Neurogenesis initiates and terminates in a spatially and temporally defined manner. Most neurogenesis occurs during development driven by the cell divisions of neural stem cells, but in some animals new neurons are also produced throughout adulthood. In adults, however, neurogenesis is rather limited; only certain neuron types are produced in only a few brain regions. Better understanding of how neurogenesis terminates once development is complete could help in devising therapies to restore neurogenesis in damaged brain regions as well as in treatment of brain tumors.
Drosophila provides a model system for understanding how neurogenesis terminates because neurogenesis patterns are rather simple and there are great genetic tools available. Like mammals, neurons in the Drosophila brain are generated either directly or indirectly from the asymmetric cell divisions of neural stem cells, known as neuroblasts in Drosophila. During development, neuroblasts divide to ‘self-renew’ and give rise to daughter cells that produce neurons and/or glia. Neuroblasts are specified during embryogenesis and proliferate throughout larval feeding stages and early pupal stages to produce an immense number of molecularly, functionally, and morphologically distinct neuron types. The adult Drosophila brain contains approximately 100,000 neurons, which are organized into functional neural circuits providing adults the ability to navigate, forage, and mate. Because neuroblasts located within the central brain region only divide asymmetrically, their numbers (approximately 100 per brain hemisphere) remain invariant over time. This feature, together with the fact that neuroblasts can be easily and unambiguously identified based on size, position, and molecular marker expression, allows neuroblasts to be tracked over time. Most neuroblasts terminate cell divisions within the first 30 h after pupal formation, except for a small subset (4 of the 100), the mushroom body (MB) neuroblasts, which generate neurons important for memory and learning. MB neuroblasts terminate their divisions approximately 2 days after the other brain neuroblasts (referred to as non-MB neuroblasts), shortly before animals eclose from their pupal case as adults. Unlike mammals and even some other insects, no new neurons are produced in the Drosophila brain during adulthood.
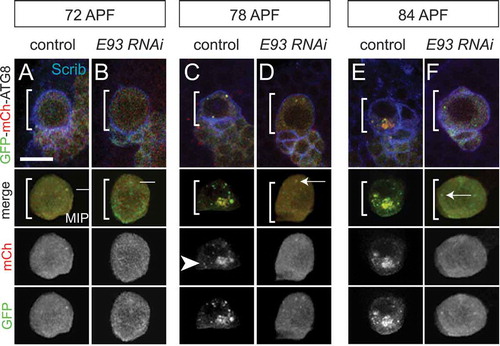
Both apoptosis and macroautophagy/autophagy are required for elimination of MB neuroblasts and termination of MB neurogenesis. If either apoptosis or autophagy is blocked alone, MB neuroblasts persist, but only transiently, and continue generating new neurons, albeit at much reduced rates. But, when both apoptosis and autophagy are blocked together, then MB neuroblasts persist long term into adulthood, most of which continue generating many new MB neurons. A reduction in phosphatidylinositol 3-kinase (PtdIns3K) levels in MB neuroblasts leads to activation of autophagy and affects timing of apoptosis, but it remained unclear how PtdIns3K levels and/or apoptosis are regulated in MB neuroblasts during late pupal stages. To gain further understanding, we carried out a large-scale RNAi screen to identify genes required for termination of MB neurogenesis [Citation1]. One of the genes identified was Eip93F, an ecdysone steroid hormone-induced gene previously shown to be required for autophagic clearance of larval tissues during metamorphosis. Prior to MB neuroblast elimination during late pupal stages, the number and size of autophagosomes and autolysosomes dramatically increases in wild-type control MB neuroblasts (). In contrast, following knockdown of Eip93F in MB neuroblasts, no autophagosomes or autolysosomes were observed and Eip9F3 knockdown MB neuroblasts continued generating new neurons into adulthood (). We further determined that Eip93F downregulates PtdIns3K levels in MB neuroblasts to induce autophagy. However, overexpression of Eip93F is not sufficient to induce autophagy in MB neuroblasts during earlier developmental stages, suggesting other factors are required. It is not yet known what the other factors are, but declining systemic nutrient levels could provide a co-factor required for termination because MB neuroblasts are eliminated during non-feeding pupal stages.
Figure 1. Eip93F is required for autophagosome and autolyosome formation in MB neuroblasts during late stages of pupal development. (a–f) Top, colored overlay of MB neuroblasts (white brackets) at the indicated times. (a,c,e) Wild-type control genotype: worGAL4,UAS-GFP-mCh-Atg8. (b,d,f) Eip93F knockdown (E93 RNAi) genotype: worGAL4,UAS-GFP-mCh-Atg8,UAS-E93 RNAi. Below, colored overlay of cropped, maximum intensity projection of MB neuroblast shown above, with single channel grayscale images below. White arrows indicate autophagomes (mCh and GFP double positive) and arrowheads indicate autolysosomes (mCh only). (a–f) Scale bar: 10 μm. scrib (scribble, blue) is a membrane marker to denote the MB neuroblast. APF, after pupal formation. Reprinted from Current Biology, 29(5), Pahl, M.C., Doyle, S.E., and Siegrist, S.E., E93 Integrates neuroblast intrinsic state with developmental time to terminate MB neurogenesis via autophagy, Figure 4A-F, 2019, with permission from Elsevier.
It also remains unknown why MB neuroblasts require both apoptosis and autophagy for elimination and whether this two-part cell elimination mechanism (autophagy and apoptosis) occurs in other stem cells during development. In MB neuroblasts, autophagy may be necessary for degradation of stem cell ‘self-renewal’ factors and/or organelles that promote proliferation and/or to degrade inhibitors of apoptosis. Important future work will include determining what exactly is inside autophagosomes of MB neuroblasts, where autophagosome formation initiates within MB neuroblasts (i.e., plasma or organelle membrane), and whether systemic and/or local extrinsic factors trigger MB neuroblast autophagy and apoptosis.
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
Reference
- Pahl MC, Doyle SE, Siegrist SE. E93 Integrates neuroblast intrinsic state with developmental time to terminate MB neurogenesis via autophagy. Curr Biol. 2019;29(5):750–762 e753.
