Abstract
Nitric oxide (NO) has emerged as an important regulator of the nitrogen assimilation pathway in plants. Nevertheless, this free radical is a double-edged sword for cells due to its high reactivity and toxicity. Hemoglobins, which belong to a vast and ancestral family of proteins present in all kingdoms of life, have arisen as important NO scavengers, through their NO dioxygenase (NOD) activity. The green alga Chlamydomonas reinhardtii has 12 hemoglobins (THB1–12) belonging to the truncated hemoglobins family. THB1 and THB2 are regulated by the nitrogen source and respond differentially to NO and the nitrate/ammonium balance. THB1 expression is upregulated by NO in contrast to THB2, which is downregulated. THB1 has NOD activity and thus a role in nitrate assimilation. In fact, THB1 is upregulated by nitrate and is under the control of NIT2, the major transcription factor in nitrate assimilation. In Chlamydomonas, it has been reported that nitrate reductase (NR) has a redox regulation and is inhibited by NO through an unknown mechanism. Now, a model in which THB1 interacts with NR is proposed for its regulation. THB1 takes electrons from NR redirecting them to NO dioxygenation. Thus, when cells are assimilating nitrate and NO appears (i.e. as a consequence of nitrite accumulation), THB1 has a double role: 1) to scavenge NO avoiding its toxic effects and 2) to control the nitrate reduction activity.
Nitrate is quantitatively the most important nitrogen source in many environments and its assimilation is a highly regulated process in photosynthetic organisms. This regulation fine-tunes nitrate assimilation to changing environmental conditions such as the nitrogen source (nitrate or ammonium) or light, among others. Nitrate reductase (NR) catalyzes the first reduction step from nitrate to nitrite and is a key enzyme to regulate nitrate assimilation. NR has to be coupled with nitrite reductase (NiR) to avoid nitrite accumulation and its undesirable side effects.
In plants, NR is regulated by phosphorylation and 14–3–3 protein binding, albeit other kinds of regulation can exist as reported in wheat or tomato where NO inhibits the enzyme.Citation1,2 In Chlamydomonas, NR is not regulated by 14–3–3 proteins and a redox mechanism of regulation has been proposed.Citation3 Additionally, it has been reported that NO inhibits NR activity. This inhibition is reversible and is not a consequence of a direct interaction NO-NR.Citation4 Thus, some cellular component has been proposed to be required. Now, this regulation starts to be revealed and seems to involve, at least partially, the interaction between NR and hemoglobins. Three types of Hbs are found in photosynthetic organisms, symbiotic, non-symbiotic (ns-Hb) and truncated (trHb). Ns-Hbs and trHbs have been described as important proteins for NO detoxification. These proteins are able to bind O2 to the reduced Fe atom of the heme group and then perform the NO dioxygenation to produce nitrate (NOD activity). This reaction is produced by almost every Hb in vitro, however, the limiting factor for NOD activity is the Hb reduction.Citation5 It has been reported that ns-Hbs from A. thaliana are able to take electrons directly from NADPHCitation6 and ns-Hbs and trHbs from L. japonicus can be reduced by NAD(P)H in the presence of FAD or FMN.Citation7 Nevertheless, a long time is required for free cofactor reduction of several of these Hbs and their slow kinetics could be useless for a fast NO scavenging in the cell. Alternatively, Hbs could be coupled to specific reductases. In some microorganisms, genes encoding globin domains are fused with reductases,Citation8 but for most Hbs there exist not this genetic association. Thus, current research on NO detoxification should focus, not only on the Hbs and their functions, but in the reductases that activate Hbs. Recently, it has been reported that GlbN from M. tuberculosis, the most studied truncated hemoglobin with NOD activity,Citation9 can be reduced efficiently by the NADH-ferredoxin/flavodoxin system from E. coli.,Citation10 but the reductase responsible for this function in Mycobacterium remains unknown. Some data point out that NR could be a reductase for particular Hbs, like the genetic evidence that NR genes are fused with a globin domainCitation11 in Raphidophyte algae, or the coordinated spatiotemporal expression of NR and Hbs described in plants.Citation12
Interestingly, it appears that NR is able to produce NO from nitrite in both plants and algae.Citation13-15 This could be a way to avoid nitrite accumulation by inhibiting nitrate reduction. But, NO production from NO2− could have 2 negative effects for cells. Firstly, NO is a toxic free radical, which has to be quickly eliminated and, secondly, diffusion and unspecific NO reactions would suppose a nitrogen loss for its assimilation. Accordingly, a strategy to avoid toxic effects and nitrogen loss is to couple NR activity with hemoglobins (Hbs) and to convert NO to NO3−, which can be accumulated.
In Chlamydomonas 12 trHbs have been reportedCitation16 and 2 of them (THB1-THB2) seem to be closely linked with nitrate assimilation. These two hemoglobins are upregulated in nitrate media (nitrate (N) and nitrate plus ammonium (NA)). We have observed that THB1 and THB2 respond to the nitrate/ammonium balance, as reported for the nitrate reductase gene (NIA1),Citation17 although they show opposite responses. THB1 expression increases when the balance is shifted to ammonium (N 4 mM and A 8 mM) and, conversely, THB2 expression is higher when nitrate is more abundant (N 4 mM and A 1mM) (). It has been described that in NA condition intracellular NO levels increase when the balance is shifted to ammonium,Citation18 and this agrees with the expression patterns of these hemoglobins in the presence of NO donors. THB1 expression is upregulated when NO is added to the medium while THB2 transcript levels decrease.
Figure 1. THB1 and THB2 expression in 704 strain (wt), in different nitrate/ammonium balances and in the presence of NO, in wt and nit2 mutant strains. (A) Wild type cells (704) were grown in minimal medium with ammonium as nitrogen source, and then incubated with a fixed nitrate concentration of 4 mM plus ammonium 1 (NA1) or 8 mM (NA8) and THB1 and THB2 expression was measured. (B) Cells were grown in the same conditions described above and then incubated in the indicated media. THB1 and THB2 expression in nitrogen free media on each strain was considered as 100% (gray line). DEANONOate (100 µM) was used as NO donor. Accession numbers of THB1 and THB2 are KC992719 and KC992720, respectively. Gene expression conditions and primers used are not included in this addendum. Error bars represent technical triplicates from at least 2 biological experiments. Observed differences in THB1 and THB2 expression between different balances (A) and NO treatments in 704 and 89.87 (B) were statistically significant with at least a p< 0,05.
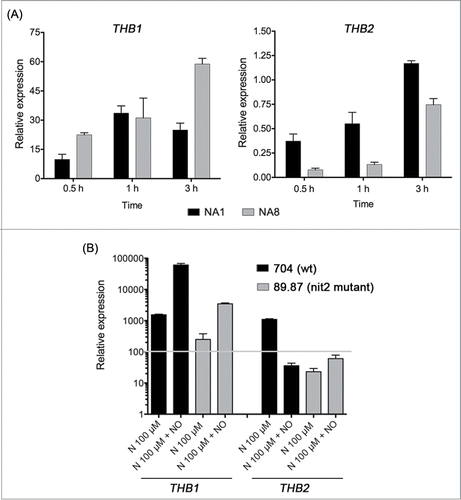
THB1 and THB2 upregulation in nitrate containing media depends on NIT2, the main transcriptional factor for nitrate assimilation genes. Nevertheless, we have observed that THB1 induction by NO was partially independent of NIT2. NO increases THB1 transcript levels in a nit2 mutant, although expression does not reach the levels observed in the wild type strain (). These results show an additive effect of nitrate and NO and arise the possibility of 2 transcriptional activators for THB1 expression. NIT2 would sense nitrate and an unknown factor could be responsible for sensing the NO signal. Nevertheless, deeper studies will be necessary. NO effects over THB2 expression in the nit2 mutant were slight and irrelevant because this strain showed a low basal expression, which cannot be indeed repressed by NO.
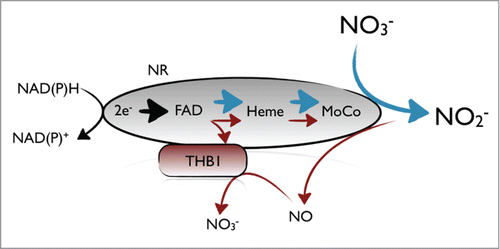
NO and nitrate-dependent THB1 overexpression highlighted this hemoglobin as a NOD enzyme involved in nitrogen assimilation. We have shown that NR was able to reduce THB1, through its diaphorase activity,Citation19 more efficiently than free cofactor (NADH and FAD) and other reductases (like Cytochrome b5 reductase (Cytb5R), which has the highest homology to NR in Chlamydomonas). In vitro experiments showed that THB1 was maintained in its reduced form by NR after several cycles of NO dioxygenation. Our data indicate that THB1 interacts with NR to scavenge NO and to inhibit nitrate reduction. A model explaining how NO inhibits NR in a THB1 dependent way is proposed in . When cells are growing in nitrate under optimal conditions for its assimilation, the electrons flux from NAD(P)H→FAD→Heme→Moco to NO3− reduction. When the environmental conditions change, i.e., the presence of ammonium or high NO2− where a NO burst is produced,Citation18 THB1 can take electrons from NR to scavenge NO and produce NO3− by its NOD activity. This bypass of electrons when THB1 is working allows less reducing power for nitrate reduction, adjusting nitrite production with the cell capability of assimilating it, and neutralizing undesirable effects of NO.
Figure 2. Schematic representation of NR regulation by THB1. In conditions where nitrite concentrations are low, all electrons go to nitrate reduction (blue arrows). If nitrite increases, NR produces NO and then THB1 can take electrons from NR to convert NO in nitrate (red arrows) and NR activity is partially inhibited.
Our In vitro and in vivo results have revealed that NR and THB1 work together to control nitrate assimilation and NO levels. However, we have also reported that THB1 is reduced by Cytb5R and by extracts of NR deficient mutants. Thus, our data have brought out new questions about the role of THB2 in nitrate assimilation and whether THB1 could regulate other processes accepting electrons from other reductases when NO appears.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Authors thank Maribel Macías for skillful technical assistance.
Funding
This work was funded by MINECO (Ministerio de Economia y Competitividad, Spain, Grant no. BFU2011–29338) with support of the European Fondo Europeo de Desarrollo Regional (FEDER) program, Junta de Andalucía (P08-CVI-04157 and BIO-502), and Plan Propio de la Universidad de Córdoba.
References
- Rosales EP, Iannone MF, Groppa MD, Benavides MP. Nitric oxide inhibits nitrate reductase activity in wheat leaves. Plant Physiol Biochem 2011; 49:124-30; PMID:21093280; http://dx.doi.org/10.1016/j.plaphy.2010.10.009
- Jin CW, Du ST, Zhang YS, Lin XY, Tang CX. Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum). Ann Bot 2009; 104:9-17; PMID:19376780; http://dx.doi.org/10.1093/aob/mcp087
- Franco AR, Cárdenas J, Fernandez E. Involvement of Reversible Inactivation in the Regulation of Nitrate Reductase Enzyme Levels in Chlamydomonas reinhardtii. Plant Physiol 1987; 84:665-9; PMID:16665499; http://dx.doi.org/10.1104/pp.84.3.665
- Sanz-Luque E, Ocana-Calahorro F, Llamas A, Galvan A, Fernandez E. Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii. J Exp Bot 2013; 64:3373-83; PMID:23918969; http://dx.doi.org/10.1093/jxb/ert175
- Smagghe BJ, Trent JT, Hargrove MS. NO Dioxygenase Activity in Hemoglobins Is Ubiquitous In Vitro, but Limited by Reduction In Vivo. PLoS ONE 2008; 3:e2039; http://dx.doi.org/10.1371/journal.pone.0002039
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 2004; 16:2785-94; PMID:15367716; http://dx.doi.org/10.1105/tpc.104.025379
- Sainz M, Pérez-Rontomé C, Ramos J, Mulet JM, James EK, Bhattacharjee U, Petrich JW, Becana M. Plant hemoglobins may be maintained in functional form by reduced flavins in the nuclei, and confer differential tolerance to nitro-oxidative stress. Plant J 2013; 76:875-87; PMID:24118423; http://dx.doi.org/10.1111/tpj.12340
- Bonamore A, Boffi A. Flavohemoglobin: Structure and reactivity. IUBMB Life 2007; 60:19-28; http://dx.doi.org/10.1002/iub.9
- Ouellet H, Ouellet Y, Richard C, Labarre M, Wittenberg B, Wittenberg J, Guertin M. Truncated hemoglobin HbN protects Mycobacterium bovis from nitric oxide. Proc Natl Acad Sci USA 2002; 99:5902-7; PMID:11959913; http://dx.doi.org/10.1073/pnas.092017799
- Singh S, Thakur N, Oliveira A, Petruk AA, Hade MD, Sethi D, Bidon-Chanal A, Martí MA, Datta H, Parkesh R, et al. Mechanistic Insight into the Enzymatic Reduction of Truncated Hemoglobin N of Mycobacterium tuberculosis: Role of the cd loop and pre-a motif in electron cycling. J Biol Chem 2014; 289:21573-83; PMID:24928505; http://dx.doi.org/10.1074/jbc.M114.578187
- Stewart JJ, Coyne KJ. Analysis of raphidophyte assimilatory nitrate reductase reveals unique domain architecture incorporating a 2/2 hemoglobin. Plant Mol Biol 2011; 77:565-75; PMID:22038092; http://dx.doi.org/10.1007/s11103-011-9831-8
- Trevisan S, Manoli A, Begheldo M, Nonis A, Enna M, Vaccaro S, Caporale G, Ruperti B, Quaggiotti S. Transcriptome analysis reveals coordinated spatiotemporal regulation of hemoglobin and nitrate reductase in response to nitrate in maize roots. New Phytol 2011; 192:338-52; PMID:21762167; http://dx.doi.org/10.1111/j.1469-8137.2011.03822.x
- Yamasaki H, Sakihama Y. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett 2000; 468:89-92; PMID:10683447; http://dx.doi.org/10.1016/S0014-5793(00)01203-5
- Sakihama Y, Nakamura S, Yamasaki H. Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: an alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol 2002; 43:290-7; PMID:11917083; http://dx.doi.org/10.1093/pcp/pcf034
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 2002; 53:103-10; PMID:11741046; http://dx.doi.org/10.1093/jexbot/53.366.103
- Hemschemeier A, Düner M, Casero D, Merchant SS, Winkler M, Happe T. Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc Natl Acad Sci USA 2013; 110:10854-9; PMID:23754374; http://dx.doi.org/10.1073/pnas.1302592110
- de Montaigu A, Sanz-Luque E, Macias MI, Galvan A, Fernandez E. Transcriptional regulation of CDP1 and CYG56 is required for proper NH4+ sensing in Chlamydomonas. J Exp Bot 2011; 62:1425-37; PMID:21127023; http://dx.doi.org/10.1093/jxb/erq384
- de Montaigu A, Sanz-Luque E, Galvan A, Fernandez E. A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 2010; 22:1532-48; PMID:20442374; http://dx.doi.org/10.1105/tpc.108.062380
- Fernandez E, Cardenas J. Regulation of the nitrate-reducing system enzymes in wild-type and mutant strains of Chlamydomonas reinhardii. Molec Gen Genet 1982; 186:164-9; PMID:6810063; http://dx.doi.org/10.1007/BF00331846
