ABSTRACT
Increasing evidence supports the bona fide function of the coat protein complex II (COPII) machinery in regulating autophagosomes biogenesis during macroautophagy/autophagy induced by nutrient starvation. However, the participation of the COPII machinery in the plant autophagy pathway remains elusive. We recently identified a unique population of COPII vesicles containing AT3G62560/AtSar1d-AT1G02130/AtRabD2a that functions in modulating autuphagosome biogenesis in Arabidopsis thaliana. Proteomic analysis identified the mechanistic connection between autophagy-related (ATG) proteins and a subset of specific COPII paralogs, including AtSar1d. Mutants of AtSar1d affect autophagosome progression and display starvation-related phenotypes. AtSar1d interacts with ATG8 by a non-canonical motif. Cellular and genetic analysis demonstrated that a plant-unique RAB1/Ypt1 homolog AtRabD2a coordinates with AtSar1d to mediate the specific COPII functions in the autophagy pathway. This study identified a plant-specific nexus in regulating autophagosome biogenesis.
KEYWORDS:
In plants, gene duplication events occurred substantially during evolution, generating multiple homologs in the genomes. Notably, the functional divergence of distinct paralogs in organelle biogenesis, plant development and stress response has been implied. For example, the dicotyledonous model plant Arabidopsis encodes a variety of paralogs for the COPII vesiculating machinery, including five Sar1, seven Sec23, three Sec24, two Sec13, and two Sec31, whose functional specificity in COPII vesicle formation and endoplasmic reticulum (ER) protein export have been uncovered. However, the functional heterogeneity of COPII paralogs in plant autophagy remains elusive. In this study [Citation1], we identified a distinct population of COPII vesicles constituting part of the AtSar1d-AtRabD2a nexus that functions in modulating autuphagosome biogenesis in Arabidopsis thaliana, with the following evidence: (1) the plant autophagy-related (ATG) protein interactome identified specific COPII paralogs, and likewise AtSar1d, as interactors under nutrient deprivation; (2) mutants of AtSar1d affect autophagosome biogenesis and display starvation-related phenotypes upon stresses and nutrient deficiency conditions; (3) the interaction between AtSar1d and ATG8 is achieved via a non-canonical motif; (4) localization-based screening identified a plant-unique RAB1/Ypt1 homolog, AtRabD2a, that colocalizes with both AtSar1d and ATG8; and (5) cellular and genetic analysis demonstrated that AtRabD2a is essential for the redirection of the AtSar1d-positive COPII vesicles to phagophores via the ATG1 complex and is indispensable for autophagosome formation.
The endoplasmic reticulum exit sites (ERESs)-derived COPII vesicles, containing five cytosolic components, namely the small GTPase Sar1, inner coat protein Sec23-Sec24, and outer coat protein Sec13-Sec31, are known to function in the classical anterograde-transport from the ER to the Golgi apparatus. Intriguingly, under nutrient deprivation, COPII vesicles delicately participate in the autophagy pathway and autophagosome biogenesis. In yeast, ERESs have been postulated to be the physical and functional core autophagosome biogenesis components. A variety of COPII proteins have been demonstrated to participate in the autophagy pathway, including Ypt1, Sec16, and Sec24. In mammals, distinct COPII homologs seem to exhibit unique functions in autophagosome biogenesis. For example, the specific function of SEC24C in reticulophagy was recently unveiled, albeit the underlying mechanism remains to be explored. The plant model Arabidopsis encodes even more COPII isoforms than mammals, and we thus hypothesized that specific COPII paralogs may be dedicated to the plant autophagy pathway. Large-scale proteomic analysis of the Arabidopsis ATGs interactome identified a variety of COPII and COPI machinery as the interaction partners of AtATG8 and AtATG6. Interestingly, among the five Arabidopsis Sar1 homologs, AtSar1d can be specifically pulled down by AtATG8 upon nitrogen starvation.
Besides AtSar1d, we also identified several other COPII components such as Sec24-like AT3G44340/CEF and AT4G14160/AtSec23f, suggesting the existence of a distinct population of COPII vesicles that function in plant autophagy. Indeed, a loss-of-function mutation of AtSar1d results in the failure of autophagosome progression and vacuolar delivery of the autophagosome. Upon nutrient deprivation, autophagy-related mutants will display early-senescence phenotypes. Consistently, AtSar1d mutants phenocopy ATG7 mutant plants and exhibit autophagy-related phenotypes, while overexpression of AtSar1d-GFP can rescue the defect. A recent study has postulated that the Arabidopsis AtSar1 may interact with ATG8 through a non-canonical motif. We performed a structure modeling prediction and identified a potential motif in AtSar1d, which is a variant region among all the Arabidopsis Sar1 homologs, to be essential for its interaction with ATG8. Although increasing evidence suggests the bona fide role of COPII machinery or COPII vesicles in autophagosome biogenesis, how the vesicles are redistributed from the ER to the phagophore rather than the Golgi apparatus under nutrient-deprivation conditions remains unknown. We hypothesized that a specific RAB GTPase, whose function is well-known for mediating vesicle docking, is essential for this process. Localization screening using an Arabidopsis protoplast system identified AtRabD2a, a close homolog of Ypt1/RAB1 in yeast and mammals, to be substantially colocalized with both AtSar1d and ATG8e. Thus, AtRabD2a bridges the AtSar1d-positive COPII vesicles with phagophores via a mechanistic connection between the COPII inner coat protein SEC23 and the ATG1 complex. Last, dominant-negative mutants of AtRabD2a perturb both autophagosome formation and vacuolar delivery of autophagosomes, indicating that AtRabD2a regulates the autophagosome biogenesis as does AtSar1d.
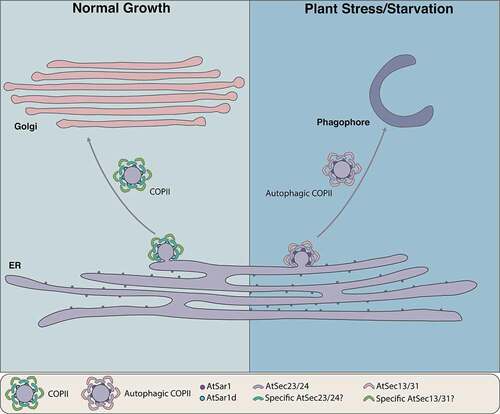
Our current working model () suggests that under normal growth conditions, plant COPII vesicles form to transport the ER-synthesized cargoes to the Golgi apparatus. Upon stresses or nutrient-deficiency conditions, a unique population of COPII vesicles containing a specific SAR1 GTPase, inner coat protein, and outer coat protein, may form and target to the phagophore through a distinct RAB GTPase and act as a membrane source for phagophore progression. Future identification and characterization of components (e.g., soluble N-ethylmaleimide–sensitive factor attachment receptor/SNARE proteins) or cargoes in this subtype of COPII vesicles will broaden our understanding about the precise roles of COPII vesicles in autophagy pathway, and the importance of functional diversification of COPII paralogs in plants.
Figure 1. Working model of the COPII vesicles function in the early secretory and autophagy pathways in plants. Under normal growth condition, the COPII vesicles, consisting of small GTPase AtSar1, inner coat AtSec23-AtSec24, and outer coat AtSec13-AtSec31, regulate the ER to Golgi trafficking. Upon plant stresses or nutrient-deficiency conditions, a AtSar1d-positive unique population of COPII vesicles forms to modulate phagophore progression
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Reference
- Zeng Y, Li B, Ji C, et al. A unique AtSar1D-AtRabD2a nexus modulates autophagosome biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2021;118(17):e2021293118.
