extant biology reveal roles for RNA-containing membraneless compartments in origins of life chemistry. Biochemistry. 2018 Mar;57(17):2509–2519. doi: 10.1021/acs.biochem.8b00081
- Abbas M, Lipiński WP, Wang J, et al. Peptide-based coacervates as biomimetic protocells. Chem Soc Rev. 2021;50(6):3690–3705. doi: 10.1039/D0CS00307G
- Donau C, Späth F, Sosson M, et al. Active coacervate droplets as a model for membraneless organelles and protocells. Nat Commun. 2020 Oct;11(1). doi: 10.1038/s41467-020-18815-9
- Zwicker D, Seyboldt R, Weber CA, et al. Growth and division of active droplets provides a model for protocells. Nat Phys. 2016 Dec;13(4):408–413. doi: 10.1038/nphys3984
- Fares HM, Marras AE, Ting JM, et al. Impact of wet-dry cycling on the phase behavior and compartmentalization properties of complex coacervates. Nat Commun. 2020 Oct;11(1). doi: 10.1038/s41467-020-19184-z
- Poudyal RR, Guth-Metzler RM, Veenis AJ, et al. Template-directed RNA polymerization and enhanced ribozyme catalysis inside membraneless compartments formed by coacervates. Nat Commun. 2019 Jan;10(1). doi: 10.1038/s41467-019-08353-4
- Le Vay K, Salibi E, Basusree Ghosh T-YDT, et al. Ribozyme activity modulates the physical properties of RNA–peptide coacervates. eLife. 2023 June;12:e83543. doi: 10.7554/eLife.83543
- Poudyal RR, Keating CD, Bevilacqua PC. Polyanion-assisted ribozyme catalysis inside complex coacervates. ACS Chem Biol. 2019 June;14(6):1243–1248. doi: 10.1021/acschembio.9b00205
- Nakashima KK, van Haren MHI, André AAM, et al. Active coacervate droplets are protocells that grow and resist Ostwald ripening. Nat Commun. 2021 Jun;12(1). doi: 10.1038/s41467-021-24111-x
- Chen IA, Roberts RW, Szostak JW. The emergence of competition between model protocells. Science. 2004 Sep;305(5689):1474–1476. doi: 10.1126/science.1100757
- Adamala K, Jack WS. Competition between model protocells driven by an encapsulated catalyst. Nat Chem. 2013 May;5(6):495–501. doi: 10.1038/nchem.1650
- Hayden EJ, Lehman N. Self-assembly of a group i intron from inactive oligonucleotide fragments. Chem Biol. 2006 Aug;13(8):909–918. doi: 10.1016/j.chembiol.2006.06.014
- Vaidya N, Manapat ML, Chen IA, et al. Spontaneous network formation among cooperative RNA replicators. Nature. 2012 Oct;491(7422):72–77. doi: 10.1038/nature11549
- Zhou L, O’Flaherty DK, Szostak JW. Assembly of a ribozyme ligase from short oligomers by nonenzymatic ligation. J Am Chem Soc. 2020 Aug;142(37):15961–15965. doi: 10.1021/jacs.0c06722
- Ameta S, Kumar M, Chakraborty N, et al. Multispecies autocatalytic RNA reaction networks in coacervates. Communications Chemistry. 2023 May;6(1). doi: 10.1038/s42004-023-00887-5
- Woese C. The universal ancestor. Proc Natl Acad Sci. 1998 June;95(12):6854–6859. doi: 10.1073/pnas.95.12.6854
