ABSTRACT
Folate is an essential mediator in one-carbon metabolism, which provides methyl groups for DNA synthesis and methylation. The availability of active methyl groups can be influenced by the uptake of folic acid. We conducted a randomized intervention trial to test the influence of folic acid supplementation on DNA methylation in an unfortified population in Germany. A total of 16 healthy male volunteers (age range 23–61 y) were randomized to receive either 400 μg (n = 9) or 800 μg (n = 7) folic acid supplements daily for 8 weeks. Infinium Human Methylation 450K BeadChip Microarrays were used to assay site-specific DNA methylation across the genome. Microarray analyses were conducted on PBL DNA. We estimated several epigenetic clocks and mean DNA methylation across all autosomal probes on the array. AgeAccel was estimated as the residual variation in each metric. In virtually all participants, both serum and red blood cell (RBC) folate increased successively throughout the trial period. Participants with a larger increase in RBC folate had a larger increase in DNAmAge AgeAccel (Spearman Rho: 0.56, p-value = 0.03). No notable changes in the methylome resulting from the folic acid supplementation emerged. In this population with adequate folate levels derived from diet, an increase in RBC folate had a modest impact on the epigenetic clock predicting chronologic age.
Introduction
Folic acid fortification of staple foods has been introduced in many countries globally to prevent neural tube defects in newborns as this birth defect results from a folate deficiency in women around the time of conception. While the mechanisms underlying the link between low folate levels in the mother and failure to close the spina bifida in the foetus have not been fully clarified, it is likely that epigenetic mechanisms play a role [Citation1]. Folate is an essential mediator in one-carbon metabolism, which provides methyl groups for DNA synthesis and methylation. The availability of active methyl groups can be influenced by diet and uptake of folic acid as well as other key players in one-carbon metabolism, such as choline and methionine. Folate deficiency may correlate with hypomethylation [Citation2]. Conversely, high folate levels may be associated with hypermethylation; however, research in this area is lacking. If folic acid fortification affects DNA methylation, this might be associated with disease initiation or promotion. Hence, it is important to understand the impact of folic acid intake on DNA methylation.
5,10-methylenetetrahydrofolate reductase (MTHFR) is a regulator enzyme of one-carbon metabolism and essential for conversion of folic acid, thus regulating folate bioavailability. The common variant (MTHFR)-C677T has reduced enzyme activity and therefore decreases blood folate concentrations, thus increasing intake requirement among carriers [Citation3]. The MTHFR genotype affects the relation of folate concentration with both DNA replication (cell division/survival) and DNA methylation (cell regulation) [Citation4].
European countries have generally not participated in fortification programmes such as fluoridation of drinking water, fortification of milk with vitamins A and D, or fortification of flour with micronutrients leaving decisions about supplementations to the consumer. Accordingly, European countries have not adopted folic acid fortification despite a high incidence of neural tube defects. In Germany, folate deficiency is common and folic acid supplementation is low. According to the German National Nutrition Survey II, the median dietary folate equivalents in Germany are 191 µg/d for men and 168 µg/d for women [Citation5]. More than half of all Germans consume less than 200 µg of folate. The German Society of Nutrition recommends a daily intake of 300 µg folate for adults [Citation6]. All women planning to get pregnant are advised in addition to a diet rich in folate-containing foods to take a supplement containing at least 400 µg of folic acid daily at least 4 weeks prior to pregnancy and to maintain this intake at least until the end of the first trimester [Citation6]. In the U.S., the respective recommended dietary allowances are 400 µg/d and, for pregnant women, 600 µg/d [Citation7]. Therefore, a folic acid supplementation trial in Germany is feasible and informative.
We conducted an intervention study in Germany to explore the impact of folic acid supplementation on DNA methylation in an unfortified population stratified by MTHFR genotype. Specifically, we evaluated associations with DNA methylation-based estimates of characteristics related to biological age termed epigenetic clocks. Epigenetic clocks refer to models that predict characteristics such as chronological age, the pace of ageing, and mortality risk using specific DNA methylation signatures (collectively referred to here as ‘epigenetic age’). Higher epigenetic age relative to those of the same chronologic age is associated with increased rates of age-related disease and mortality [Citation8]. Several large studies indicate that accelerated epigenetic age is associated with greater cardiovascular disease risk [Citation9–13]. Both observational studies and randomized controlled trials suggest that folic acid supplementation and higher folate levels are associated with reduced risk of cardiovascular disease [Citation14,Citation15]. Folate is the cofactor for methionine synthase, which catalyzes the conversion of homocysteine, corresponding to in higher levels of homocysteine with lower folate levels. It is hypothesized that hyperhomocysteinemia may contribute to cardiovascular disease by causing damage to endothelial cells and leading to atherosclerosis [Citation16]. Our study will evaluate whether changes in folate levels are correlated with changes in epigenetic age, both of which are indicators of cardiovascular disease risk. We expect that higher folate levels will be associated with lower epigenetic age, given their respective relations to cardiovascular health.
Methods
Study design and population
We conducted a randomized intervention trial to test the influence of folic acid supplementation on DNA methylation at the University of Freiburg, Germany. A total of 16 healthy male volunteers (age range 23–61 y) were randomized to receive either 400 μg (n = 9) or 800 μg (n = 7) folic acid supplements daily for 8 weeks. Prior to the start of the intervention and after 2, 4, and 8 weeks of supplementation, blood and oral mucosa samples were collected and analysed. Blood samples were processed immediately following collection. Serum and red blood cells were isolated from ETDA blood samples and stored until further analyses. Peripheral blood leukocytes (PBLs) were isolated from the blood samples, and DNA was isolated from the PBLs and the mucosa samples. All samples yielded sufficient materials of good quality for further analyses. The study was approved by the committee for research ethics of the University of Freiburg.
Folate and vitamin B12 assessment
Serum folate and red blood cell (RBC) folate were measured in hemolyzed EDTA blood samples using a chemiluminescent assay. Vitamin B12 levels were assessed to exclude a vitamin B12 deficiency. Vitamin B12 was measured by an ElektroChemiLumineszenz ImmunoAssay “ECLIA“. Serum folate and vitamin B12 analyses were performed in the Clinical Laboratory of the University Medical Center, University of Freiburg, Germany. Red blood cell folate was measured at the Clotten Laboratory, Freiburg, Germany. Assessments of all analytes were adapted on the Roche Cobas e801, Germany.
MTHFR genotyping
MTHFR genotyping was performed in our laboratory using Realtime PCR via TaqMan (LC480 from Roche) following the PCR-RFLP method described by Frosst et al. [Citation17].
Genome-wide methylation assessment
Infinium Human Methylation 450K BeadChip Microarrays were used to assay site-specific DNA methylation across the genome. Microarray analyses were conducted on PBL DNA. We analysed individual samples (1 μg) from each of the 16 participants collected at baseline prior to the intervention and at the end of the intervention at week 8 (week 4 for one participant). We also pooled samples across all participants by combining 2.5 μl DNA solution standardized at 25 ng/μl from each of the 16 participants at time point zero and vortexing the pooled sample for 2 minutes; a second pooled sample for time point week 8 was combined in the same way. The 32 individual samples and 2 pooled samples were shipped on dry ice to the University of Southern California Epigenome Center in Los Angeles, California, for Methylation Microarray Analysis. We pre-processed raw signal intensities by performing dye bias correction followed by single-sample background correction based on Normal-exponential convolution using out-of-band Infinium I probes (ssNoob) [Citation18]. Site-specific DNAm levels (β-values) were estimated based on the ratio of methylated (M) and unmethylated (U) fluorescence intensities, given by β=M/(M+U + 100), and ranging from 0 (unmethylated) to 1 (methylated).
DNA methylation-based biomarkers
We estimated several epigenetic clocks, including those trained to predict chronologic age (DNAmAge [Citation19], DNAmAgeHannum [Citation20]), telomere length (DNAmTL) [Citation21], and characteristics associated with time-to-death (PhenoAge [Citation22], GrimAge [Citation23], and GrimAge2 [Citation24]) and pace of ageing (DunedinPACE) [Citation25]. Description of the training sets, the specific characteristics used to develop the model, and validation of the estimates are included in the specified references. We additionally estimated mean DNA methylation across all autosomal probes on the array (‘Genome-Wide Mean’). AgeAccel was estimated as the residual variation in each metric after adjusting for participants’ chronological age at sample collection and DNA methylation-based estimates of cell composition (CD8+ T cells, CD4+ T cells, natural killer cells, B-cells, monocytes, neutrophils). Cell composition was estimated using reference-based constrained projection [Citation26], and the Identifying Optimal Libraries (IDOL) [Citation27,Citation28] probes to distinguish cell types for 450K arrays.
Statistical analysis
Linear mixed-effects models were used to model the change in serum and RBC folate over follow-up (week), incorporating a random intercept for participant. Effect modification by folic acid supplement formulation (400 µg vs 800 µg) and MTHFR genotype was assessed based on the significance (Wald test) of a product term between time and each characteristic individually. For analyses integrating the DNA methylation data, the longitudinal change in AgeAccel was estimated as the difference between AgeAccel at follow-up (week 8; week 4 for one participant) and baseline (week 0). The longitudinal change in folate measures was similarly estimated. RBC and serum folate was missing at week 8 for one participant, and imputed with the values at week 4. A paired non-parametric Wilcoxon signed-rank test was used to evaluate longitudinal change in AgeAccel across individuals. The non-parametric Spearman’s rank correlation was used to evaluate the relation between AgeAccel and folate measures at baseline, as well as the correlation between the longitudinal change in the AgeAccel and folate measures. The benefit of this approach is that our analysis is robust to influential observations, which are more likely to impact significance in a small study. As a supplementary analysis, we evaluate these associations using linear models, adjusting for age, to estimate effect size, corresponding 95% confidence interval (CI) and significance (Wald test). The alpha-level for all tests was 0.05. All analyses were performed in R v 4.2.0.
Results
Serum and RBC folate levels
Prior to intervention, all trial participants had normal serum and RBC folate level and normal vitamin B12 levels (). In almost all participants, both serum and RBC folate increased successively throughout the trial period (). There was a significant 0.98 ng/ml (95% confidence interval (CI): 0.67,1.29) mean increase in serum folate per week. There was a significant 22.16 ng/ml (95% CI: 16.81, 27.52) mean increase in RBC folate per week. The increase in serum and RBC folate over time was not significantly modified by MTHFR genotype or folic acid supplement formulation. RBC and serum folate were highly correlated at baseline (Spearman Rho: 0.83, p < 0.001). By week 8, this correlation was attenuated and no longer significant (Spearman Rho: 0.44, p = 0.12).
Figure 1. Graphic representation of a) RBC and b) serum folate levels of the 16 study participants after daily consumption of 400 or 800 µg folic acid. Normal range of RBC folate: 263–1028 ng/ml; serum folate: 4.6–18.7 ng/ml. Two participants had serum folate values >40 ng/ml, above the detection limit.
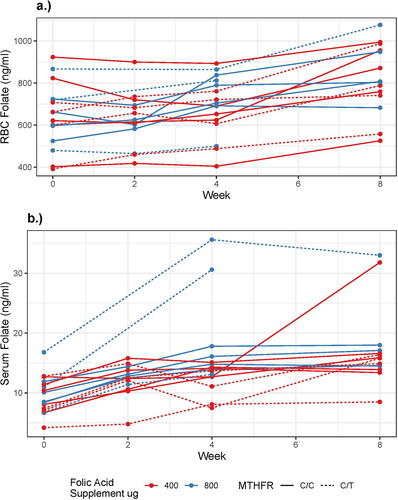
Table 1. Study participant characteristics at blood draws.
DNA methylation-based biomarkers
At baseline, RBC and serum folate were not correlated with any of our estimates of AgeAccel (). The average change in AgeAccel between baseline and follow-up across participants was not significant for any measure (). However, the longitudinal change in DNAmAge estimates of AgeAccel was modestly correlated with the longitudinal change in RBC folate (, ). Participants with a larger increase in RBC folate had a larger increase in DNAmAge AgeAccel (Spearman Rho: 0.56, p-value = 0.03); this relation was not observed with serum folate. Based on linear models, a 100 ng/ml increase in RBC folate was associated with a 2.33 (95% CI: −0.35, 5.00) year increase in DNAmAge AgeAccel over follow-up. Increases in RBC and serum folate were not significantly correlated with changes in any other AgeAccel measures (, ). Hence, no notable changes in the methylome resulting from the folic acid supplementation were observed.
Figure 2. Correlation between AgeAccel and a) RBC folate, and b) serum folate at baseline. Point colour corresponds to participant.
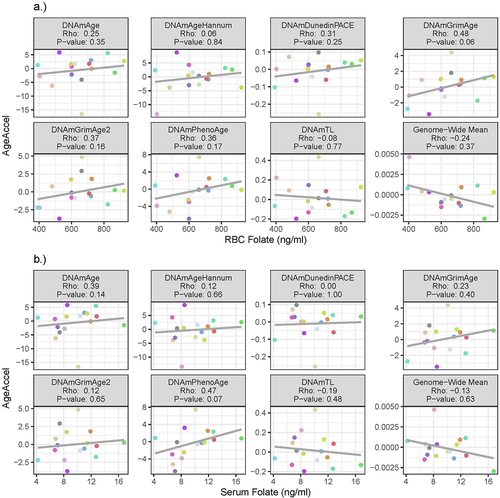
Figure 3. The longitudinal change in AgeAccel a) over follow-up, and in relation to the change in b) RBC folate, and c) serum folate. Point colour corresponds to participant.
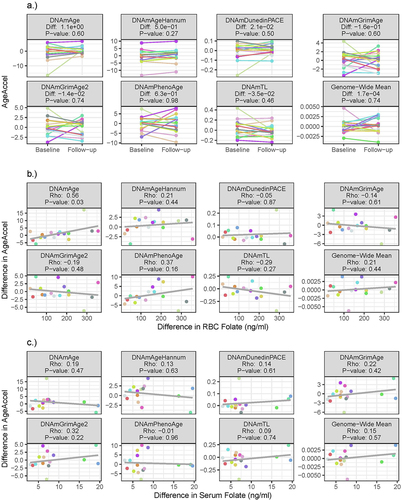
Table 2. Association between AgeAccel and folate levels.
Discussion
In this randomized intervention trial, we aimed to elucidate whether folic acid supplementation in an unfortified population with adequate folate blood levels affects DNA methylation status. The folic acid supplementation influenced RBC folate and serum folate levels over the course of 2 months in all participants; however, DNA methylation status remained unaffected. We observed a modest correlation between an increase in RBC folate levels and increase in DNAmAge AgeAccel.
Studies on the impact of folic acid supplementation are sparse but important in light of safety consideration given the folic acid fortification programmes in many countries around the globe. In the Aspirin‐Folate Polyp Prevention Study in the U.S., folic acid supplementation with 1 mg/d for up to 6 y in subjects with previous colorectal adenomas increased the number of new adenomas [Citation29]. In Norway, where there is no folic acid fortification, the combined supplementation of folic acid and vitamin B12 resulted in an increase in cancer incidence among patients with ischaemic heart disease [Citation30]. It has been suspected that folic acid supplementation might promote the progression of underlying preneoplastic lesions [Citation29,Citation31]; however, it remains unclear whether DNA methylation may play a role in this observation.
Previous studies have reported mixed results of folic acid intake on DNA methylation. A study in colorectal cancer patients found an increase in global methylation [Citation32], while another study observed hypomethylation [Citation33], yet a third study found no methylation changes [Citation34]. Van den Donk et al. [Citation35] observed an increase in DNA methylation in promoters of six tumour suppressor genes after folic acid supplementation. Kok and colleagues examined the effects of long-term (104 weeks) daily folic acid (400 µg/d) and vitamin B 12 (500 µg/d) supplementation on genome- wide DNA methylation assessed with the 450K array in adult leukocyte samples of elderly subjects and found six significant DMRs between the intervention and placebo groups [Citation36]. The amount of folic acid intake ranging from 400 µg/d to 5 mg/d as well as the duration of intervention ranging between 2 and 6 months varied considerably across studies. These differences may explain some of the diverging results observed since folate may have dual effects [Citation28,Citation37,Citation38]. A recent review of studies addressing the association between nutrients involved in one-carbon metabolism and DNA methylation supported this heterogeneity of findings, which may also have been due to differences in the assessment of methylation status [Citation39]. The majority of intervention studies examined the effect of one-carbon metabolism nutrients on global methylation, while only a few investigated gene-specific or genome-wide methylation [Citation39].
The potential impact of folic acid supplementation on age acceleration assessed by the epigenetic clock has been little studied. Sae-Lee et al. used data from a folic acid intervention trial with publicly available Illumina Infinium 450K methylation datasets to explore the impact on the epigenetic clock [Citation40]. In this study, 44 participants were randomized to supplementation with folic acid (400 µg/d) and vitamin B12 (500 µg/d) for 2 y [Citation36]. Age acceleration did not significantly change following the intervention, however, decreased epigenetic ageing was observed among women with the MTHFR 677CC genotype [Citation40].
There are a number of possible explanations why we did not observe more substantial changes in DNA methylation despite continuously rising folate levels among all participants throughout the trial. Far and foremost, none of the participants had a folate deficit prior to supplementation. DNA methylation may be relatively stable as long as one-carbon metabolism is sufficiently catalysed by all important mediators and therefore generates adequate methyl groups. Had we selected a deficient population, DNA methylation profiles may have been more variable. Moreover, while a deficit in the availability of methyl groups is strongly correlated with hypomethylation, excess availability does not necessarily translate into hypermethylation [Citation41]. An alternative explanation may have been our intervention and observation period which spanned 2 months and may not have been sufficient to alter the methylation pattern more substantially. DNA methylation has been considered a relatively stable mechanism [Citation42]. However, there may be differences in DNA methylation stability depending on the genomic location. Moreover, studies have identified certain stressors that may induce fairly rapid changes in DNA methylation. Incubation with a methyltransferase inhibitor may result in a rapid turnover of DNA methylation [Citation43]. Immune response to infection also induces fast active demethylation of many CpG sites [Citation44]. However, our intervention may not represent a sufficiently strong impact to influence DNA methylation. Furthermore, we only measure methylation in PBLs, hence changes in methylation patterns in other tissues would have escaped our assessment. Moreover, synthetic folic acid and folate from diet may have different effects.
Previous studies have pointed to an increased need for folate among carriers of the (MTHFR)-C677T variant. We did not detect differences according to the MTHFR variants; however, our study participants had adequate folate blood levels prior to intervention despite the lack of folic acid fortification programmes in Germany.
Our study was modest in sample size and duration limiting statistical power and generalizability of the results. However, we were able to test two regimens of folic acid interventions, 400 and 800 µg. Our modest study size precluded adjustment for potential confounding of the association between AgeAccel and folate levels, and between changes in these measures over follow-up. While the observed associations may be driven by common causes of change in AgeAccel and folate levels, our study does characterize shared longitudinal variation. Future investigations may illuminate what factors contribute to the observed correlations.
In summary, we observe few and select differences in PBL DNA methylation patterns following supplementation with folic acid in a population with adequate folate levels.
Author contributions
K.B. Michels designed the research; A.M. Binder analysed the data; K.B. Michels wrote the manuscript; A.M. Binder provided critical scientific input on the manuscript; both authors approved the final version of the manuscript.
Acknowledgments
We thank Mara Waldschmidt for help with conducting the trial and collecting the samples. We are grateful to the 16 trial participants for their time and compliance with the study protocol.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available upon request from the corresponding author. The data are not publicly available due to privacy restrictions.
Additional information
Funding
References
- Chang H, Zhang T, Zhang Z, et al. Tissue-specific distribution of aberrant DNA methylation associated with maternal low-folate status in human neural tube defects. J Nutr Biochem. 2011;22(12):1172–10. doi: 10.1016/j.jnutbio.2010.10.003
- Choi S-W, Friso S. Nutrients and Epigenetics [Internet]. 1st. CRC Press; 2009 . Available from10.1201/9781420063561
- Bailey LB. Folate in Health and disease [Internet]. 2nd. CRC Press; 2009 . Available from10.1201/9781420071252
- Crider KS, Yang TP, Berry RJ, et al. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr. 2012;3(1):21–38. doi: 10.3945/an.111.000992
- Martiniak Y, Heuer T, Hoffmann I. Intake of dietary folate and folic acid in Germany based on different scenarios for food fortification with folic acid. Eur J Nutr. 2015;54(7):1045–1054. doi: 10.1007/s00394-014-0781-1
- Folat [Internet]. https://www.dge.de/wissenschaft/referenzwerte/folat/?L=0 [cited 2023 Mar 3]; Available from: https://www.dge.de/wissenschaft/referenzwerte/folat/?L=0
- Dietary Supplement Fact Sheets [Internet]. https://ods.od.nih.gov/factsheets/list-all/[cited 2023 Mar 3]; Available from: https://ods.od.nih.gov/factsheets/list-all/
- Fransquet PD, Wrigglesworth J, Woods RL, et al. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin Epigenetics. 2019;11(1):62. doi: 10.1186/s13148-019-0656-7
- Lind L, Ingelsson E, Sundström J, et al. Methylation-based estimated biological age and cardiovascular disease. Eur J Clin Invest. 2018;48(2):48. doi: 10.1111/eci.12872
- Roetker NS, Pankow JS, Bressler J, et al. Prospective study of epigenetic age acceleration and incidence of cardiovascular disease outcomes in the ARIC study (atherosclerosis risk in communities). Circ Genom Precis Med. 2018;11(3):e001937. doi: 10.1161/CIRCGEN.117.001937
- Si J, Chen L, Yu C, et al. Healthy lifestyle, DNA methylation age acceleration, and incident risk of coronary heart disease. Clin Epigenetics. 2023;15(1):52. doi: 10.1186/s13148-023-01464-2
- Ammous F, Zhao W, Ratliff SM, et al. Epigenetic age acceleration is associated with cardiometabolic risk factors and clinical cardiovascular disease risk scores in African Americans. Clin Epigenetics. 2021;13(1):55. doi: 10.1186/s13148-021-01035-3
- Joyce BT, Gao T, Zheng Y, et al. Epigenetic Age Acceleration Reflects Long-Term Cardiovascular Health. Circ Res. 2021;129(8):770–781. doi: 10.1161/CIRCRESAHA.121.318965
- Li Y, Huang T, Zheng Y, et al. Folic acid supplementation and the risk of cardiovascular diseases: a meta-analysis of randomized controlled trials. J Am Heart Assoc. 2016;5(8):e003768. doi: 10.1161/JAHA.116.003768
- Wang Z-M, Zhou B, Nie Z-L, et al. Folate and risk of coronary heart disease: a meta-analysis of prospective studies. Nutr Metab Cardiovasc Dis. 2012;22(10):890–899. doi: 10.1016/j.numecd.2011.04.011
- Ganguly P, Alam SF. Role of homocysteine in the development of cardiovascular disease. Nutr J. 2015;14(1):6. doi: 10.1186/1475-2891-14-6
- Frosst P, Blom HJ, Milos R, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10(1):111–113. doi: 10.1038/ng0595-111
- Fortin J-P, Triche TJ, Hansen KD, et al. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. doi: 10.1093/bioinformatics/btw691
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115
- Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016
- Lu AT, Seeboth A, Tsai P-C, et al. DNA methylation-based estimator of telomere length. Aging. 2019;11(16):5895–5923. doi: 10.18632/aging.102173
- Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. doi: 10.18632/aging.101414
- Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–327. doi: 10.18632/aging.101684
- Lu AT, Binder AM, Zhang J, et al. DNA methylation GrimAge version 2. Aging. 2022;14:9484–9549. doi: 10.18632/aging.204434
- Belsky DW, Caspi A, Corcoran DL, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife. 2022;11:e73420. doi: 10.7554/eLife.73420
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13(1):86. doi: 10.1186/1471-2105-13-86
- Koestler DC, Jones MJ, Usset J, et al. Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL). BMC Bioinf. 2016;17(1):120. doi: 10.1186/s12859-016-0943-7
- Osterhues A, Holzgreve W, Michels KB. Shall we put the world on folate? Lancet. [cited 2012 Oct 12] 2009;374(9694):959–961. Available from http://www.ncbi.nlm.nih.gov/pubmed/19766871 10.1016/S0140-6736(09)61646-9
- Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297(21):2351–2359. doi: 10.1001/jama.297.21.2351
- Ebbing M, Bønaa KH, Nygård O, et al. Cancer incidence and mortality after treatment with folic acid and vitamin B12. JAMA. 2009;302(19):2119–2126. doi: 10.1001/jama.2009.1622
- Kim Y-I. Folate: a magic bullet or a double edged sword for colorectal cancer prevention? Gut. 2006;55(10):1387–1389. doi: 10.1136/gut.2006.095463
- Pufulete M. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005;54(5):648–653. doi: 10.1136/gut.2004.054718
- Cravo M, Fidalgo P, Pereira AD, et al. DNA methylation as an intermediate biomarker in colorectal cancer: modulation by folic acid supplementation. Eur J Cancer Prev. 1994;3(6):473–480. doi: 10.1097/00008469-199411000-00004
- Figueiredo JC, Grau MV, Wallace K, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1041–1049. doi: 10.1158/1055-9965.EPI-08-0926
- van den Donk M, van Engeland M, Pellis L, et al. Dietary folate intake in combination with MTHFR C677T genotype and promoter methylation of tumor suppressor and DNA repair genes in sporadic colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2007;16(2):327–333. doi: 10.1158/1055-9965.EPI-06-0810
- Kok DEG, Dhonukshe-Rutten RAM, Lute C, et al. The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin Epigenetics. 2015;7(1):121. doi: 10.1186/s13148-015-0154-5
- Kim Y-I. Folate and colorectal cancer: an evidence-based critical review. Mol Nutr Food Res. 2007;51(3):267–292. doi: 10.1002/mnfr.200600191
- Smith AD, Kim Y-I, Refsum H. Is folic acid good for everyone? Am J Clin Nutr. 2008;87(3):517–533. doi: 10.1093/ajcn/87.3.517
- Amenyah SD, Hughes CF, Ward M, et al. Influence of nutrients involved in one-carbon metabolism on DNA methylation in adults—a systematic review and meta-analysis. Nutr Rev. 2020;78(8):647–666. doi: 10.1093/nutrit/nuz094
- Sae-Lee C, Corsi S, Barrow TM, et al. Dietary intervention modifies DNA methylation age assessed by the epigenetic clock. Mol Nutr Food Res. 2018;62(23):e1800092. doi: 10.1002/mnfr.201800092
- In: Michels KB. editor. Epigenetic epidemiology. 2nd. Cham, Switzerland: Springer International Publishing; 2022. [cited 2023 Mar 3]. Available from https://link.springer.com/10.1007/978-3-030-94475-9
- Allis CD, Jenuwein T, Reinberg D, et al. , editors. Epigenetics. 1st ed. Cold Spring Harbour, New York: Cold Spring Harbor Laboratory Press; 2007. Overview and Concepts; p. 23–61.
- Yamagata Y, Szabó P, Szüts D, et al. Rapid turnover of DNA methylation in human cells. Epigenetics. 2012;7(2):141–145. doi: 10.4161/epi.7.2.18906
- Pacis A, Mailhot-Léonard F, Tailleux L, et al. Gene activation precedes DNA demethylation in response to infection in human dendritic cells. Proc Natl Acad Sci U S A. 2019;116(14):6938–6943. doi: 10.1073/pnas.1814700116
