ABSTRACT
General autophagy is an evolutionarily conserved process in eukaryotes, by which intracellular materials are transported into and degraded inside lysosomes or vacuoles, with the main goal of recycling those materials during periods of starvation. The molecular bases of autophagy have been widely described in Saccharomyces cerevisiae, and the specific roles of Atg proteins in the process were first characterized in this model system. Important contributions have been made in Schizosaccharomyces pombe highlighting the evolutionary similarity and, at the same time, diversity of Atg components in autophagy. However, little is known regarding signals, pathways and role of autophagy in this distant yeast. Here, we undertake a global approach to investigate the signals, the pathways and the consequences of autophagy activation. We demonstrate that not only nitrogen but several nutritional deprivations including lack of carbon, sulfur, phosphorus or leucine sources, trigger autophagy, and that the TORC1, TORC2 and MAP kinase Sty1 pathways control the onset of autophagy. Furthermore, we identify an unexpected phenotype of autophagy-defective mutants, namely their inability to survive in the absence of leucine when biosynthesis of this amino acid is impaired.
Abbreviations: ATG: autophagy-related; cAMP: cyclic adenosine monophosphate; cDNA: complementary deoxyribonucleic acid; GFP: green fluorescence protein; Gluc: glucose; Leu: leucine; MAP: mitogen-activated protein; MM: minimal medium; PI: propidium iodine; PKA: protein kinase A; RNA: ribonucleic acid; RT-qPCR: real time quantitative polymerase chain reaction; S. cerevisiae: Saccharomyces cerevisiae; S. pombe: Schizosaccharomyces pombe; TCA: trichloroacetic acid; TOR: target of rapamycin; TORC1: target of rapamycin complex 1; TORC2: target of rapamycin complex 2; YE5S: yeast extract 5 amino acid supplemented.
Introduction
Macroautophagy, hereafter referred to as autophagy, has an important role in differentiation, and it is an evolutionarily conserved process that degrades cellular organelles as well as proteins. It requires rearrangement of membranes to create vesicles that sequester cytosolic lumen containing the organelles/proteins to be degraded: the autophagosome. Its outer membrane will fuse to lysosomes/vacuoles (in yeast and plants), to create the autophagic bodies, to be released and degraded by the proteases of these compartments (for recent reviews, see [Citation1–3]).
Using Saccharomyces cerevisiae (S. cerevisiae) as a model system, a set of genes, termed ATG, were discovered that participate in autophagy. All the yeast mutants lacking Atg proteins lose viability during nitrogen starvation, with 80% viability loss after 5 days of nutrient depletion, and they cannot sporulate. In S. cerevisiae, both the PKA (protein kinase A) and the TOR (Target of Rapamycin) pathways are involved in the regulation of autophagy during conditions of nutrient scarcity (for reviews, see [Citation4–7]).
Regarding the biological roles of autophagy, defects in this process are clearly linked to aging and to diseases (for reviews, see [Citation8,Citation9]). This degradative pathway causes turnover of organelles or proteins, and this autophagic degradation may breakdown proteins into amino acids, to use them again in subsequent protein synthesis pathways when environmental supplies are limiting.
Schizosaccharomyces pombe (S. pombe) has been used in the last years as a new model system to study autophagy. This process was first studied in the fission yeast through the characterization of vacuolar functions, with the identification and characterization of vacuolar proteases [Citation10–16]. With the isolation of the first atg mutants [Citation17], it was early discovered that autophagy is required during nitrogen starvation to supply enough nitrogen intracellularly, explaining the mating defects of these mutants. In general, during nutritional starvation cells exit the normal growth cycle to undertake alternative paths such as sexual differentiation in fission yeast. In particular, nitrogen depletion causes a G1 arrest and triggers sexual differentiation, and in parallel, protein degradation linked to autophagy occurs as well [Citation13]. It was important to demonstrate that, even though autophagy promotes the entry into the sexual differentiation fate, atg mutants are not sterile in the presence of nitrogen [Citation17]. Therefore, the major role of autophagy is to provide a nitrogen supply in the absence of external sources. Furthermore, the group of Takegawa demonstrated that the mating defects of some atg mutants are not complete, proposing that fission yeast is capable of storing a large enough intracellular nitrogen pool to allow partial sporulation under nitrogen-limiting conditions even in the absence of autophagy, which provides the majority of the nitrogen source for protein synthesis during sporulation [Citation18].
Due to all the above, most atg mutants of fission yeast have been identified through screenings of deletion mutant collections searching for defects in mating [Citation19]. Data from the Du lab has shed light on the conservation but at the same time diversity of the autophagy components, demonstrating the usefulness of fission yeast as a unique model for studying autophagy. In fact, the characterization of the molecular bases by which the Atg proteins rule general and/or selective autophagy are currently being unraveled by the Du laboratory.
For instance, they have identified a selective autophagy pathway for the import of soluble cargos into the vacuole which, surprisingly, does not require core Atg proteins for function, contrary to the system described in budding yeast, confirming the unexpected versatility of eukaryotic autophagy mechanisms [Citation20]. Regarding the engulfment of bulky organelles instead of soluble proteins, specific Atg proteins such as Atg20, Atg24 and Atg24b/SPB1711.11 have been shown to participate in the nitrogen starvation-induced autophagy of endoplasmic reticulum and mitochondria [Citation21]. The Du laboratory has also provided mechanistic details on the biochemical principles of the onset of autophagy [Citation22].
Even though most Atg components ruling autophagy in S. pombe have been identified and the participation of autophagy in mating has been largely demonstrated, little is known about the pathways driving the onset of autophagy in this model organism, nor on the role of this degradation system in other physiological processes. Here, we have performed a global study on the signals, the pathways and the consequences of activated autophagy in fission yeast. Using a modified version of the GFP (green fluorescent protein)-Atg8 reporter of autophagy, we show that not only nitrogen depletion but also glucose, sulfate, phosphate or leucine deprivation can trigger autophagy to different extents. We also reveal the unexpected participation of the MAP (mitogen-activated protein) kinase Sty1-driven stress pathway in autophagy regulation, in concert with the well characterized target of rapamycin (TOR) cascades. We also demonstrate that mutants auxotrophic for leucine are extremely dependent on autophagy to survive in front of leucine deprivation, suggesting that the intracellular pools of this amino acid are very limiting.
Results
General autophagy can be triggered not only by nitrogen depletion but also by phosphate, sulfate or glucose starvation
In most organisms, from yeast to human, autophagy has been monitored using the GFP-tagged Atg8 protein as reporter, which allows to locate autophagosomes by microscopy and to quantify autophagy by western blot [Citation18,Citation23]. The reporter designed in Takegawa’s laboratory to study autophagy in fission yeast presents some limitations: the GFP-Atg8 fusion protein is expressed from an episomal plasmid, with a ura4 selection marker under the control of the nmt (no-message in thiamine) promoter. Therefore, it can only be used in MM (minimal media) cultures of strains auxotrophic for uracil under non-repressive conditions, so that the plasmid copy number and the concentration of the reporter (which can change depending on the number of duplications after thiamine withdrawal) vary depending on strains and experimental conditions. We designed a new plasmid to express the fusion protein GFP-Atg8 under the control of the constitutive sty1 promoter from a chimeric gene integrated at the leu1-32 locus. Immunoblot analysis showed the formation of a free GFP band, due to the autophagic delivery of GFP-Atg8 into vacuoles and the subsequent proteolysis that releases the protease-resistant GFP moiety, few hours after nitrogen withdrawal (, without [w/o] N).
Figure 1. Autophagy is triggered not only by nitrogen starvation, but also by depletion of sulfur, phosphorus of carbon sources. (A) GFP-Atg8 cleavage is caused by nitrogen, sulfur or phosphorus starvation. MM cultures from the wild-type strain CC10, expressing GFP-Atg8 under the control of the constitutive sty1 promoter from an integrative plasmid, were collected at the logarithmic phase (-) and 4-8-24 h after switching cells to media without nitrogen (w/o N), sulfur (w/o S) or phosphorus (w/o P) sources. Native extracts were analyzed by western blot using anti-GFP antibody. Ponceau staining of the membrane after immuno-detection is shown as a loading control. Numbers below the panels represent the amount of cleaved GFP (quantified as indicated in Materials and Methods) relative to cleaved GFP after 4 h without nitrogen (with an assigned value of 1). (B) GFP-Atg8 cleavage is also induced by glucose exhaustion. The wild-type strain CC10 was grown in standard YE5S (3% glucose) and modified YE5S (1% glucose). Cells were collected at the logarithmic phase and at the indicated times after reaching an OD600 of 0.5. For glucose-starvation in YE5S 0.08% of glucose, cells were switched at the logarithmic phase from standard YE5S (3%) to YE5S 0.08%. Samples were treated and analyzed as in (A). Numbers below the panels represent the amount of cleaved GFP relative to the amount after 1 day in standard YE5S (3%) (with an assigned value of 1). (C) Scheme of the different cellular pathways studied in this work, related to sensing and responding to nitrogen and glucose deprivation in fission yeast. See text for details.
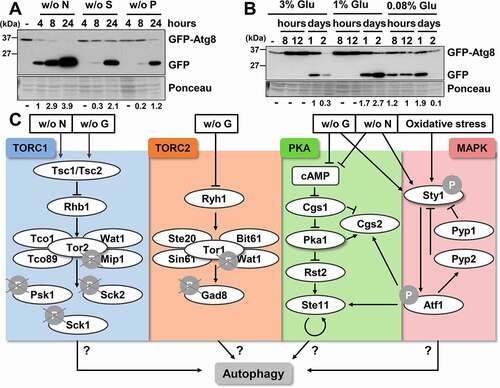
It has been proposed that the conditions that induce autophagy in fission yeast are not the same as those in budding yeast. In S. cerevisiae, the induction of autophagy has been described in response to a variety of nutrient starvation conditions, such as the depletion of nitrogen, carbon, sulfur, phosphorus and zinc [Citation10,Citation24,Citation25]. However, in S. pombe, autophagy is mainly related with nitrogen starvation, and no evidence of induction has been detected upon glucose depletion or through rapamycin treatment [Citation18]. We decided to test, using our new integrative reporter, whether other nutrient deprivation conditions could trigger autophagy in fission yeast. As shown in , we also detected GFP-Atg8 cleavage after depriving the sulfur or phosphorus sources, although with slower kinetics and less intensity than in the absence of nitrogen. We then tested whether glucose depletion can trigger autophagy. We have previously demonstrated that glucose concentrations below 1%, which do not allow maximal optical densities of cultures due to glucose exhaustion, promote longer lifespan probably due to higher respiratory rates [Citation26]. As shown in , when cells were grown in standard YE, with 3% glucose, a weak but reproducible GFP-Atg8 cleavage could be observed at the stationary phase; this cleavage was exacerbated (1% glucose) or occured earlier (0.08% glucose) in cultures grown in respiratory-prone conditions. In fact, processing of the reporter started so early in 0.08% glucose that free GFP was rather undetectable at long times (day 2 in ). Similar results were obtained in cells grown in minimal medium (MM) with different concentrations of the carbon source (Fig. S1A).
We discarded that autophagy induction upon depletion of the sulfur, phosphorus or carbon sources was caused by simultaneous nitrogen depletion in the media: further addition of a nitrogen supply 5 h after starvation did not cause any significant decrease in GFP-Atg8 cleavage (Fig. S1B). We conclude that not only nitrogen starvation but also depletion of sulfur or phosphorus sources and, importantly, low glucose conditions known to contribute to longer lifespan [Citation26] (Fig. S1C) can trigger the onset of autophagy in fission yeast with different kinetics and to different extents.
Role of the TORC1 complex in autophagy
Once established that any type of nutritional starvation can trigger autophagy in S. pombe, we tested the participation of several nutrient-sensing cascades in the regulation of autophagy: the TORC1 (target of rapamycin complex 1), TORC2, PKA and MAP kinase pathways (). These regulatory routes, which respond to nitrogen and/or glucose deprivation, are involved in G1 arrest, sexual development, amino acid uptake, cell growth, meiosis, lifespan and stress response (Fig. S1D).
Budding and fission yeast have two TOR kinases whereas higher eukaryotes generally have only one [Citation27,Citation28]. In fission yeast, TORC1 kinase complex is composed of the essential catalytic subunit Tor2 and Mip1, and the non-essential Tco89, Wat1 and Tco1 [Citation29–31] (). Tuberous sclerosis complex, the hererodimer Tsc1-Tsc2, acts as an upstream negative regulator of TORC1 during starvation [Citation32–34] (). Tor2 is essential during physiological conditions in S. pombe, and its inactivation by nitrogen starvation induces cell cycle arrest in the G1 phase, initiation of alternative cellular programs such as sexual development and induction of the nitrogen starvation gene expression program [Citation29,Citation30,Citation35,Citation36]. When TORC1 is active under nutrient-rich conditions, it phosphorylates members of the AGC (protein kinase A/protein kinase G/protein kinase C) kinase family: Psk1, Sck1 and Sck2. Psk1 is an RPS6KB1/S6K1 homolog, which has as a substrate Rps6 (ribosomal protein S6; for a review see [Citation4]) (). Tsc1-Tsc2 senses nitrogen depletion, and causes dephosphorylation/inactivation within minutes of TORC1 complex, Psk1 and Rps6, leading to translation inhibition, among other effects [Citation37,Citation38].
To test whether autophagy is one of the alternative programs activated through the TORC1 pathway upon nutrient depletion, we studied the phosphorylation state of Psk1 and Rps6 in response to not only to nitrogen, but also to glucose, sulfur or phosphorus deprivation, all capable of inducing autophagy. As shown in and previously reported [Citation37,Citation38], not only nitrogen depletion but also glucose starvation triggered Psk1 and Rps6 dephosphorylation. In addition, a moderate dephosphorylation of Psk1 was observed 30 min after removing the sulfur or phosphorus sources, which was exacerbated after 8 h of nutrient depletion ().
Figure 2. TORC1 negatively regulates autophagy. (A) TORC1 kinase activity and phosphorylation of its downstream consecutive targets, Psk1 and Rps6, are inhibited upon nutrient deprivation. MM cultures of strain CC10 (expressing GFP-Atg8) were collected at the logarithmic phase (-) and 30 min or 8 h after switching to modified MM with low glucose (0.08% Gluc), or without nitrogen (- N), sulfur (- S), and phosphorus (- P) sources. TCA extracts were analyzed by western blot with anti-phospho-Psk1 (anti-RPS6KB/p70) and anti-phospho-Rps6 (anti-AKT) antibodies. (B) Effect of TORC1 pathway mutants on the inhibition of Psk1 and Rsp6 phosphorylation after nitrogen depletion. MM cultures of strains CC10 (WT), JUP1350 (tor2L1310P), CC31 (tsc1Δ tsc2Δ), CC18 (tco89Δ) and CC21 (sck2Δ) were shifted to nitrogen depleted media for 10–30 min and analyzed as in (A). (C) Effect of TORC1 pathway mutants on autophagy activation upon nitrogen depletion. MM cultures of strains CC10 (WT), CC14 (tsc1Δ), CC31 (tsc1Δ tsc2Δ), CC18 (tco89Δ), CC66 (psk1Δ), CC89 (sck1Δ) and CC21 (sck2Δ), all expressing GFP-Atg8, were shifted or not at the logarithmic phase (unt) to nitrogen-free medium for 1–3 h, and were processed and analyzed as in . Accelerated GFP-Atg8 cleavage is indicated with green circles, while deficient cleavage is labeled with a red circle. Numbers below the panels represent the amount of cleaved GFP relative to the amount after 2 h without nitrogen in WT (with an assigned value of 1). (D) Atg1 dephosphorylation after nitrogen depletion is defective in tor2L1310P and tsc1Δ mutants. MM cultures of strains CC99 (WT), CC115 (tor2L1310P), CC101 (tsc1Δ), RB15 (tco89Δ), RB5 (psk1Δ), RB13 (sck1Δ), and RB6 (sck2Δ), all expressing Atg1-HA, were shifted or not to nitrogen-free medium, and processed and analyzed as in (A), using anti-HA antibody. (E) Scheme depicting the nutrient depletion-dependent antagonistic regulation by TORC1 pathway of growth-related functions and of autophagy. Deletion or point mutants used in this study are represented in red (low autophagy) and green (high autophagy). See text for details.
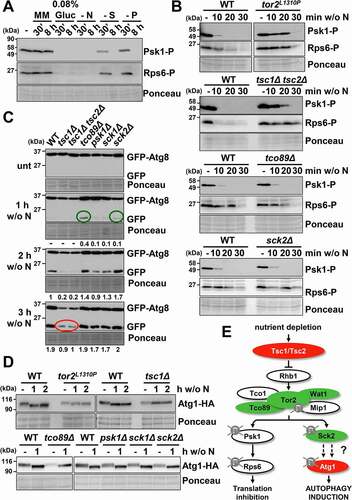
In order to test the role of TORC1 pathway in autophagy, we first searched for mutants in which the inactivation of downstream kinases, i.e. Psk1 and Rps6, was altered in response to nutrient deprivation. As previously reported by the group of Tamanoi [Citation37,Citation39], cells carrying the tor2L1310P allele were largely blind to nitrogen depletion and displayed very slow dephosphorylation of the Psk1 and Rps6 kinases; very similar behavior was seen in cells lacking the upstream sensors Tsc1 and Tsc2 (, upper panels). The TORC1 component Tco89 is dispensable for Psk1 and Rps6 dephosphorylation upon nitrogen starvation, and as expected cells lacking this subunit had slightly but reproducibly lower levels of phosphorylated Rps6 during nitrogen-rich growth (, third panel). As expected, the lack of Sck2 did not have any impact on Rps6 phosphorylation (, bottom panel).
We used these mutants to test the role of the TORC1 pathway in autophagy. As shown in and Fig. S2, tsc1Δ and tsc1Δ tsc2Δ mutants showed lower levels of GFP-Atg8 cleavage after nitrogen depletion (red circles in ), while the tco89Δ and sck2Δ mutants displayed an slight but significant faster induction of GFP release (green circles in ). These results indicate that TORC1 cascade plays a role in regulating autophagy as previously suggested [Citation40].
The direct autophagy substrate of TORC1 in budding yeast is Atg13, which is hyper-phosphorylated under nutrient-rich conditions and is dephosphorylated during starvation facilitating the dephosphorylation of downstream Atg1 (for a review, see [Citation41]). The electrophoretic mobility shift upon phosphorylation of Atg13 or Atg1 has proven to be a good indicator of autophagy in S. pombe [Citation17]. As shown in , Atg1 was dephosphorylated 1 h after nitrogen depletion in wild-type cells, while this mobility shift was not observed in mutants defective in TORC1 inhibition, tor2L1310P and tsc1Δ (, upper panel). However, Atg1 dephosphorylation did not seem to discriminate a moderate gain of TORC1 kinase activity: tco89Δ and sck2Δ displayed Atg1 dephosphorylation kinetics similar to those of the wild type strain (, bottom panel). On the basis of these evidences we propose that during nutrient depletion Tsc1-Tsc2 inhibits the TORC1 pathway, triggering the dephosphorylation of Sck2 that would stop Atg1 phosphorylation, inducing autophagy ().
The TORC2 pathway also modulates autophagy in fission yeast
In fission yeast TORC2 complex responds to low glucose conditions, but it is also related to processes that are triggered during nitrogen starvation. Thus, it is required for G1 arrest, sexual development and for resistance to osmotic and oxidative stresses [Citation42–46]. The TORC2 complex is composed of the kinase Tor1 and the regulatory subunits Ste20, Sin1, Wat1 (also present in TORC1) and Bit61 [Citation30,Citation31,Citation46] (). Tor1 recruits and phosphorylates the fission yeast AGC kinase Gad8 [Citation47] to regulate cell proliferation, the switch to bipolar cell growth, cell fusion during mating, and the meiotic program [Citation42]. As previously shown [Citation48], Gad8 was dephosphorylated in a TORC2 dependent manner during glucose starvation, but not after nitrogen depletion, which instead triggered a slower mobility shift in gels ().
Figure 3. TORC2 interacts with TORC1 under basal condition and affects autophagy activation. (A) Inhibition of TORC2 kinase activity by glucose, but not nitrogen, depletion causes Gad8 dephosphorylation. MM cultures of strain AN0175 (WT), expressing Gad8-HA, were shifted or not to nitrogen-free or glucose-free medium, and processed and analyzed as in , using anti-HA antibody. (B) Effect of TORC2 pathway mutants on the inhibition of Psk1 and Rsp6 phosphorylation, downstream of TORC1, after nitrogen depletion. MM cultures of strains AN0179 (WT), RB17 (tor1Δ) and RB18 (gad8Δ), all expressing Psk1-Myc, were shifted or not to nitrogen depleted media for 10–30 min and analyzed as in . Anti-Myc was used as a loading control to demonstrate that Psk1 phosphorylation but not Psk1 levels are altered in the TORC2 mutants (Psk1). (C) Effect of TORC2 pathway mutants on autophagy activation upon nitrogen depletion. MM cultures of strains CC10 (WT), CC92 (wat1Δ), CC70 (gad8Δ), CC16 (tor1Δ) and RB4 (gad8Δ tor1Δ), all expressing GFP-Atg8, were shifted or not to nitrogen-free medium, and processed and analyzed as in . Numbers below the panels represent the amount of cleaved GFP relative to the amount after 2 h without nitrogen in WT (with an assigned value of 1). (D) Atg1 dephosphorylation after nitrogen depletion is unaffected in strain gad8Δ. MM cultures of strains CC99 (WT) and CC116 (gad8Δ), both expressing Atg1-HA, were shifted or not to nitrogen-free medium, and processed and analyzed as in . (E) Synthetic interaction between TORC1 and TORC2 mutants on TORC1 kinase activity. MM cultures of strains CC10 (WT), CC70 (gad8Δ), CC128 (tor2L1310P gad8Δ) and JUP1350 (tor2L1310P) were shifted or not to nitrogen-free medium, and processed and analyzed as in . (F) Synthetic interaction between TORC1 and TORC2 mutants on autophagy. MM cultures of strains CC10 (WT), CC70 (gad8Δ), RB32 (tor2L1310P gad8Δ) and CC112 (tor2L1310P), all expressing GFP-Atg8, were shifted or not to nitrogen-free medium, and processed and analyzed as in . Numbers below the panels represent the amount of cleaved GFP relative to the amount after 2 h without nitrogen in WT (with an assigned value of 1). (G) Scheme depicting the positive regulation of TORC1 kinase activity by the TORC2 components Tor1 and Gad8 under nutrient-rich conditions. Deletion or point mutants used in this study are represented in red (low autophagy) and green (high autophagy). See text for details.
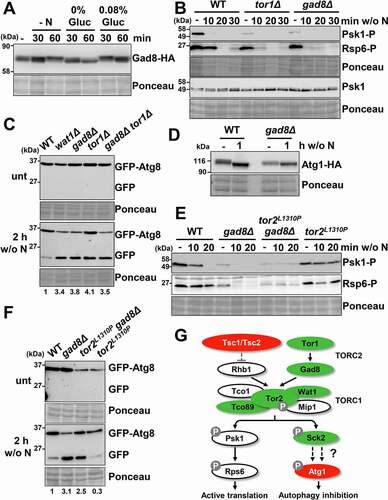
To test a possible cross-talk between the TORC1 and the TORC2 pathways, we tested Psk1 and Rps6 dephosphorylation, located downstream of TORC1, after nitrogen depletion in TORC2 mutants (). In untreated conditions, tor1Δ and gad8Δ showed significantly lower Psk1 or Rps6 phosphorylation levels compared to wild type cells, whereas there were no differences in the concentration of total Psk1-(Psk1 in ). Dephosphorylation of Psk1 and Rsp6 upon nitrogen depletion in tor1Δ and gad8Δ seemed as fast as that of a wild-type strain. These results suggest a connection between both cascades under nutrient-rich conditions. To test whether this cross-talk also affects autophagy induction upon nitrogen starvation, we monitored GFP-Atg8 cleavage in the mutants wat1Δ, gad8Δ, tor1Δ and tor1Δ gad8Δ after nitrogen depletion. As shown in all mutants exhibited higher induction of autophagy than wild-type cells 2 h after nitrogen depletion. We conclude that mutants of the TORC2 cascade display enhanced levels of autophagy upon nutrient deprivation, and we propose that this probably occurs by modulation of starting TORC1 activity, which was partially inhibited in gad8Δ and tor1Δ mutants even prior to nutrient depletion (). Furthermore, no cumulative effect was observed in the double deletion of tor1 and tco89: tor1Δ tco89Δ mutant exhibited an increase in autophagy similar to the single deletes tor1Δ or tco89Δ, supporting the theory that both TORC1 and TORC2 complexes cooperate modulating autophagy (Fig. S3A). As shown above for the partially active tco89Δ and sck2Δ mutants (), dephosphorylation of Atg1-HA was not significantly different from wild-type cells ().
In order to determine whether Tor1 and Gad8 work upstream of Tsc1 or converge synergistically with Tsc1-Rhb1 on the TORC1 kinase complex, we analyzed the genetic interactions between tsc1Δ or tor2L1310P (mutants of TORC1 blinded to nutrient depletion; and Fig. S2) and gad8Δ (Gad8 is downstream of TORC2, but gad8Δ displays decreased levels of TORC1 kinase activity during nutrient-rich conditions). Thus, the double mutant gad8Δ tor2L1310P displayed a combination of phenotypes upon nitrogen depletion regarding phosphorylation of Psk1 and Rsp6, with lower starting levels and sustained phosphorylation upon nitrogen depletion (). A similar intermediate phenotype was observed in the double mutant tsc1Δ gad8Δ (Fig. S3B). Regarding GFP-Atg8 cleavage, gad8Δ tor2L1310P displayed an intermediate behavior compared with the single deletes (); the same occured with strain tsc1Δ gad8Δ (Fig. S3C). We also discarded an effect of TORC1 over Gad8 phosphorylation, since the electrophoretic mobility of Gad8-HA in response to starvation in tsc1Δ was identical to that of a wild-type strain (Fig. S3D). Thereby, we propose that TORC2 positively regulates TORC1 activity even in basal conditions, and that the lack of Tor1 or Gad8 mimics a mild nutrient deprivation decreasing growth and promoting autophagy ().
While mutations in the Pka1 pathway do not affect autophagy induction, cells lacking the MAP kinase Sty1 are severely defective in the onset of autophagy
Once the involvement of TORC1 and TORC2 in autophagy have been established, we compared the autophagy levels in mutants from other nutrients and stress response pathways: those ruled by Pka1 and by the MAP kinase Sty1 (). In S. cerevisiae, high levels of cAMP (cyclic adenosine monophosphate) or constitutive activation of PKA prevents autophagy in response to rapamycin or nutrient-depletion, indicating that PKA negatively regulates autophagy [Citation49–51]. On the contrary, the fission yeast mutants pka1Δ and cgs1Δ, which display opposite global effects on the activity of the cascade [Citation52], displayed kinetics of GFP-Atg8 cleavage after nitrogen depletion undistinguishable from those of wild type background (), indicating that the PKA cascade does not participate in autophagy regulation in S. pombe.
Figure 4. Comparative regulation of autophagy through the TORC1, TORC2 and MAP kinase pathways. (A) Effect of TORC1, TORC2, PKA and MAP kinase pathway mutants on autophagy activation upon nitrogen depletion. MM cultures of strains CC10 (WT), CC31 (tsc1Δ tsc2Δ), CC18 (tco89Δ), CC21 (sck2Δ), CC66 (psk1Δ), CC16 (tor1Δ), CC33 (cgs1Δ), CC22 (pka1Δ), CC13 (sty1Δ) and CC23 (atf1Δ), all expressing GFP-Atg8, were shifted or not at the logarithmic phase (unt) to nitrogen-free medium for 1–3 h, and were processed and analyzed as in . Accelerated GFP-Atg8 cleavage is indicated with green circles, while deficient cleavage is labeled with red circles. Numbers below the panels represent the amount of cleaved GFP relative to the amount after 2 h without nitrogen in WT (with an assigned value of 1). (B) Effect of TORC1, TORC2 and MAP kinase pathway mutants on autophagy activation upon depletion of the glucose, sulfur or phosphorus sources. MM cultures of strains CC10 (WT), CC31 (tsc1Δ tsc2Δ), CC18 (tco89Δ), CC21 (sck2Δ) CC16 (tor1Δ), CC70 (gad8Δ) and CC13 (sty1Δ), all expressing GFP-Atg8, were shifted at the logarithmic phase (unt) to modified MM with low glucose (0.08% Gluc), or without sulfur (w/o S) or phosphorus (w/o P) sources for 8 h, and were processed and analyzed as in . A sample of extracts from CC10 (WT) after 2 h of nitrogen-starvation was used as an autophagy control. Numbers below the panels represent the amount of cleaved GFP relative to the amount in WT after 8 h under indicated conditions (0.08% Gluc, without [w/o] S and without [w/o] P) (with assigned values of 1). (C) Effect of TORC1, TORC2 and MAP kinase pathway mutants on bulk autophagy activation upon nitrogen depletion. MM cultures of strains RB44 (WT), RB40 (atg1Δ), RB45 (tsc1Δ), RB41 (tor2L1310P), RB48 (tco89Δ), RB39 (gad8Δ) and RB38 (sty1Δ), all expressing Pgk1-GFP, were shifted or not at the logarithmic phase (unt) to nitrogen-free medium for 1–2 h, and were processed and analyzed as in . Numbers below the panels represent the amount of cleaved GFP relative to the amount in WT after 1 h under nitrogen deprivation (with an assigned value of 1). (D) Effect of TORC1, TORC2 and MAP kinase pathway mutants on bulk autophagy activation upon depletion of the glucose, sulfur or phosphorus sources. MM cultures of strains strains RB44 (WT), RB40 (atg1Δ), RB45 (tsc1Δ), RB41 (tor2L1310P), RB48 (tco89Δ), RB39 (gad8Δ) and RB38 (sty1Δ), all expressing Pgk1-GFP, were shifted at the logarithmic phase (unt) to modified MM with low glucose (0.08% Gluc), or without sulfur (w/o S) or phosphorus (w/o P) sources for 8 h, and were processed and analyzed as in . Numbers below the panels represent the amount of cleaved GFP relative to the amount in WT after 8 h under each condition (0.08% Gluc, without [w/o] S and without [w/o] P) (with assigned values of 1). (E) Scheme depicting the impact of mutations at the TORC1, TORC2 or MAP kinase pathways on autophagy induction. Accelerated nutrient starvation-dependent autophagy of some mutants is indicated in green, while deficient autophagy is indicated in red. See text for details.
![Figure 4. Comparative regulation of autophagy through the TORC1, TORC2 and MAP kinase pathways. (A) Effect of TORC1, TORC2, PKA and MAP kinase pathway mutants on autophagy activation upon nitrogen depletion. MM cultures of strains CC10 (WT), CC31 (tsc1Δ tsc2Δ), CC18 (tco89Δ), CC21 (sck2Δ), CC66 (psk1Δ), CC16 (tor1Δ), CC33 (cgs1Δ), CC22 (pka1Δ), CC13 (sty1Δ) and CC23 (atf1Δ), all expressing GFP-Atg8, were shifted or not at the logarithmic phase (unt) to nitrogen-free medium for 1–3 h, and were processed and analyzed as in Figure 1A. Accelerated GFP-Atg8 cleavage is indicated with green circles, while deficient cleavage is labeled with red circles. Numbers below the panels represent the amount of cleaved GFP relative to the amount after 2 h without nitrogen in WT (with an assigned value of 1). (B) Effect of TORC1, TORC2 and MAP kinase pathway mutants on autophagy activation upon depletion of the glucose, sulfur or phosphorus sources. MM cultures of strains CC10 (WT), CC31 (tsc1Δ tsc2Δ), CC18 (tco89Δ), CC21 (sck2Δ) CC16 (tor1Δ), CC70 (gad8Δ) and CC13 (sty1Δ), all expressing GFP-Atg8, were shifted at the logarithmic phase (unt) to modified MM with low glucose (0.08% Gluc), or without sulfur (w/o S) or phosphorus (w/o P) sources for 8 h, and were processed and analyzed as in Figure 1A. A sample of extracts from CC10 (WT) after 2 h of nitrogen-starvation was used as an autophagy control. Numbers below the panels represent the amount of cleaved GFP relative to the amount in WT after 8 h under indicated conditions (0.08% Gluc, without [w/o] S and without [w/o] P) (with assigned values of 1). (C) Effect of TORC1, TORC2 and MAP kinase pathway mutants on bulk autophagy activation upon nitrogen depletion. MM cultures of strains RB44 (WT), RB40 (atg1Δ), RB45 (tsc1Δ), RB41 (tor2L1310P), RB48 (tco89Δ), RB39 (gad8Δ) and RB38 (sty1Δ), all expressing Pgk1-GFP, were shifted or not at the logarithmic phase (unt) to nitrogen-free medium for 1–2 h, and were processed and analyzed as in Figure 1A. Numbers below the panels represent the amount of cleaved GFP relative to the amount in WT after 1 h under nitrogen deprivation (with an assigned value of 1). (D) Effect of TORC1, TORC2 and MAP kinase pathway mutants on bulk autophagy activation upon depletion of the glucose, sulfur or phosphorus sources. MM cultures of strains strains RB44 (WT), RB40 (atg1Δ), RB45 (tsc1Δ), RB41 (tor2L1310P), RB48 (tco89Δ), RB39 (gad8Δ) and RB38 (sty1Δ), all expressing Pgk1-GFP, were shifted at the logarithmic phase (unt) to modified MM with low glucose (0.08% Gluc), or without sulfur (w/o S) or phosphorus (w/o P) sources for 8 h, and were processed and analyzed as in Figure 1A. Numbers below the panels represent the amount of cleaved GFP relative to the amount in WT after 8 h under each condition (0.08% Gluc, without [w/o] S and without [w/o] P) (with assigned values of 1). (E) Scheme depicting the impact of mutations at the TORC1, TORC2 or MAP kinase pathways on autophagy induction. Accelerated nutrient starvation-dependent autophagy of some mutants is indicated in green, while deficient autophagy is indicated in red. See text for details.](/cms/asset/624a8212-070f-45e9-804a-8f35bfd4f043/kaup_a_1935522_f0004_oc.jpg)
In S. cerevisiae, MAP kinases control specific autophagy programs in response to starvation; thus, Hog1 is required for mitochondrial autophagy (mitophagy) after nitrogen depletion [Citation53]. The fission yeast Sty1 cascade is activated by environmental stresses, but also by nutrient deprivation (). We tested the role of the MAP kinase Sty1 and of its main downstream transcription factor, Atf1, in autophagy activation. We found that sty1Δ cells exhibited a dramatic reduction in autophagy during nitrogen starvation whereas atf1Δ presents a wild type pattern (). Therefore, the MAP kinase cascade induces autophagy through Sty1 in an Atf1-independent manner.
We have shown above that the TORC1, TORC2 and Sty1 pathways regulate the onset of autophagy in response to lack of nitrogen (); the effect of mutants of these cascades could be observed 1–3 h after nitrogen withdrawal, while all of them ended up displaying maximal GFP-Atg8 cleavage 24 h after nitrogen depletion (data not shown). We have shown above that not only nitrogen depletion but also the lack of glucose, sulfur or phosphorus sources could drive autophagy activation (). As shown in , the three cascades shown to affect autophagy in response to nitrogen depletion also differentially responded to the other three types of stimuli, with cells lacking Sty1 displaying the more dramatic defects.
To confirm that these signals and pathways modulate the onset of autophagy, we have also used cytosolic Pgk1-GFP as a reporter of bulk autophagy, as previously described in budding yeast and, very recently, in fission yeast [Citation54,Citation55]. Pgk1-GFP cleavage occurred in response to nitrogen () or glucose, sulfur or phosphorus deprivations (), and this cleavage was accelerated or impaired in mutants of the TORC1, TORC2 and Sty1 cascades, mimicking the results obtained with GFP-Atg8.
We conclude that, to different extents, TORC1, TORC2 and MAP kinase pathways are involved in the onset of autophagy in response to different types of nutrient depletions; namely, cells unable to fully inhibit TORC1 in response to nutrient deprivation (tor2L1310P or tsc1Δ tsc2Δ, among others) or lacking the MAP kinase Sty1 displayed impaired efficiency to induce autophagy ().
Sty1 does not control autophagy through the TORC1 pathway, but rather at the level of transcription of atg genes
We first confirmed that Sty1 was activated/phosphorylated not only by environmental stresses such as hydrogen peroxide but also by the signals triggering autophagy, such as nitrogen and glucose starvation (). To test if the role of Sty1 on autophagy is due to a cross-talk with the TORC1 pathway, we tested whether the lack of Sty1 affects the basal phosphorylation levels of Psk1 and Rps6, or impairs their dephosphorylation during nitrogen depletion. As shown in , sty1Δ and wild type cells displayed undistinguishable Psk1 and Rps6 phosphorylation patterns before and after nitrogen deprivation. Indeed, the absence of Sty1 did not exacerbate the defective dephosphorylation of Psk1 and Rps6 of tsc1Δ cells, indicating that the MAP kinase does not regulate the TORC1 pathway (Fig. S4A). Furthermore, Sty1 does not seem to have an effect on the TORC2 pathway, since Gad8 phosphorylation levels upon nutrient starvation were unaffected in sty1Δ cells (Fig. S4B). Sty1 and TORC1 seem to affect autophagy independently. Indeed, GFP-Atg8 cleavage upon nitrogen withdrawal was significantly slower in the double tsc1Δ sty1Δ mutant than in the single mutants at all the time points tested (), supporting the idea that Sty1 and TORC1 activate the onset of autophagy through independent molecular events.
Figure 5. The MAP kinase Sty1 does not regulate TORC1-dependent growth promotion functions, nor affects phosphorylation levels of Atg1 and Atg2, but positively regulates autophagy. (A) Sty1 is phosphorylated upon nitrogen or glucose starvation. MM cultures of wild-type strain CC10 were grown and shifted or not from the logarithmic phase (-) to modified MM without nitrogen (w/o N) or with low glucose (0.08%). As a control, logarithmic cultures were treated 10 min with 1 mM H2O2. TCA extracts were analyzed by western blot using anti-phospho-MAPK/p38 (Sty1-P) and anti-Sty1 antibodies. (B) Lack of Sty1 does not impair dephosphorylation of Psk1 and Rsp6 after nitrogen depletion. MM cultures of strains CC10 (WT) and CC13 (sty1Δ) were shifted or not to nitrogen depleted media for 10–30 min and analyzed as in . (C) Synergistic role of TORC1 and MAP kinase pathways on autophagy activation. MM cultures of strains CC10 (WT), CC14 (tsc1Δ), CC13 (sty1Δ) and CC111 (tsc1Δ sty1Δ), all expressing GFP-Atg8, were shifted or not to nitrogen-free medium for 2–7 h, and processed and analyzed as in Figure 2 C. Numbers below the panels represent the amount of cleaved GFP relative to the amount after 2 h without nitrogen in WT (with an assigned value of 1). (D) Lack of Sty1 does not impair dephosphorylation of Atg1 after nitrogen depletion. MM cultures of strains CC99 (WT) and CC110 (sty1Δ), both expressing Atg1-HA, were shifted or not to nitrogen depleted media for 1–2 h and analyzed as in Figure 2A, using anti-HA antibody. Numbers below the panels represent the amount of Atg1 relative to the amount in WT under physiological conditions (-) (with an assigned value of 1). (E) Lack of Sty1 does not impair phosphorylation of Atg2 after nitrogen depletion. MM cultures of strains CC122 (WT) and CC123 (sty1Δ), both expressing Atg2-HA, were shifted or not to nitrogen depleted media for 1–2 h and analyzed as in , using anti-HA antibody.
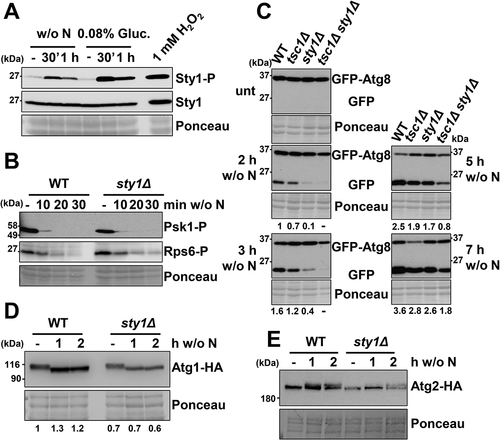
tsc1Δ cells displayed defects in Atg1 dephosphorylation after depletion of nitrogen (), but the absence of Sty1 did not jeopardize Atg1 dephosphorylation/activation (, Fig. S4C). During autophagy, Atg13 is dephosphorylated and mediates Atg1 dephosphorylation, which then triggers by an unknown mechanism the phosphorylation of Atg2 (for a review, see [Citation56]). We tested whether Sty1 could directly promote Atg2 phosphorylation in response to nitrogen starvation. We detected a mobility shift, indicative of phosphorylation, in both wild-type and sty1Δ cell extracts obtained after nitrogen depletion (), indicating that Sty1 does not directly affect the phosphorylation state of Atg1 nor Atg2 ().
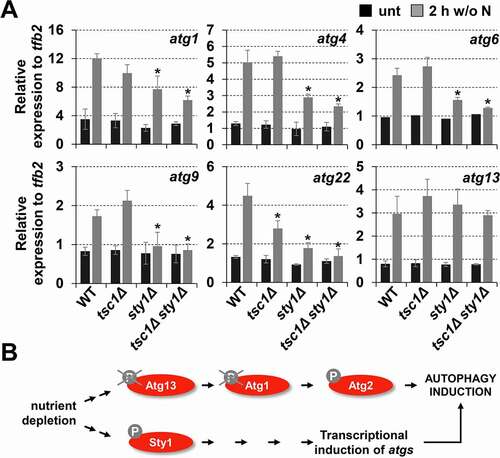
The Sty1 cascade is strongly activated by environmental stressors, and massive changes in the gene expression program occur to promote survival [Citation57]. Hundreds of genes are upregulated in a Sty1-dependent manner, and many of the atg genes are triggered by some but not all the signals used [Citation57]. We decided to test whether the induction of some atg genes could occur upon nitrogen starvation in a Sty1-dependent manner, to explain the role of the MAP kinase in autophagy activation. As shown in , many autophagy genes such as atg1, atg4, atg6, atg9, atg22 and atg13 were strongly induced during nitrogen starvation. Importantly, the expression pattern in wild type strain and tsc1Δ was similar for most genes, suggesting that TORC1 does not regulate autophagy at the transcriptional level. On the contrary, sty1Δ strain exhibited a significantly reduced upregulation of atg transcripts upon nitrogen depletion, except for atg13 (), indicating that activated Sty1 contributes to the onset of autophagy indirectly by upregulating the expression of atg mRNAs (). It is worth noting that the upregulation of atg genes was Atf1-independent (Fig. S5), explaining why atf1Δ triggered autophagy upon nutrient deprivation similarly to wild-type cells ().
Figure 6. Sty1 plays a role in transcriptional upregulation of atg genes after nitrogen starvation. (A) Expression of some atg genes is dependent on Sty1. MM cultures of strains CC10 (WT), CC14 (tsc1Δ), CC13 (sty1Δ) and CC111 (tsc1Δ sty1Δ) were collected at the logarithmic phase (unt) and 2 h after switching to nitrogen-starved medium; total RNA was isolated and the transcripts specific for atg1, atg4, atg6, atg9, atg22 and atg13 were quantified by RT-qPCR. Amplification with tfb2 primers was used as a normalization control. Each column represents the mean value and SD (standard deviation), calculated from four biological replicates. Asterisks represent significant differences, *p˂0.01 (Welch’s t-test). (B) Scheme suggesting that Sty1 does not induce autophagy through a direct effect on Atg13-Atg1-Atg2 phosphorylation levels, but rather through the transcriptional upregulation of atg genes. See text for details.
Leucine starvation is a potent activator of autophagy in cells auxotrophic for leucine
Regarding the biological role of autophagy in fission yeast, it has only been reported that it contributes to maintain cell viability during very long periods, more than 20–25 days, of nitrogen starvation [Citation17]. As shown in , wild-type cells deprived of nitrogen, sulfur or phosphorus sources displayed different degrees of viability 25 days after nutrient withdrawal, suggesting that the reservoirs for each essential nutrient are quantitatively different. Importantly, the atg1Δ and atg2Δ strains displayed decreased viability in all conditions, highlighting the role of this degradation pathway in providing nutrients from the digestion of non-essential molecular components.
Figure 7. Biological role of autophagy in cells auxotrophic for leucine. (A) The negative effect on lifespan of atg mutants can only be observed 25 days after nutrient deprivation. MM cultures of strains CC10 (WT), CC11 (atg1Δ) and CC20 (atg2Δ) grown in MM until logarithmic phase (day 0) were switched to modified MM without nitrogen (w/o N), sulfur (w/o S) or phosphorus (w/o P) sources during 25 days. Serial dilutions of the cultures were plated onto YE5S plates. (B) The negative effect on lifespan of atg mutants auxotrophic for leucine can be observed 12 h after nitrogen or leucine deprivation. Strains HM123 (WT) and SK1 (atg1Δ), both carrying the leu1-32 allele, were grown in leucine-supplemented MM until logarithmic phase (day 0) and switched to MM without leucine (MM), MM with leucine (MM +Leu), MM without nitrogen (w/o N) or MM without nitrogen with leucine (w/o N +Leu), for 12 or 24 h. Serial dilutions of the cultures were plated onto YE5S plates. (C, D) Microscopy images (C) and percent of survival (D) of HM123 (WT leu1-32) and SK1 (atg1Δ leu1-32) after 48 h in MM and leucine-supplemented MM (+ Leu). For survival measurements, cells were incubated with the permeability-dependent dye PI, and the percentage of non-fluorescent cells, indicative of viability, was determined by flow cytometry. (E) TORC1 kinase activity is unaffected in an autophagy-proficient leu1-32 background. MM cultures of strains CC10 (WT) and CC88 (WT leu1-32) were shifted or not to nitrogen depleted media for 10–30 min and analyzed as in Figure 2A. (F) Leucine deprivation can trigger autophagy in leu1-32 auxotrophic strains. MM cultures of strains CC10 (WT) and CC88 (WT leu1-32), both expressing GFP-Atg8, were shifted or not to leucine (2 h) or nitrogen (3 h) depleted MM and processed and analyzed as in Figure 2 C. Numbers below the panels represent the amount of cleaved GFP relative to the amount after 3 h without nitrogen in WT (with an assigned value of 1). (G) Leucine deprivation can inhibit TORC1 kinase activity in auxotrophic strains. Strains and conditions as in (F) were processed and analyzed as in . (H) The fast negative effect on lifespan of atg mutants auxotrophic for leucine is also displayed by TORC1 mutants with impaired autophagy activation. MM cultures of strains HM123 (WT) and atg1Δ (SK1), CC129 (tsc1Δ), CC134 (tco89Δ) and CC135 (atg1Δ tco89Δ), all carrying the leu1-32 allele, were grown and processed as in (B).
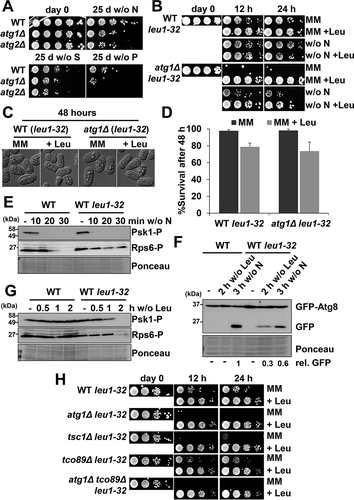
It has been previously shown that the combination of atg deletions with auxotrophic mutations, especially those affecting leucine synthesis, exacerbates the viability defects upon nitrogen depletion [Citation17]. We studied the viability of atg1Δ leu1-32 during nitrogen starvation and after leucine depletion. Surprisingly, the effect of leucine withdrawal was even more dramatic than nitrogen depletion on atg1Δ leu1-32 cells: only 12 h after leucine withdrawal the culture lost completely its colony-forming capacity (). The morphology of the starved atg1Δ leu1-32 cells was undistinguishable from wild-type cells, suggesting that they are not dead but rather arrested (). Indeed, both wild-type leu1-32 and atg1Δ leu1-32 cells were refractory to propidium iodide internalization, as determined by flow cytometry, a classical reporter of viability (). We propose that autophagy is essential to escape from the leucine-induced arrest in cells that are unable to synthesize the amino acid [Citation58]. We tested the role of autophagy on other amino acid deficiencies such as arginine or lysine in auxotrophic strains, to demonstrate that deletion of atg1 did not significantly affect cell viability in those cases (Fig. S6A).
Upon nitrogen depletion, cells auxotrophic and prototrophic for leucine followed very similar kinetics regarding TORC1 inactivation () and autophagy induction ( and Fig. S6B and C). On the contrary, leucine withdrawal induced Psk1 and Rps6 dephosphorylation () and GFP-Atg8 cleavage ( and Fig. S6C and D) only in wild-type leu1-32 strain barely 2 h after amino acid starvation. We also checked the effect of leucine withdrawal in leu1-32 TORC1 mutants: tsc1Δ, tco89Δ and atg1Δ tco89Δ (). Similar to atg1Δ leu1-32, 12 h after leucine depletion tsc1Δ leu1-32 and atg1Δ tco89Δ leu1-32 did not recover mitotic competence. However, tco89Δ leu1-32 presented similar pattern that wild-type leu1-32 strain: these mutant does not impair autophagy activation (see ). Our experiments suggest that leucine depletion itself is a potent inducer of autophagy in cells auxotrophic for this amino acid, and that the requirement for leucine regulates and/or contributes to autophagy probably through TORC1 activity, revealing itself as an amino acid of great importance in cell survival.
Discussion
Even though the molecular components that govern autophagy upon nitrogen deprivation have been identified in fission yeast, little is known about the pathways that modulate its activation or the signals and biological roles linked to the onset of autophagy. Here, using an optimized reporter system based on the vacuolar cleavage of GFP-Atg8, we demonstrate that autophagy occurs not only in response to low nitrogen levels, but also in response to the depletion of a variety of nutrients, including carbon starvation, and that this activation is mediated through pathways related to cell growth, G1 arrest, meiosis, lifespan and stress response. As in other eukaryotes, inactivation of the TORC1 pathway is an important contributor to the onset of autophagy, but unexpectedly we show here that activation of the stress-dependent MAP kinase pathway governed by Sty1 is crucial to promote full autophagy induction in response to nutrient depletion.
Thus, we have uncovered here new signals, all related to the depletion of essential nutrients, which are able to induce autophagy in fission yeast: not only nitrogen deprivation but also depletion of sulfur, phosphorus, glucose, or leucine sources were all capable of promoting GFP-Atg8 cleavage. Importantly, addition of nitrogen to cultures deprived of these other alternative nutrients did not prevent autophagy activation, dismissing that nitrogen is also becoming limiting in our experimental conditions. Thus, similar to nitrogen deprivation, depletion of sulfur or phosphorus sources induced autophagy () and extended lifespan (). Supporting our results, in S. cerevisiae autophagy is induced during sulfur or phosphorus scarcity [Citation10,Citation24,Citation25] and, in fission yeast, depletion of a sulfur supply extends chronological lifespan [Citation59].
Regarding carbon depletion-mediated autophagy, previous studies suggested that the levels of glucose do not regulate autophagy in S. pombe [Citation13,Citation18], contrary to the classical autophagy activation upon carbon starvation described in budding yeast [Citation10]. We report here the early cleavage of our sensitive reporter GFP-Atg8 in stationary phase cultures grown in low glucose media, conditions linked to elongated lifespan (, Fig. S1A and C). We had previously demonstrated that the growth in low (1%) glucose media promotes respiratory metabolism and longer lifespan [Citation26]. In addition, it has been also shown that S. pombe cells grown in media containing 0.08% glucose display hallmarks of respiratory metabolism, while fermentation is repressed [Citation60]. We propose that autophagy mediated by glucose deprivation could be caused by an increase in respiratory metabolism, and that elimination of damaged mitochondria could be the biological goal linked to this activation.
We have here dissected the signaling cascades controlling autophagy activation in fission yeast. As expected, the TORC1 is crucial for the onset of autophagy, and mutants of the pathway controlling normal growth have the opposite impact on autophagy activation. Thus, cells lacking Tsc1 or expressing the signal blinded kinase tor2L1310P could not trigger autophagy, and mutants decreasing TORC1 kinase activity (tco89Δ) had defects in cell growth (i.e. Psk1 phosphorylation) and were faster triggering GFP-Atg8 cleavage. We have also shown that not only TORC1 but also the TORC2 pathway had a significant impact on the onset of autophagy. As in other organisms, S. pombe has two types of TOR kinase complexes, TORC1 and TORC2, and interestingly the two have opposite roles in sexual differentiation, which is induced by nutrient starvation [Citation61]. Nevertheless, we show here that both TORC1 and TORC2 kinases negatively regulate autophagy: mutants decreasing the kinase activity of the complexes (tco89Δ and tor1Δ) displayed an earlier induction of autophagy not only after nitrogen depletion but also upon low glucose, or upon depletion of sulfur or phosphorus source. Our data suggest that the direct regulator of cell growth promotion during nutrient-rich conditions and of autophagy activation during nutrient depletion is TORC1, but there is a positive crosstalk between both branches: the TORC2 mutants gad8Δ and tor1Δ displayed under nutrient-rich conditions impaired phosphorylation of substrates downstream of the TORC1 kinase (Psk1 and Rps6), and accelerated autophagy during nutrient depletion (). A similar crosstalk has been proposed in mammalian cells: TSC2 is directly phosphorylated and inhibited by the action of AKT, a homolog of Gad8 [Citation33,Citation62]. In fission yeast, our genetic interaction analysis suggests that both branches impinge on the same substrates regarding autophagy: both tor1Δ tco89Δ and tco89Δ mutants showed similar accelerated autophagy upon lack of nutrients compared to wild-type cells (Fig. S3A). Furthermore, the double mutant tor2L1310P gad8Δ displayed a mixed phenotype regarding basal phosphorylation of Psk1 and Rps6 and autophagy induction (), and tsc1Δ gad8Δ cells had an intermediate effect on GFP-Atg8 cleavage compared to the single deletes (). We propose that TORC2 kinase positively regulates TORC1 kinase independently of Tsc1-Tsc2 ().
Once established that carbon starvation induces autophagy, we investigated the participation of the PKA and the MAP kinase Sty1 pathways, since both unambiguously respond to glucose exhaustion. The fission yeast mutant pka1Δ and its antagonist, cgs1Δ, did not display significant defects in autophagy activation, even though the cAMP-dependent PKA cascade regulates autophagy from yeast to mammals [Citation49,Citation51,Citation63,Citation64]. Our genetic evidences are in accordance with previous reports indicating that autophagy is unaffected by cAMP levels [Citation18]. We demonstrate here, however, that cells lacking Sty1 had striking defects in GFP-Atg8 cleavage, which are quantitatively more dramatic that those of TORC1 mutants ().
In S. pombe, the MAP kinase Sty1 is essential for survival under diverse environmental conditions such as oxidative stress, it is required for the initiation of the mating and meiosis program and is activated at the onset of stationary phase and during calorie restriction [Citation26,Citation65,Citation66]. Our results reveal that Sty1 is activated after nitrogen or carbon depletion and is a main activator of bulk autophagy independently of its main effector Atf1 (). The other two MAP kinases cascades in S. pombe, driven by Pek1 and Pmk1, have not yet been connected to autophagy, but it is worth to point out that cross-talks between the MAP kinase cell wall integrity and stress pathways have been described in plant pathogenic fungi [Citation67,Citation68], and a combination of both may affect the onset of autophagy. In S. cerevisiae, the stress MAP kinase Hog1 does not have a role in bulk autophagy, but it controls specific autophagy programs in response to starvation such as mitophagy [Citation53]. In mammalian cells, the participation of the Sty1 ortholog MAPK/p38 is controversial, since it has a dual role in autophagy both as a positive and negative regulator [Citation69].
We have shown here that the Sty1 kinase is not required for the phosphorylation of Atg1 or Atg2 (). We then tested whether the transcriptional autophagy program was regulated by the MAP kinase, since in budding yeast a transcriptional signature of atg genes has been reported in response to starvation (for a review, see [Citation5]). Indeed, we show that several autophagy genes were induced upon nitrogen starvation, confirming a former genome-wide analysis [Citation70], and this upregulation was, in most cases, strongly dependent on Sty1 but not on Atf1 ( and Fig. S5). For some genes such as atg1, even the basal levels were decreased in a sty1Δ background (see ), as well as the encoded protein Atg1 (see ). Regarding nutrient-dependent autophagy activation, we propose that an alternative transcription factor downstream of Sty1 could trigger atg transcription. The zinc finger transcription factor Rsv2 was a likely candidate, since it is required for cell viability during stationary phase, its over-expression elongates lifespan and induces stress-related genes during sporulation [Citation71–73], it is upregulated at the transcriptional level by environmental stresses [Citation57], nitrogen starvation [Citation70], and during stationary phase in a Sty1-dependent manner (our own unpublished data). However, GFP-Atg8 cleavage after nitrogen starvation was unaffected in rsv2Δ cells (our own unpublished data).
In budding yeast and in mammalian cells, autophagy is required to maintain viability under starvation conditions [Citation74–76]. In S. pombe, the lack of viability of atg mutants is only observed after ≥ 25 days of growth in the absence of a nitrogen source, while few hours of nitrogen or leucine starvation impairs mitotic competence in auxotrophic autophagy mutants [Citation17] (). We do not know why atg mutants depleted for nitrogen or leucine for only 12 h were then unable to grow on plates, since the hallmarks of viability were unperturbed: the atg mutant cells displayed good microscopic morphological fitness, and propidium iodide staining, as an indicator of membrane integrity, did not differ from that of wild-type cells. Remarkably, we detected similar loss of colony-forming units upon nitrogen or leucine depletion in tsc1Δ leu1-32 mutants (). As suggested by others [Citation58], we believe that leucine or nitrogen depletion in cells auxotrophic for the amino acid causes severe cell arrest, and that the autophagy cascade is required for exiting the arrest and/or for the restoration of mitotic competence. Further studies will be required to elucidate the relationship between TORC1 and autophagy in leucine auxotrophic strains, and to decipher the interplay between growth and alternative gene programs regulated by the TORC1 pathway.
Materials and methods
Yeast strains and plasmids
Genotypes and origins of the strains used in this study are described in Table S1. Deleted and tagged strains including Atg1-HA, Atg2-HA and Pgk1-GFP were generated by homologous recombination using PCR fragments, amplified from plasmids derived from pFA6a derivatives [Citation77] or by crossing deletion mutants from the Bioneer collection [Citation78]. We PCR-amplified atg8 from plasmid GFP-Atg8+ pREP41 [Citation18], kindly provided by Kaoru Takegawa (Kyushu University, Japan), and cloned in plasmid p541ʹ (pJK148 derivative [Citation79], expressing GFP under the control of the constitutive sty1 promoter [Citation80]), yielding the integrative plasmid p750′, allowing the integration of the sty1ʹ:GFP-atg8::leu1 gene and the expression of GFP-Atg8. Strains with a chimeric gene sty1ʹ:GFP-atg8::natMX6 were obtained by substituting the leu1 locus of strain CC10 (wild-type, sty1ʹ:GFP-atg8::leu1) by homologous recombinant using a PCR-amplified fragment obtained with a pair of primers flanking leu1 using as a template a pFA-derivative plasmid containing nourseothricin resistance (natMX6). When required, leucine auxotrophy was recovered by crossing with leu1-32 strains.
Experimental conditions
Cells were grown in rich medium YE5S (yeast extract 5 amino acid supplemented; Sunrise, 2011–500, with the standard glucose concentration (3%) or, when indicated, with only 1% or 0.08% glucose, or in synthetic minimal medium MM (Sunrise, 2005–250; 2% glucose) at 30°C as described previously [Citation81]. Unless indicated in the figure legend, autophagy was induced by shifting logarithmic phase cells growing in MM (OD600 of 0.5) to MM containing different concentrations of glucose (1% or 0.08%), or lacking the nitrogen source (NH4Cl; Merck, 12,125–02-5), the phosphorus source (Na2HPO4; Sigma Aldrich, S-0876), or the sulfur source (Na2SO4; Sigma, S-5640). When indicated, MM was supplemented with leucine, lysine, or arginine. Culture media were pre-warmed to 30°C prior to media shift.
Stationary phase conditions and survival assays
To test lifespan in different glucose concentrations, wild type was grown at 30°C in YE5S or MM to the logarithmic phase (OD600 of 0.5), and 1/10 serial dilutions (from 10 to 105 cells) in 3 µl were spotted onto plates of rich media to measure viability. To test survival of auxotrophic strains in starved media, cells were grown at 30°C in supplemented MM media (with leucine, arginine or lysine, as required) to the logarithmic phase (OD600 of 0.5). Then cells were washed twice and transferred to MM without amino acids and/or nitrogen. Again, colony forming units were determined by spotting 1/10 serial dilutions. Viability was measured using dye permeability of PI (propidium iodine) analyzed by flow cytometry, FACS Calibur (Becton Dickinson) at low flow, as described before [Citation26]. Briefly, diluted cells at OD600 of 0.1 were stained during 30 min at 30°C with 2 µg/mL of PI. PI was monitored in channel FL3 (detecting red fluorescence). Percent of survival was obtained comparing color-positive population with unstained cells.
Microscopy
Images were acquired using a Nikon Eclipse 90i microscope equipped with differential interference contrast optics. Images shown were taken by using PLAN APO VC 100 × 1.4 oil immersion objective, and an ORCA-II-ERG camera (Hamamatsu). Processing of all images was performed using Fiji (ImageJ, National Institutes of Health).
Protein extracts and immunoblot analysis
For immunoblot analysis of phosphorylated proteins and immunoblot analysis of GFP-Atg8 from Figures S3A and C, modified TCA (trichloroacetic acid) extracts from 5 ml cultures were prepared as previously described [Citation82]. For most immunoblots of GFP-Atg8, 10 ml of cell cultures at an OD600 of 0.5 were pelleted, and cells were resuspended in 200 µl of PBS with appropriate protease inhibitors. Cells were disrupted using a vortex (Vortex-Genie 2, Scientific Industries) with 0.5 mm glass beads. The membrane fraction was discarded by centrifugation and whole cell extracts were quantified by Bradford assay (Bio-Rad, 5,000,006), to load 30 µg of protein sample per lane. All samples were separated by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and proteins were immuno-detected using the following antibodies: anti-GFP (Takara, 632,381), anti-HA (monoclonal homemade;12 C5A), anti-MYC (SIGMA, C3956), anti-RPS6KB/p70 (Cell Signaling Technology, 9206), anti-AKT (Cell Signaling Technology, 9611), anti-phosphorylated MAPK/p38 (Cell Signaling Technology, 9215) and anti-Sty1 [Citation83]. Protein loading was determined by staining membranes with ATX Ponceau S (Sigma-Aldrich, 09276) in a dilution of Ponceau five-fold in 1% of acetic acid. Visualization, image processing and relative protein quantification were performed using ChemiDoc MP Imaging System (Bio-Rad, 17,001,402) and Fiji (ImageJ, National Institutes of Health).
Gene expression analysis
Total RNA (ribonucleic acid) extraction was performed from logarithmically growing cells and after nitrogen depletion. DNase I treatment of RNA, cDNA (complementary deoxyribonucleic acid) synthesis and RT-qPCR (real time quantitative polymerase chain reaction) were performed as previously described [Citation84,Citation85]. Briefly, reverse transcription was made using 1 μg of RNA using Reverse Transcription System (ThermoFisher, 4,374,967) following the manufacturer’s instructions. Quantification of cDNA was analyzed by using real-time qPCR on LightCycler II using Light Cycler 480 SYBR Green I Master (Roche, 04887352001). Sequence and information of the oligonucleotides used are listed in Table S2.
Statistical analysis
Statistical analysis of gene expression was performed using R Studio software.
Data from four (main figures) or three (supplementary figures) biological replicates are represented as ± standard deviation (SD). We used unpaired Welch’s t-test to calculate P-values and to determine significant differences between deletion strains and wild-type strain. P-values < 0.01 (main figures) or <0.05 (supplementary figures) were considered as significant.
Disclosure of potential conflicts of interest
The authors declare no competing interests.
Supplemental Material
Download MS Word (2.5 MB)Acknowledgments
We thank Kaoru Takegawa for kindly providing plasmid pGFP-Atg8 in pREP41 and strain SK1, and Akio Nakashima and Fuyuhiko Tamanoi for providing AN0175, AN0179 and JUP1350 strains. We thank Li-Lin Du for helpful discussions. This work was supported by the Ministerio de Ciencia, Innovación y Universidades, PLAN E and FEDER (Spain) (PGC2018-093920-B-I00 to E.H.). The Oxidative Stress and Cell Cycle group is also supported by Generalitat de Catalunya (Spain) (2017-SGR-539) and by Unidad de Excelencia María de Maeztu, funded by the AEI (Spain) (CEX2018-000792-M). R.B. is recipient of a FPI contract from the Ministerio de Ciencia, Innovación y Universidades (Spain). E.H. is recipient of an ICREA Academia Award (Generalitat de Catalunya, Spain).
Supplementary material
Supplemental data for this article can be accessed here.
Data and code availability
All images included in the main and supplemental figures are available as Mendeley dataset (doi: 10.17632/wpvb37w474.1).
Additional information
Funding
References
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27(1):107–132.
- Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9–23.
- Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14(2):207–215.
- Otsubo Y, Nakashima A, Yamamoto M, et al. TORC1-Dependent Phosphorylation Targets in Fission Yeast. Biomolecules. 2017;3:7. DOI:https://doi.org/10.3390/biom7030050
- Delorme-Axford E, Klionsky DJ. Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J Biol Chem. 2018;293(15):5396–5403.
- Corona Velazquez AF, Jackson WT. So Many Roads: the Multifaceted Regulation of Autophagy Induction. Mol Cell Biol. 2018;21:38. DOI:https://doi.org/10.1128/MCB.00303-18
- MAA A-B, Xu P. 2020. Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann N Y Acad Sci. 2020; 1467(1):3–20.
- Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19(6):365–381.
- Levine B, Kroemer G. Biological Functions of Autophagy Genes: a Disease Perspective. Cell. 2019;176:11–42.
- Takeshige K, Baba M, Tsuboi S, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311.
- Imai Y, Yamamoto M. Schizosaccharomyces pombe sxa1+ and sxa2+ encode putative proteases involved in the mating response. Mol Cell Biol. 1992;12(4):1827–1834.
- Iwaki T, Osawa F, Onishi M, et al. Characterization of vps33+, a gene required for vacuolar biogenesis and protein sorting in Schizosaccharomyces pombe. Yeast. 2003;20(10):845–855.
- Nakashima A, Hasegawa T, Mori S, et al. A starvation-specific serine protease gene, isp6+, is involved in both autophagy and sexual development in Schizosaccharomyces pombe. Curr Genet. 2006;49(6):403–413.
- Mikawa T, Kanoh J, Ishikawa F. Fission yeast Vps1 and Atg8 contribute to oxidative stress resistance. Genes Cells. 2010;15(3):229–242.
- Stephan J, Franke J, Ehrenhofer-Murray AE. Chemical genetic screen in fission yeast reveals roles for vacuolar acidification, mitochondrial fission, and cellular GMP levels in lifespan extension. Aging Cell. 2013;12(4):574–583.
- Nunez A, Dulude D, Jbel M, et al. Calnexin Is Essential for Survival under Nitrogen Starvation and Stationary Phase in Schizosaccharomyces pombe. PLoS One. 2015;10(3):e0121059.
- Kohda TA, Tanaka K, Konomi M, et al. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells. 2007;12(2):155–170.
- Mukaiyama H, Kajiwara S, Hosomi A, et al. Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology. 2009;155(12):3816–3826.
- Sun LL, Li M, Suo F, et al. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 2013;9(8):e1003715.
- Liu XM, Sun LL, Hu W, Ding YH, Dong MQ, Du LL. ESCRTs Cooperate with a Selective Autophagy Receptor to Mediate Vacuolar Targeting of Soluble Cargos. Mol Cell. 2015;59(6):1035–1042.
- Zhao D, Liu XM, Yu ZQ, et al. Atg20- and Atg24-family proteins promote organelle autophagy in fission yeast. J Cell Sci. 2016;129(22):4289–4304.
- Yu ZQ, Sun LL, Jiang ZD, et al. Atg38-Atg8 interaction in fission yeast establishes a positive feedback loop to promote autophagy. Autophagy. 2020;16(11):2036–2051.
- Tanida I, Ueno T, Kominami E. LC3 and Autophagy. Methods Mol Biol. 2008;445:77–88.
- Yokota H, Gomi K, Shintani T. Induction of autophagy by phosphate starvation in an Atg11-dependent manner in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2017;483(1):522–527.
- Kawamata T, Horie T, Matsunami M, et al. Zinc starvation induces autophagy in yeast. J Biol Chem. 2017;292(20):8520–8530.
- Zuin A, Carmona M, Morales-Ivorra I, et al. Lifespan extension by calorie restriction relies on the Sty1 MAP kinase stress pathway. Embo J. 2010;29(5):981–991.
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484.
- Avruch J, Long X, Ortiz-Vega S, et al. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296(4):E592–602.
- Otsubo Y, Yamamato M. TOR signaling in fission yeast. Crit Rev Biochem Mol Biol. 2008;43(4):277–283.
- Matsuo T, Otsubo Y, Urano J, et al. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27(8):3154–3164.
- Hayashi T, Hatanaka M, Nagao K, et al. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells. 2007;12(12):1357–1370.
- Gao X, Zhang Y, Arrazola P, et al. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4(9):699–704.
- Inoki K, Li Y, Zhu T, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4(9):648–657.
- Tee AR, Fingar DC, Manning BD, et al. Tuberous sclerosis complex-1 and −2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99(21):13571–13576.
- Alvarez B, Moreno S. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J Cell Sci. 2006;119(21):4475–4485.
- Uritani M, Hidaka H, Hotta Y, et al. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells. 2006;11(12):1367–1379.
- Nakashima A, Otsubo Y, Yamashita A, et al. Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase. J Cell Sci. 2012;125(Pt 23):5840–5849.
- Nakashima A, Sato T, Tamanoi F. Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J Cell Sci. 2010;123(5):777–786.
- Urano J, Sato T, Matsuo T, et al. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc Natl Acad Sci U S A. 2007;104(9):3514–3519.
- Ma N, Liu Q, Zhang L, et al. TORC1 signaling is governed by two negative regulators in fission yeast. Genetics. 2013;195(2):457–468.
- Noda T. Regulation of Autophagy through TORC1 and mTORC1. In: Biomolecules. 2017;7(3):52.
- Du W, Forte GM, Smith D, et al. Phosphorylation of the amino-terminus of the AGC kinase Gad8 prevents its interaction with TORC2. In: Open biology. 2016; 6(3):150189.
- Hilti N, Baumann D, Schweingruber AM, et al. Gene ste20 controls amiloride sensitivity and fertility in Schizosaccharomyces pombe. Curr Genet. 1999;35(6):585–592.
- Kawai M, Nakashima A, Ueno M, et al. Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr Genet. 2001;39(3):166–174.
- Weisman R, Choder M. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J Biol Chem. 2001;276:7027–7032.
- Ikeda K, Morigasaki S, Tatebe H, et al. Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle. 2008;7(3):358–364.
- Matsuo T, Kubo Y, Watanabe Y, et al. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 2003;22(12):3073–3083.
- Hatano T, Morigasaki S, Tatebe H, et al. Fission yeast Ryh1 GTPase activates TOR Complex 2 in response to glucose. Cell Cycle. 2015;14(6):848–856.
- Budovskaya YV, Stephan JS, Reggiori F, et al. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279(20):20663–20671.
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966.
- Schmelzle T, Beck T, Martin DE, et al. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 2004;24(1):338–351.
- Hoffman CS. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem Soc Trans. 2005;33(1):257–260.
- Mao K, Wang K, Zhao M, et al. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193(4):755–767.
- Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010;6:794–797.
- Fukuda T, Ebi Y, Saigusa T, et al. Atg43 tethers isolation membranes to mitochondria to promote starvation-induced mitophagy in fission yeast. In: eLife. 2020; 9. doi:https://doi.org/10.7554/eLife.61245.
- Osawa T, Noda NN. Atg2: a novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci. 2019;28(6):1005–1012.
- Chen D, Toone WM, Mata J, et al. Global transcriptional responses of fission yeast to environmental stress. MolBiolCell. 2003;14:214–229.
- Ohtsuka H, Kato T, Sato T, et al. Leucine depletion extends the lifespans of leucine-auxotrophic fission yeast by inducing Ecl1 family genes via the transcription factor Fil1. Mol Genet Genomics. 2019;294(6):1499–1509.
- Ohtsuka H, Takinami M, Shimasaki T, et al. Sulfur restriction extends fission yeast chronological lifespan through Ecl1 family genes by downregulation of ribosome. Mol Microbiol. 2017;105(1):84–97.
- Takeda K, Starzynski C, Mori A, et al. The critical glucose concentration for respiration-independent proliferation of fission yeast, Schizosaccharomyces pombe. Mitochondrion. 2015;22:91–95.
- Weisman R, Roitburg I, Schonbrun M, et al. Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics. 2007;175(3):1153–1162.
- Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183–203.
- Furuta S, Hidaka E, Ogata A, et al. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23(22):3898–3904.
- Mavrakis M, Lippincott-Schwartz J, Stratakis CA, et al. Depletion of type IA regulatory subunit (RIalpha) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Hum Mol Genet. 2006;15(19):2962–2971.
- Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10(18):2276–2288.
- Wilkinson MG, Samuels M, Takeda T, et al. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10(18):2289–2301.
- Yun Y, Liu Z, Zhang J, et al. The MAPKK FgMkk1 of F usarium graminearum regulates vegetative differentiation, multiple stress response, and virulence via the cell wall integrity and high-osmolarity glycerol signaling pathways. Environ Microbiol. 2014;16(7):2023–2037.
- Joubert A, Bataille-Simoneau N, Campion C, et al. Cell wall integrity and high osmolarity glycerol pathways are required for adaptation of Alternaria brassicicola to cell wall stress caused by brassicaceous indolic phytoalexins. Cell Microbiol. 2011;13(1):62–80.
- Webber JL. Regulation of autophagy by p38alpha MAPK. Autophagy. 2010;6(2):292–293.
- Mata J, Bahler J. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc Natl Acad Sci U S A. 2006;103(42):15517–15522.
- Hao Z, Furunobu A, Nagata A, et al. A zinc finger protein required for stationary phase viability in fission yeast. J Cell Sci. 1997;110(Pt 20):2557–2566.
- Mata J, Wilbrey A, Bahler J. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 2007;8(10):R217.
- Ohtsuka H, Azuma K, Kubota S, et al. Chronological lifespan extension by Ecl1 family proteins depends on Prr1 response regulator in fission yeast. Genes Cells. 2012;17(1):39–52.
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–174.
- Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036.
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7(2):167–178.
- Bahler J, Wu JQ, Longtine MS, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14(10):943–951.
- Kim DU, Hayles J, Kim D, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28(6):617–623.
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136(3):849–856.
- Fernandez-Vazquez J, Vargas-Perez I, Sanso M, et al. Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013;9(7):e1003647.
- Alfa C, Fantes P, Hyams J, et al. Experiments with Fission Yeast: a Laboratory Course Manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1993.
- Sanso M, Gogol M, Ayte J, et al. Transcription factors Pcr1 and Atf1 have distinct roles in stress- and Sty1-dependent gene regulation. Eukaryot Cell. 2008;7(5):826–835.
- Jara M, Vivancos AP, Calvo IA, et al. The Peroxiredoxin Tpx1 Is Essential as a H2O2 Scavenger during Aerobic Growth in Fission Yeast. Mol Biol Cell. 2007;18(6):2288–2295.
- Gonzalez-Medina A, Hidalgo E, Ayte J. Gcn5-mediated acetylation at MBF-regulated promoters induces the G1/S transcriptional wave. Nucleic Acids Res. 2019;47(16):8439–8451.
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823.
