ABSTRACT
Macroautophagy/autophagy has classically been recognized for its vital role in supporting cellular survival during various stresses. However, emerging work has demonstrated that selective autophagy has an impact on diverse cell biological processes by mediating the degradation of various cellular contents during normal cellular homeostasis. We recently established that selective autophagy supports cell migration by promoting the turnover of integrin-based cell-matrix adhesion sites, or focal adhesions (FAs). The autophagy cargo receptor NBR1 acts as a critical mediator of this pathway by promoting targeting of autophagosomes to FAs, leading to their disassembly via the sequestration of FA proteins. Our results demonstrate FAs as a new cellular target for selective autophagy.
Selective autophagy mediates the turnover of large cellular components, organelles and macromolecular assemblies in a tightly coordinated fashion. Initially identified as a fundamental pathway in protein and organelle homeostasis, selective autophagy is becoming increasingly recognized for its ability to fine-tune diverse cell biological processes. We recently have discovered a new role for selective autophagy in facilitating cell migration by supporting the turnover of FAs.
Cell migration is a highly integrated process requiring complex spatiotemporal coordination of multiple pathways throughout the cell. Notable among these are FAs, which consist of multiple scaffolding and signaling proteins that bridge the extracellular matrix to the actin cytoskeleton. FAs have a critical role in generating traction required for forward cellular movement, but they must also disassemble to enable productive displacement of the cell body. Multiple studies have demonstrated that autophagy supports the motility of diverse cell types, but the underlying mechanisms have remained largely unknown. To expand on these previous findings we first sought to better characterize the precise nature of the motility defect in autophagy-deficient cells. Using live-cell imaging to perform single-cell tracking studies of migrating cells, we demonstrated that autophagy-deficient cells migrate at a significantly slower rate than autophagy-competent cells. In addition, our immunofluorescence studies showed that autophagy-deficient cells possess enlarged FAs. Because FAs are key determinants of migratory speed, these findings led to the hypothesis that autophagy inhibition stabilizes FAs, hence leading to decreased forward migration rates. High-resolution, live-cell confocal microscopy was used to directly image dynamic FAs in migrating cells and further dissect how autophagy regulates the assembly and disassembly of these cell-matrix contacts. These studies revealed that autophagy-defective cells exhibit decreased rates of FA assembly and disassembly, as well as increased FA lifetime, overall demonstrating that FAs are stabilized by autophagy inhibition.
To determine if autophagy affects FA turnover by promoting disassembly, we then asked if and when autophagosomes associate with dynamic FAs in migrating cells. Using multiple imaging methods including high-resolution spinning-disk confocal microscopy, total internal reflection fluorescence microscopy, and super-resolution structured illumination microscopy, we found that autophagosomes localize with high spatial resolution to FAs. Detailed quantification of the kinetics of this targeting revealed that autophagosomes and/or phagophores (i.e., autophagosome precursors) localize to FAs primarily during the disassembly phase of FA turnover. In addition to the striking temporal specificity with which autophagosomes target FAs, autophagosomes are significantly enriched at FAs compared to non-FA areas in the leading edge region of migrating cells, indicative of selective autophagic targeting. Moreover, multiple FA components localize within the autophagosomes of migrating cells, further corroborating that phagophores actually capture FA components (). Thus, autophagy specifically facilitates FA disassembly by promoting the sequestration of FA proteins, resulting in the destabilization of cell-matrix adhesions.
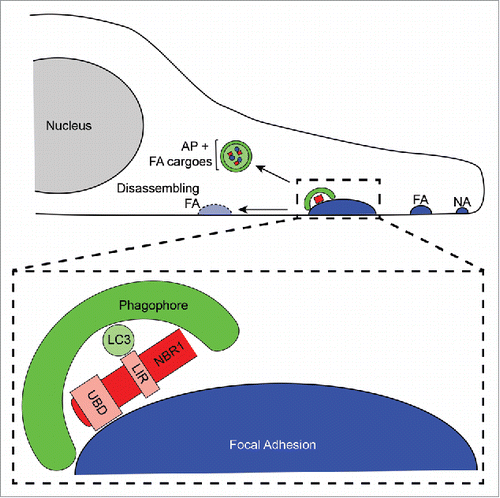
Figure 1. Selective autophagy promotes focal adhesion disassembly during cell migration. Forward, leading edge protrusions in migrating cells are stabilized by the formation of nascent adhesions (NA), which bind the extracellular matrix. NAs grow into mature, stable FAs through the addition of numerous scaffolding and signalling proteins. Subsequently, FAs must disassemble to allow the cell body to productively move forward, and NBR1-dependent selective autophagy is required for this process. NBR1 interacts with FAs via its ubiquitin binding domain (UBD), and this binding enables targeting of phagophores to FAs through interaction of the LC3 interacting region (LIR) of NBR1 with LC3 on the phagophore membrane. Autophagic targeting of FAs results in sequestration of FA proteins within autophagosomes (AP) and consequent destabilization of FAs leading to their disassembly.
The highly selective capture of specific autophagy substrates is mediated by autophagy cargo receptor proteins, which simultaneously interact with LC3 on the phagophore membrane and with cargoes. The observed specificity of phagophore targeting to FAs and the functional role of autophagy in supporting FA disassembly led us to hypothesize that specific autophagy cargo receptors enable autophagy-dependent FA turnover. We first investigated whether the depletion of the 4 main autophagy cargo receptors, SQSTM1/p62, OPTN (optineurin), CALCOCO2/NDP52, and NBR1, phenocopies the loss of autophagy function during cell motility. Among these, NBR1 was unique in its ability to support motility. Likewise, NBR1 depletion also resulted in impaired FA turnover, and, similar to phagophores and autophagosomes, NBR1 is highly enriched at FAs in the leading edge regions of migrating cells. Collectively, these results indicate that NBR1 and autophagy coordinately function in the same pathway to facilitate FA turnover during cell migration.
Expanding on these results, we obtained several additional lines of evidence substantiating a distinct and specific role for NBR1-mediated selective autophagy in promoting FA turnover. First, NBR1 loss of function disrupts the efficient targeting of phagophores to FAs. Second, gain-of-function studies demonstrate that the ability of NBR1 to enhance FA disassembly requires its LC3 interacting region, a motif crucial for its interaction with phagophore membranes. Third, consistent with its autophagy cargo receptor function, NBR1 biochemically interacts with multiple FA proteins that are also localized to phagophores. Finally, the ability of NBR1 to enhance FA disassembly requires its ubiquitin-binding domain, which is critical for autophagy cargo recognition. Overall, these results show that NBR1 interacts with FAs to facilitate the localized targeting of phagophores to FAs, resulting in disassembly of cell-matrix adhesion sites in migrating cells ().
In summary, our findings reveal a new pathway of selective autophagy that targets leading edge FAs in migrating cells, thus expanding the list of fundamental cellular processes affected by autophagy. Going forward, it will be necessary to dissect and elaborate the molecular and biochemical underpinnings by which NBR1 promotes autophagy-dependent FA turnover. Emerging work showing phosphorylation and ubiquitination as key mediators of substrate-cargo receptor interactions suggests that similar mechanisms may regulate precise spatiotemporal autophagic targeting of FA components to support optimal motility. Studies uncovering the factors that modulate these potential layers of regulation in autophagy-dependent FA disassembly will provide new mechanistic insight into the control of both cell migration and selective autophagy pathways.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Grant support to J.D. includes the NIH (CA126792, CA188404), DOD BCRP (W81XWH-11-1-0130 and W81XWH-12-1-0505), and Samuel Waxman Cancer Research Foundation. C.M.K. was supported by NIH pre-doctoral fellowship F31CA167905.
