ABSTRACT
Pathogen-associated molecular pattern-triggered immunity (PTI) builds one of the first layers of plant disease resistance. In susceptible plants, PTI is overcome by adapted pathogens. This can be achieved by suppression of PTI with the help of pathogen virulence effectors. However, effectors may also contribute to modification of host metabolism or cell architecture to ensure successful pathogenesis. Barley responds to treatment with the pathogen-associated molecular patterns flg22 or chitin with phosphorylation of mitogen-activated protein kinases and an oxidative burst. RAC/ROP GTPases can act as positive or negative modulators of these plant immune responses. The RAC/ROP GTPase RACB is a powdery mildew susceptibility factor of barley. However, RACB apparently does not negatively control early PTI responses but functions in polar cell development during invasion of the pathogen into living host epidermal cells. Here, we further discuss the incomplete picture of PTI in Triticeae.
Abbreviations
| At | = | Arabidopsis thaliana |
| Bgh | = | Blumeria graminis f.sp. hordei |
| CA | = | constitutively activated |
| MPK | = | mitogen-activated protein kinase |
| PTI | = | pattern-triggered immunity |
| Hv | = | Hordeum vulgare |
| PRR | = | pattern recognition receptor |
Little is known about mechanisms of pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) in Triticeae.Citation1 This is astonishing regarding the fact that a large part of human and livestock nutrition depends on Triticeae crops. Additionally, literature suggested for a long time that Triticeae react to exogenous elicitors considered as PAMPs with typical defense reactions and induced resistance (e.g. Citation2-4).
Pattern recognition receptors in Triticeae
PAMPs, such as fungal chitin oligomers, bacterial lipopolysaccharides, or bacterial flagellin peptides (flg22), are recognized in plants by cell surface pattern recognition receptors (PRRs).Citation5 However, Triticeae PRRs remain largely unidentified although candidates can be identified in the corresponding genomes. First steps for functional characterization of barley and wheat chitin receptors were made. Transient silencing of chitin PRRs CeBIP or CERK1 rendered silenced tissue more susceptible to Magnaporthe oryzae or Mycosphaerella graminicola.Citation6,7 Transfer of the EF-Tu peptide (elf18) receptor EFR from Arabidopsis thaliana to Triticum aestivum led to enhanced defense gene expression and callose deposition upon elicitation with elf18. Importantly, EFR-transgenic wheat plants showed enhanced basal resistance to the bacterial pathogen Pseudomonas syringae pv. oryzae. Together this shows functionality of PRRs in and transferability of PRRs on Triticeae crops.Citation8
Early PTI responses in Triticeae
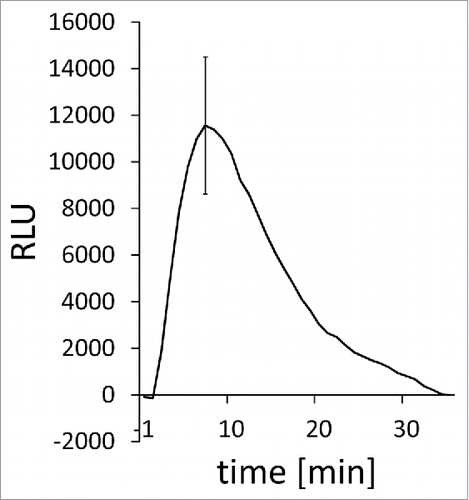
Few data are available on bona fide early PTI responses of Triticeae to microbial patterns. The barley leaf apoplast shows a rapid alkalization in response to injection of chitooctaose.Citation9 Chitin or flg22 induce early receptor-gene expression and chitin induces late callose depositions after infiltration into barley leaves.Citation1 Proels and co-workers reported on an early flg22-elicited oxidative burst in barley. This burst was unchanged in transgenic barley with silenced expression of barley NADPH oxidases RBOHF1 and RBOHF2.Citation10 Hence, the barley enzyme responsible for this oxidative burst remains unidentified. Similarly, barley leaf discs react with transient ROS production to chitin elicitors within minutesCitation11 (). A putative barley ortholog of AtRBOHD, the major ROS-producing enzyme of Arabidopsis in PTI,Citation12 was not yet described. Additionally, data from Zea mays question whether NADPH oxidases alone are responsible for the oxidative burst in Poaceae. Peroxidases appear to additionally contribute to the chitosan-triggered oxidative burst in maize.Citation13 Transient elevations of cytosolic Ca2+ concentrations are another typical early PTI response of Arabidopsis.Citation14 For Triticeae, little is known about fluxes of calcium or other ions during PTI. Calcium spiking in the barley apoplast was observed during the compatible interaction with the powdery mildew fungus Blumeria graminis f.sp hordei (Bgh).Citation9 Ion channel regulation and stomata closure is also observed as an early PTI response of dicots.Citation15 Barley stomata close or do not open in light when barley is infected by Bgh, and guard cell S-type anion channels are apparently stimulated in interaction with Bgh and by chitosan.Citation16,17 In this case, non-specific elicitors of Bgh may cause the response because Bgh does not penetrate through stomata. Together this suggests that canonical early PTI-associated ion fluxes do operate in Triticeae-pathogen interactions.
Figure 1. Chitin-triggered ROS production in barley. (A) Typical ROS production measurement in response to chitin in barley. Normalized relative light units (RLU) are shown over the measured time course. Chitin elicitors were added to leaf discs at 0 min and ROS-dependent luminol luminescence recorded over time. Data show relative light units (RLU) corrected after subtraction of leaf disc-specific background (recorded for 5 minutes before elicitation) and average mock treatment associated blanks (n = 8). Error bars show standard error over the mean (n = 8). For methodology, see Scheler et al. (2016).Citation11
Cytoplasmic kinases in pathogen-interactions of Triticeae
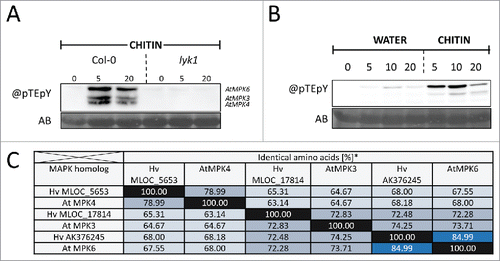
Cytoplasmic kinases such as receptor-like cytoplasmic kinases, mitogen-activated protein kinases (MPK) and calcium-dependent protein kinases are also typical signal transducing and signal modulating kinases in PTI. In Triticeae, calcium-dependent protein kinases are transcriptionally regulated in response to fungal infection and can regulate basal resistance positively or negatively depending on the individual family member.Citation18,19 The barley receptor-like cytoplasmic kinase HvRBK1 (ROP binding kinase 1) is involved in basal resistance and in controlling protein abundance of the barley susceptibility factor HvRACB.Citation20 However, a connection of those kinases to early PTI responses has not been shown. The barley MPKs HvPWMK1 and HvMPK4 are involved in biotic stress responses. However, HvPWMK1 and HvMPK4 are phylogenetically not closely related to Arabidopsis AtMPK3, AtMPK4 and AtMPK6, which are activated during Arabidopsis PTI.Citation21-23 No direct MPK activation in response to microbial patterns was shown in Triticeae until recently. We have now used an anti-pTEpY antibody that detects the phosphorylated TEY motif in MPKs typically involved in the early PTI response.Citation24,25 This showed phosphorylation of 2 barley MPKs after elicitation with flg22 or chitin with a similar pattern as seen in Arabidopsis.Citation11 Further western blotting of protein extracts from barley leaves after chitin treatment showed sometimes even 3 bands of presumably activated MPKs in barley (). However, despite the similarity of the activation pattern to that of AtMPK6, AtMPK3 and AtMPK4 in Arabidopsis, the nature of those kinases remains unknown because no corresponding mutants of barley MPK genes are available. Similarity of barley MPKs, which were predicted from genomic and cDNA sequences of barley, to AtMPK6, AtMPK3 and AtMPK4 suggests presence of highly conserved barley orthologs with corresponding TEY motifs (). We hence hypothesize that the anti-pTEpY antibody detects PAMP-activated barley orthologs of AtMPK6, AtMPK3 and AtMPK4.
Figure 2. Comparison of Arabidopsis and barley MAP kinases. (A) Arabidopsis MPKs activated in response to chitin. MAPK activation of wild type and lyk1 (synonym cerk1) mutants was analyzed by immunoblotting using an antibody against phosphorylated TEY-motif containing MPKs (@pTEpY). For methodology see Ranf et al. (2015)Citation25 and Scheler et al. (2016).Citation11 (B) Barley MPKs activated in response to chitin were detected by @pTEpY immunoblotting. AB, amidoblack total protein staining as loading control. (C) Percent identity matrix of Arabidopsis TEY-MPKs activated in PTI and related barley proteins. Arabidopsis proteins (AtMPKs) were compared by reciprocal BLAST searches to high confidence predicted barley proteins and most similar barley (Hv) proteins containing a TEY-motif were selected as putative orthologs.
Defense gene expression in elicitor response of Triticeae
Early defense gene expression in general is considered to be at least in part a PTI response.Citation1 Both early (starting from 15´ after treatment) and late defense gene expression after elicitor treatment has been reported for wheat and barley.Citation1,4,8,26 Little information is currently available about Triticeae transcription factors that activate defense gene expression in response to PAMPs but candidates are available.Citation27-29 In barley, a glucomannan-based elicitor from B. graminis activates expression of the defense gene Germin4, which has a function in basal penetration resistance to Bgh.Citation30 The Germin4c promoter is negatively regulated by WRKY transcription factors of subgroup IIaCitation27, which also function as negative regulators of powdery mildew resistance.Citation21,31
RAC/ROP GTPases in plant immunity
In the Poaceae crop plant Oryza sativa the RAC/ROP GTPase OsRAC1 functions in PTI downstream of the PRR OsCERK1 and in basal and effector-triggered immunity to M. oryzae. OsRAC1 supports among other responses MAP kinase activity and the oxidative burst.Citation32 Hardly anything is known about RAC/ROP functions in PTI of Triticeae.Citation11 Individual members of the family of RAC/ROP GTPase can have opposite functions or act antagonistically in plant immunity or polar cell development.Citation32,33 Host disease susceptibility factors can function in negative control of plant immunity or serve other demands of successful pathogenesis. We therefore analyzed whether the barley powdery mildew susceptibility factor HvRACB would possibly control early PTI responses in barley negatively. HvRACB appears also to be a target of a Bgh virulence effector that can be detected in the barley host cytoplasm (Mathias Nottensteiner TU München, Bernd Zechmann, University of Graz, R.H. unpublished results). The oxidative burst and MPK phosphorylation was similar in wild type barley, in super-susceptible barley expressing constitutively activated (CA) HvRACB and in little susceptible barley silenced for expression of HvRACB. Cell biological studies instead strongly suggest that HvRACB has a physiological function in polar cell development during formation of root hairs and stomata subsidiary cells. Nucleus positioning is also affected by silencing HvRACB and was suggested as a common factor of early fungal interaction with leaf epidermal cells and development of root hairs and stomata subsidiary cells.Citation11 Because cell polarity is crucial for basal defense of fungal penetration,Citation34 HvRACB could have a function in regulating cellular defense responses, which block fungal entry. However, HvRACB´s function in susceptibility might also organize polar growth processes required for ingrowth of the haustorial complex into intact epidermal cells. In such a scenario Bgh might use HvRACB in a hostile takeover of a cell development program. HvRACB does not suppress early PTI responses and even supports expression of defense genes when stably expressed in a CA version.Citation11 It was not yet investigated, whether HvRACB can be considered as a positive regulator of defense gene expression or whether expression of CA HvRACB and silencing of HvRACB indirectly provoke altered defense gene expression in barley. Nevertheless, Bgh appears not to be affected by enhanced expression of defense genes in this case because CA HvRACB-expressing barley is super-susceptible to both penetration by and reproduction of Bgh. Together, individual Triticeae RAC/ROP proteins could be linked to early PTI responses or basal defense reactions but HvRACB´s function in susceptibility to powdery mildew appears to be independent of this.
Conclusions
There is increasing evidence for canonical PTI responses of Triticeae crops. This is per se not surprising. Genetic evidence identifying the key components of sensing pathogen-associated molecular patterns and signal transduction is largely lacking. With the recent availability of barley and wheat genome sequences and new tools for genome editing, the community can now find and exploit key genes of Triticeae crops acting in PTI. PTI in rice is already understood to a higher extent, which was recently reviewed.Citation35 This possibly provides an interesting starting point for identification of candidate genes from Triticeae and for comparative research on PTI in grasses.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Stefanie Ranf for critical reading and to Milena Schäffer and Alexander Coleman for technical support and to Vera Schnepf for identification of barley TEY-MPKs. We apologize to colleagues whose work on elicitor activated responses or PTI in Triticeae could not be referenced due to space limitation.
Funding
Experimental work was supported by a grant from the German Research Foundation to R.H. within the collaborative research center SFB924.
References
- Boyd LA, Ridout C, O'Sullivan DM, Leach JE, Leung H. Plant-pathogen interactions: disease resistance in modern agriculture. Trends in genetics 2013; 29:233-40; PMID:23153595; http://dx.doi.org/10.1016/j.tig.2012.10.011
- Faoro F, Maffi D, Cantu D, Iriti M. Chemical-induced resistance against powdery mildew in barley: the effects of chitosan and benzothiadiazole. Biocontrol 2008; 53:387-401; http://dx.doi.org/10.1007/s10526-007-9091-3
- Vander P, KM Vr, Domard A, Eddine El Gueddari N, Moerschbacher BM. Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol 1998; 118:1353-9; PMID:9847109; http://dx.doi.org/10.1104/pp.118.4.1353
- Shetty NP, Jensen JD, Knudsen A, Finnie C, Geshi N, Blennow A, Collinge DB, Jørgensen HJ. Effects of beta-1,3-glucan from Septoria tritici on structural defence responses in wheat. J Exp Bot 2009; 60:4287-300; PMID:19880540; http://dx.doi.org/10.1093/jxb/erp269
- Macho AP, Zipfel C. Plant PRRs and the Activation of Innate Immune Signaling. Molecular Cell 2014; 54:263-72; PMID:24766890; http://dx.doi.org/10.1016/j.molcel.2014.03.028
- Tanaka S, Ichikawa A, Yamada K, Tsuji G, Nishiuchi T, Mori M, Koga H, Nishizawa Y, O'Connell R, Kubo Y. HvCEBiP, a gene homologous to rice chitin receptor CEBiP, contributes to basal resistance of barley to Magnaporthe oryzae. BMC Plant Biol 2010; 10:288; PMID:21190588; http://dx.doi.org/10.1186/1471-2229-10-288
- Lee WS, Rudd JJ, Hammond-Kosack KE, Kanyuka K. Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol Plant-Microbe Interact 2014; 27:236-43; PMID:24073880; http://dx.doi.org/10.1094/MPMI-07-13-0201-R
- Schoonbeek HJ, Wang HH, Stefanato FL, Craze M, Bowden S, Wallington E, Zipfel C, Ridout CJ. Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol 2015; 206:606-13; PMID:25760815; http://dx.doi.org/10.1111/nph.13356
- Felle HH, Herrmann A, Hanstein S, Hückelhoven R, Kogel KH. Apoplastic pH signaling in barley leaves attacked by the powdery mildew fungus Blumeria graminis f. sp hordei. Mol Plant-Microbe Interact 2004; 17:118-23; PMID:14714875; http://dx.doi.org/10.1094/MPMI.2004.17.1.118
- Proels RK, Oberhollenzer K, Pathuri IP, Hensel G, Kumlehn J, Hückelhoven R. RBOHF2 of Barley Is Required for Normal Development of Penetration Resistance to the Parasitic Fungus Blumeria graminis f. sp hordei. Mol Plant-Microbe Interact 2010; 23:1143-50; PMID:20687804; http://dx.doi.org/10.1094/MPMI-23-9-1143
- Scheler B, Schnepf V, Galgenmuller C, Ranf S, Hückelhoven R. Barley disease susceptibility factor RACB acts in epidermal cell polarity and positioning of the nucleus. J Exp Bot 2016; 67(11):3263-75; PMID:27056842; http://dx.doi.org/10.1093/jxb/erw141
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. Direct Regulation of the NADPH Oxidase RBOHD by the PRR-Associated Kinase BIK1 during Plant Immunity. Molecular Cell 2014; 54:43-55; PMID:24630626; http://dx.doi.org/10.1016/j.molcel.2014.02.021
- Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, Doehlemann G. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog 2012; 8:e1002684; PMID:22589719; http://dx.doi.org/10.1371/journal.ppat.1002684
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant Journal 2011; 68:100-13; PMID:21668535; http://dx.doi.org/10.1111/j.1365-313X.2011.04671.x
- McLachlan DH, Kopischke M, Robatzek S. Gate control: guard cell regulation by microbial stress. New Phytol 2014; 203:1049-63; PMID:25040778; http://dx.doi.org/10.1111/nph.12916
- Koers S, Guzel-Deger A, Marten I, Roelfsema MR. Barley mildew and its elicitor chitosan promote closed stomata by stimulating guard-cell S-type anion channels. Plant J 2011; 68:670-80; PMID:21781196; http://dx.doi.org/10.1111/j.1365-313X.2011.04719.x
- Prats E, Gay AP, Mur LAJ, Thomas BJ, Carver TLW. Stomatal lock-open, a consequence of epidermal cell death, follows transient suppression of stomatal opening in barley attacked by Blumeria graminis. J Exp Bot 2006; 57:2211-26; PMID:16793847; http://dx.doi.org/10.1093/jxb/erj186
- Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, Zhang ZL, Chen XB, Zhao GY, Kong XY, et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol 2008; 66:429-43; PMID:18185910; http://dx.doi.org/10.1007/s11103-007-9281-5
- Freymark G, Diehl T, Miklis M, Romeis T, Panstruga R. Antagonistic control of powdery mildew host cell entry by barley calcium-dependent protein kinases (CDPKs). Mol Plant-Microbe Interact 2007; 20:1213-21; PMID:17918623; http://dx.doi.org/10.1094/MPMI-20-10-1213
- Reiner T, Hoefle C, Hückelhoven R. A barley SKP1-like protein controls abundance of the susceptibility factor RACB and influences the interaction of barley with the barley powdery mildew fungus. Mol Plant Pathol 2016; 17:184-95; PMID:25893638; http://dx.doi.org/10.1111/mpp.12271
- Eckey C, Korell M, Leib K, Biedenkopf D, Jansen C, Langen G, Kogel KH. Identification of powdery mildew-induced barley genes by cDNA-AFLP: functional assessment of an early expressed MAP kinase. Plant Mol Biol 2004; 55:1-15; PMID:15604661; http://dx.doi.org/10.1007/s11103-004-0275-2
- Abass M, Morris PC. The Hordeum vulgare signalling protein MAP kinase 4 is a regulator of biotic and abiotic stress responses. J Plant Physiol 2013; 170:1353-9; PMID:23702246; http://dx.doi.org/10.1016/j.jplph.2013.04.009
- Lee J, Eschen-Lippold L, Lassowskat I, Bottcher C, Scheel D. Cellular reprogramming through mitogen-activated protein kinases. Front Plant Sci 2015; 6:940; PMID:26579181; http://dx.doi.org/10.3389/fpls.2015.00940
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Haweker H, Dong X, Robatzek S, Schulze-Lefert P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. Embo J 2009; 28:3439-49; PMID:19763087; http://dx.doi.org/10.1038/emboj.2009.263
- Ranf S, Gisch N, Schaffer M, Illig T, Westphal L, Knirel YA, Sánchez-Carballo PM, Zähringer U, Hückelhoven R, Lee J, et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol 2015; 16:426-33; PMID:25729922; http://dx.doi.org/10.1038/ni.3124
- Kervinen T, Peltonen S, Teeri T, Karjalainen R. Differential expression of phenylalanine ammonia‐lyase genes in barley induced by fungal infection or elicitors. New Phytol 1998; 139:293-300; http://dx.doi.org/10.1046/j.1469-8137.1998.00202.x
- Liu D, Leib K, Zhao P, Kogel KH, Langen G. Phylogenetic analysis of barley WRKY proteins and characterization of HvWRKY1 and -2 as repressors of the pathogen-inducible gene HvGER4c. Mol Genetic Genomics 2014; 289:1331-45; http://dx.doi.org/10.1007/s00438-014-0893-6
- Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen QH. Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 2013; 25:1158-73; PMID:23532068; http://dx.doi.org/10.1105/tpc.113.109942
- Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu H, Harter K, Jansson C, Wanke D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 2008; 9:1-17; http://dx.doi.org/10.1186/1471-2164-9-194
- Himmelbach A, Liu L, Zierold U, Altschmied L, Maucher H, Beier F, Müller D, Hensel G, Heise A, Schützendübel A, et al. Promoters of the barley germin-like GER4 gene cluster enable strong transgene expression in response to pathogen attack. Plant Cell 2010; 22:937-52; PMID:20305123; http://dx.doi.org/10.1105/tpc.109.067934
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007; 315:1098-103; PMID:17185563; http://dx.doi.org/10.1126/science.1136372
- Kawano Y, Kaneko-Kawano T, Shimamoto K. Rho family GTPase-dependent immunity in plants and animals. Front Plant Sci 2014; 5:522; PMID:25352853; http://dx.doi.org/10.3389/fpls.2014.00522
- Yang Z, Lavagi I. Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Curr Opin Plant Biol 2012; 15:601-7; PMID:23177207; http://dx.doi.org/10.1016/j.pbi.2012.10.004
- Hückelhoven R, Panstruga R. Cell biology of the plant-powdery mildew interaction. Curr Opin Plant Biol 2011; 14:738-46; PMID:21924669; http://dx.doi.org/10.1016/j.pbi.2011.08.002
- Liu W, Liu J, Triplett L, Leach JE, Wang GL. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol 2014; 52:213-41; PMID:24906128; http://dx.doi.org/10.1146/annurev-phyto-102313-045926
