ABSTRACT
Mitochondrial autophagy (mitophagy) selectively degrades mitochondria and plays an important role in mitochondrial homeostasis. In the yeast Saccharomyces cerevisiae, the phosphorylation of the mitophagy receptor Atg32 by casein kinase 2 is essential for mitophagy, whereas this phosphorylation is counteracted by the protein phosphatase Ppg1. Although Ppg1 functions cooperatively with the Far complex (Far3, Far7, Far8, Vps64/Far9, Far10 and Far11), their relationship and the underlying phosphoregulatory mechanism of Atg32 remain unclear. Our recent study revealed: (i) the Far complex plays its localization-dependent roles, regulation of mitophagy and target of rapamycin complex 2 (TORC2) signaling, via the mitochondria- and endoplasmic reticulum (ER)-localized Far complexes, respectively; (ii) Ppg1 and Far11 form a subcomplex, and Ppg1 activity is required to assemble the sub- and core-Far complexes; (iii) association and dissociation between the Far complex and Atg32 are crucial determinants for mitophagy regulation. Here, we summarize our findings and discuss unsolved issues.
Mitochondrial autophagy (mitophagy) contributes to maintaining mitochondrial quality and quantity via selective degradation of damaged or excessive mitochondria. The budding yeast Saccharomyces cerevisiae has served as a model organism for mitophagy as well as autophagy. Recent studies have expanded our understanding of the molecular mechanisms of mitophagy in yeast. Upon mitophagy induction (i.e., a shift from respiratory medium to nitrogen starvation or continuous culture in respiratory medium), the mitophagy receptor Atg32 is phosphorylated at Ser114 by casein kinase 2 (CK2), which is an essential trigger for the interaction between Atg32 and the scaffold/adaptor protein Atg11. The autophagy core machinery is then recruited to initiate autophagosome formation that delivers mitochondria to the vacuole for degradation. Atg32 phosphorylation is counteracted by the yeast equivalent of the mammalian striatin-interacting phosphatase and kinase (STRIPAK) complex consisting of the PP2A-like protein phosphatase Ppg1 and the Far complex (Far3, Far7, Far8, Vps64/Far9, Far10 and Far11). The Far complex was previously shown to be localized to the endoplasmic reticulum (ER) and be involved in regulation of TORC2 signaling. However, the relationship between Ppg1 and the Far complex, and the mechanism by which the ER-localized Far complex regulates dephosphorylation of the mitochondria-resident Atg32 has remained largely unclear.
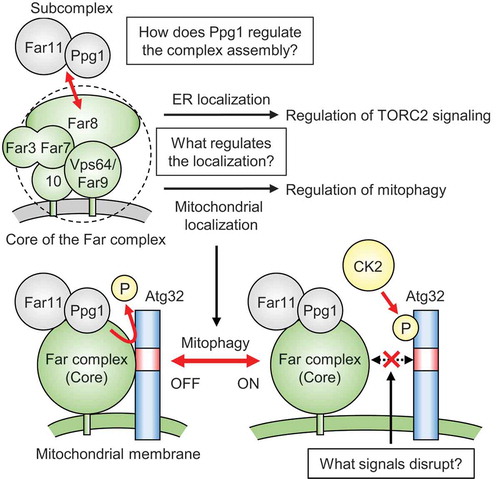
Initially, we reinvestigated the cellular localization of the Far complex using GFP-fused Far proteins [Citation1]. We found that a substantial portion of the Far complex is localized also to the mitochondria and that the tail-anchor (TA) domains of Far9 and Far10 are required for both mitochondrial and ER localization. To distinguish the roles of the mitochondria- and ER-localized Far complexes, we constructed two Vps64/Far9 mutants carrying the TA domains of Tom5 (mitochondrial protein) and Cyb5 (ER protein), which localize specifically to the mitochondria and ER, respectively. This analysis revealed that the former inhibits mitophagy via Ppg1-dependent dephosphorylation of Atg32, and the latter regulates the TORC2 signaling pathway. Thus, the Far complex has distinct and localization-dependent functions.
Next, we systematically analyzed the interaction between Ppg1 and the Far components using immunoprecipitation assays. Ppg1 co-immunoprecipitates with all six Far components, whereas Ppg1-Far11 co-immunoprecipitation is independent of the other components, suggesting that Ppg1 and Far11 form a subcomplex. Although this subcomplex also contains the PP2A scaffolding protein Tpd3, it likely does not contribute to the role of the Far complex at least in dephosphorylation of Atg32. We also found that Ppg1 activity is required for the Far8-Far11 interaction and mitochondria and ER localization of Far11, indicating that Ppg1 regulates not only Atg32 dephosphorylation but also assembly of the sub- and core-Far complexes.
We further explored whether mitophagy-inducing conditions would disrupt the interaction between Ppg1 and the Far complex or among Far components and thereby stimulate Atg32 phosphorylation by CK2. However, we found that such interactions and localization of the Far components are not altered before and after mitophagy induction. These results prompted us to investigate the interaction between the Far complex and Atg32 as an alternative candidate for the key determinant of mitophagy regulation. Several approaches, including in vivo and in vitro assays, demonstrated that Far8 directly interacts with Atg32, and this interaction is weakened under mitophagy-inducing conditions. Moreover, forced interaction between Far8 and Atg32 by their artificial tethering strongly inhibited Atg32 phosphorylation and mitophagy. Taken together, these results suggest that the association and dissociation between the Far complex and Atg32 are crucial determinants for mitophagy regulation.
Our study uncovered several mechanistic issues regarding the Far complex-mediated mitophagy regulation in yeast (). However, the following issues remained unclear. First, what regulates the mitochondria or ER localization of the Far complex? Second, how does Ppg1 regulate the Far complex formation? Third, what signals disrupt the interaction between the Far complex and Atg32 upon mitophagy induction? We speculate that mitophagy induction stimulates dissociation between the Far complex and Atg32 through (de)phosphorylation or other post-translational modifications of the Far components. Future studies on the regulatory mechanisms of the Far complex will elucidate these issues and provide insights into how the mitophagy-inducing signal is sensed and transmitted to Atg32 and the evolutionarily conserved mechanisms of the mammalian STRIPAK complex.
Figure 1. Model for the phosphoregulatory mechanism of Atg32. The Far complex localizes to both the ER and mitochondria, and the tail-anchor domain of Vps64/Far9 determines the localization. The Far complex exhibits localization-dependent regulation of TORC2 signaling at the ER and regulation of mitophagy at the mitochondria. Ppg1 phosphatase activity is required to assemble the Ppg1-Far11 subcomplex and the core complex (Far3, Far7, Far8, Vps64/Far9, Far10). Without stimuli, the mitochondria-localized Far complex mediates Ppg1-dependent Atg32 dephosphorylation via interaction with Atg32. Upon mitophagy stimuli, the interaction between the Far complex and Atg32 is impaired, allowing Atg32 to be phosphorylated by CK2. Unsolved issues are shown in the boxes
Disclosure statement
All authors declare no conflict of interest.
Additional information
Funding
Reference
- Innokentev A, Furukawa K, Fukuda T, et al. Association and dissociation between the mitochondrial Far complex and Atg32 regulate mitophagy. eLife. 2020;9:e63694.
