ABSTRACT
Background
The discovery of circulating cell-free fetal DNA (cff-DNA) in maternal plasma has inspired the noninvasive prenatal testing (NIPT) approaches for various genetic fetal screening including rhesus D typing, sex determination, aneuploidies, and single-gene disorders.
Objective
Noninvasive determination of paternally inherited beta-thalassemia mutations in maternal total cell-free DNA (cf-DNA) by using allele-specific amplification refractory mutation system (ARMS) real-time PCR (RT-PCR) in concordance with the conventional invasive method.
Methods
An observational study was conducted at the Armed Forces Institute of Blood Transfusion in collaboration with the genetics resource center from March 2021 to August 2021. A total number of 26 couples were selected having a history of previously affected children with beta-thalassemia. A routine chorionic villus sampling (CVS) invasive procedure was carried out, and the mutation analysis was done using conventional PCR. To assess NIPT, a total cf-DNA was also extracted from maternal plasma and analyzed using allele-specific ARMS RT-PCR.
Results
Based on conventional PCR testing, 13 of 26 couples were found having beta-thalassemia carriers with homozygous mutation, and 13 couples were carriers with heterozygous mutations. Further to assess NIPT, the cf-DNA of 13 pregnant females among the couples with different mutational patterns was analyzed by allele-specific ARMS RT-PCR to detect paternally inherited mutations. In comparison with conventional PCR, 11 cases (84.6%) were matched successfully, while two cases (15.4%) had no concordance with conventional invasive prenatal testing (IPT).
Conclusion
NIPT using maternal cf-DNA by allele-specific ARMS RT-PCR can be feasible to screen paternal inherited mutant alleles to rule out pregnant women from invasive procedures where the test would be negative for paternal inheritance. However, a low amount of fetal DNA in maternal plasma is a limiting factor and required further improvement to enrich fetal cf-DNA for complete concordance with conventional IPT.
Introduction
Thalassemia is one of the most common single-gene inherited disorders [Citation1] leading to major health concerns in Pakistan, and beta-thalassemia is the commonest genetically transmitted disorder worldwide [Citation2]. Every year in Pakistan, approximately 5000 children are born with thalassemia major [Citation1]. Thalassemia patients are managed by frequent blood transfusions and chelation therapy, but bone marrow transplantation and stem cell therapy from a related compatible donor are the only curative options, which are highly risk-based and expensive [Citation3]. Preventive measures are the best solution to prevent the further spread of this genetic disorder.
Prenatal genetic testing for pregnant women is being done to diagnose the thalassemia in the fetus, where both partners are thalassemia carriers. This genetic test is usually done by routine chorionic villus sampling (CVS) and amniocentesis where a tiny piece of the placenta and a fluid that surrounds the fetus are taken, respectively. Both CVS and amniocentesis are invasive procedures for fetal sampling, and chances of miscarriage are 1/200–400 and 1/100–200, respectively [Citation4] with a 98–99% accuracy rate [Citation5]. Due to the lack of effective treatment, beta-thalassemia is putting a major financial and psychological adverse effect on parents, society, and increasing patient burden on the national health system. Up to now, prevention is the best gold standard way to reduce this genetic disorder. A couple where both partners are having beta-thalassemia carrier is advised to the prenatal screening of fetus usually on 10–12 weeks of pregnancy and opt to terminate the pregnancy in case of an affected fetus. To address this issue, invasive prenatal testing (IPT) was initiated for the first time in Pakistan by Ahmed et al. in 1994 [Citation6].
The discovery of fetus-originated cell-free fetal DNA (cff-DNA) in maternal plasma of pregnant females has opened a new approach for noninvasive prenatal testing (NIPT). In the last decade, NIPT has become an emerging technique in prenatal care to diagnose common aneuploidies and some monogenic disorders or direct paternally inherited mutation using maternal cell-free DNA (cf-DNA) [Citation7]. Currently, in Pakistan, CVS-based IPT and conventional PCR techniques are being used as gold standard methods for DNA testing using cellular DNA, which is laborious, time-consuming, and involves a long delay in reporting. Additionally, due to the lack of CVS procedure expertise, only two private laboratories are offering this facility as a routine public service for clinical reporting. The study aimed to develop a NIPT approach in comparison with IPT to screen paternally inherited mutations using cf-DNA from maternal plasma by using allele-specific amplification refractory mutation system (ARMS) real-time PCR (ARMS RT-PCR) which is more sensitive, accurate, and a rapid testing technique.
Methodology
Study samples
Informed consent for blood samples was obtained from 26 singleton pregnant women who arrived for invasive procedures at the Genetics Resource Centre and Armed Force Institute of Pathology. The study was approved by the Ethical Review Committee (appl. # Riphah/IRC/20/250) of Islamic International Medical College, Riphah International University, Pakistan. Inclusion criteria were both partners must be a carrier with different mutant alleles, women with 10–14 weeks of pregnancy, and informed consent. Gestational age was confirmed by ultrasound at the time of CVS sampling for all samples. The exclusion criteria were, if one of the partners was not a carrier of betathalassemia, and those who were not willing to participate in the study.
Blood collection and processing
Whole maternal blood was collected into 2 × 3 ml EDTA tubes and processed within 4–6 h to separate plasma. The blood samples were centrifuged at 1600 g for 10 min at 4°C and transferred plasma into 1.5 ml tubes. Subsequently, plasma was further centrifuged at 16,000× g for 10 min at 4°C. The supernatant was then transferred into aliquots and stored at −80°C until further processing to isolate total cf-DNA.
Total cf-DNA extraction from maternal plasma
The cf-DNA from maternal plasma was extracted under manufacturer instructions using a commercial kit (cat # A29319, applied biosystems, Thermo Fisher Scientific). The extracted cf-DNA was stored into aliquots for subsequent PCR analysis of RASSF1A and beta-thalassemia mutant alleles.
Confirmation of the presence of cff-DNA in maternal plasma
A universal RASSF1A gene fetal marker was used to confirm the presence of cff-DNA in the total cf-DNA of maternal plasma. RASSF1 gene promoter sequence is hypermethylated in fetal DNA but hypomethylated in maternal DNA [Citation8]. The total cf-DNA was treated with methylated sensitive 5UBstUI restriction enzyme (New England Biolabs, England) to digest the hypomethylated maternal RASSF1 gene and leaves behind hypermethylated fetal RASSF1 gene at 60°C for 2 h before to run the subsequent RT-PCR.
RT-PCR for RASSF1A fetal marker
To confirm the presence of cff-DNA in all the samples, 05 µl of cf-DNA or digested cf-DNA was added to a 20 µl SYBR green master mix (cat # K1081, Thermofisher) containing 10 pmol for each forward and reverse primers for RASSF1A gene (). Amplification was done using PCR machine (Applied biosystems 7500) with the following thermal conditions: holding denaturation step for 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 55°C for 60 s with signal acquiring on green channel filter simultaneously. Amplified products were then melted from 60 to 95°C with a 1°C increment at each step while acquiring on the green channel. The threshold limit was set at 0.02 for all PCR runs. No template control and cf-DNA extracted from nonpregnant women (n = 2) plasma (2 replicates of each) were included as controls for RASSF1A gene amplifications. The No template control and cf-DNA extracted from a nonpregnant female, treated with RASSF1A methylation-sensitive enzyme, were used as a negative digest control which showed no amplification for RASSF1A gene products, while undigested cf-DNA showed RASSF1A gene amplification was used as a positive control.
Table 1. Primers for RASSF1A gene marker and beta-thalassemia alleles.
Allele-specific ARMS RT-PCR analysis of beta-thalassemia mutations
Five 05 µl of tcf-DNA was added to a 20 µl SYBR green master mix (cat # K1081, Thermofisher) containing mutant or normal primers () with common primer (10 pmol for each) in separate reaction PCR tubes. Amplification was done using a PCR machine (Applied biosystems7500) with the following thermal conditions; initial denaturation for 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 55°C for 60 s with simultaneously signal acquiring using a green channel. The melting curves were obtained for all amplified products from 65 to 95°C with a 1°C increment with acquiring green signals at each step using a green filter. The threshold limit was set at 0.02 for all PCR runs.
Data analysis
The presence of hyper or hypomethylated RASSF1A promoter sequence was determined by RT-PCR amplification plots represented by Ct values and a correct melt profile showing a specific peak at 86.1°C. At least two independent experiments were run for the RASSF1A gene using tcf-DNA (digested and/or undigested). Two independent ARMS RT-PCR runs were analyzed, and Ct values data were represented as means ± SD. The correct melting curves for specific peaks were also analyzed at 87.1°C.
Results
Sample cohort and blood collection
A total of 26 beta-thalassemia carrier couples who visited for routine CVS procedure were included in the study. The maternal ages ranged from 22 to 40 years (mean = 30 years), and the gestational ages ranged from 10 to 14 weeks (mean = 11.7 weeks). All maternal blood samples were taken in the laboratory, and plasma samples were stored at −80°C within 48 h.
RT-PCR analysis for RASSF1A gene
All cases showed positive amplification of RASSF1A in the undigested sample indicating that total cf-DNA had been extracted successfully in all cases. Hypermethylated RASSF1A gene was amplified in duplicates in 13 digests by the methylation-sensitive restriction enzyme indicating the presence of cff-DNA in maternal plasma. The total cf-DNAs (n = 2) were extracted from nonpregnant females; one treated with RASSF1A gene methylation-sensitive enzyme and the other without treatment were run as negative and positive controls, respectively. There was no RASSF1A gene amplification been observed in the treated tcf-DNA sample, while the RASSF1A gene was amplified in the untreated tcf-DNA sample.
Allele-specific ARMS RT-PCR analysis for beta-thalassemia
A routine conventional invasive prenatal testing (IPT) was done in a cohort of 26 pregnant women. Cellular DNAs extracted from maternal or paternal blood cells and CVS tissues were analyzed by conventional PCR and gel method to detect common Pakistani beta-thalassemia. Out of 26 couples, 13 were found carriers with the homozygous mutations, and 13 couples were carriers with different alleles heterozygous mutations. Mutational data of all cases including maternal, paternal, and fetuses are given in . Further to assess the feasibility of NIPT, 13 pregnant women (median gestational age 11, and average 12.3) among the heterozygous couples were selected for total cf-DNA isolation to detect paternally inherited mutations. Total cf-DNA was analyzed by allele-specific amplification refractory mutation system (ARMS) real-time PCR-PCR method. The overall paternally inherited mutation (PIM) cycle threshold using cf-DNA was 31 ± 2.6, while the cycle thresholds for maternal and normal alleles were 28.2 ± 1.8 and 25.7 ± 1.05, respectively. Of 13 cases, 11 (84.6%) cases had complete concordance with conventional IPT, of which six cases were detected positive, and five cases were negative for PIM, while two of 13 cases had no concordance with IPT ( and ).
Figure 1. Results interpretation of fetal inheritance of paternally inherited mutation (Cd 5 (-CT)) pattern. (a) Cellular DNA (fetal, father, and mother) analysis by conventional ARMS-PCR: Mutant bands on lanes 1 and 2 represent paternal inherited mutation in fetal samples; lane 3 shows the same paternal allele; and lane 4 shows negative for the paternal mutation. (b) RASSF1A gene amplification plot of RT-PCR using cell-free DNA from maternal plasma: Amplification curves indicate the confirmation of cf-DNA in undigested DNA of nonpregnant plasma sample and cff-DNA in digested DNA plasma sample of pregnant women, while no amplification curve appeared in DNA digests of nonpregnant women plasma sample. NTC, not template control. (c) Identification of fetal inheritance using cf-DNA by ARMS RT-PCR using allele-specific primers: amplification curves show the identification of maternal normal and mutant alleles and paternal inherited mutant allele in maternal plasma.
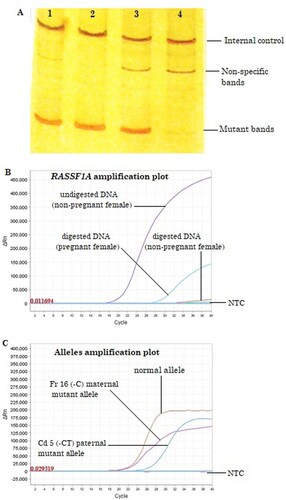
Table 2. Mutational data deduced by invasive prenatal testing (IPT) by conventional ARMS-PCR using cellular DNA.
Table 3. Noninvasive prenatal testing (NIPT) in heterozygous cases by allele-specific ARMS RT-PCR in cell-free DNA of maternal plasma.
Discussion
The discovery of blood circulating cff-DNA in maternal plasma by Lo et al. in 1997 [Citation7,Citation10] has opened a noninvasive prenatal procedure for screening of beta-thalassemia mutations [Citation3,Citation11,Citation12] which reduces the chance of miscarriage by 1:100. Currently, the presence of fetal DNA (f-DNA) along with maternal cf-DNA in maternal plasma is an alternative source to detect the paternally inherited mutations in maternal blood circulation where both carriers are with different mutations [Citation13]. Studies have shown that the concentration of cff-DNA in maternal plasma ranges from 3.4 to 10% which can be detected as early at the 7th week of gestation [Citation7,Citation14,Citation15], while in the present study, total cell-free DNA (tcf-DNA) was extracted at 10–14th week of gestation. This low concentration of f-DNA in maternal circulation is not detectable by the conventional ARMS-PCR and gel electrophoresis method. In the present study, we transformed the conventional PCR method into more sensitive and allele-specific ARMS RT-PCR SYBR green method using the same set of primers. Furthermore, the detection of hypermethylated RASSF1A fetal-promoter sequence was used as a universal fetal marker [Citation8,Citation9] to confirm the presence of methylated cff-DNA in tcf-DNA of pregnant maternal plasma, while hypomethylated maternal RASSF1A could not be detected by RT-PCR after digestion with the methylation-sensitive enzyme. The detection of methylated cff-DNA by RT-PCR is an accurate method for fetal disorders [Citation16]. Additionally, the sizes of cff-DNA fragments had been reported as detectable by RT-PCR in maternal plasma [Citation17,Citation18]. Considering all these factors, we successfully applied ARMS RT-PCR using allele-specific primers for common Pakistani mutations, specifically Fr 8-9 (+G), VSI-5 (G-C), Fr 41-42 (-TTCT), Cd 15 (G-A), Cd 5 (-CT), Cd 30 (G-C), Fr 16 (-C), and Cap+1 in 13 couples at risk with beta-thalassemia mutations. In Pakistan at present, the prenatal diagnosis of beta-thalassemia relies on conventional invasive procedures including chorionic villus sampling or amniocentesis, and the method of detection to screen thalassemia mutations is being done by using conventional PCR. In the current study, common Pakistani mutations were screened using allele-specific primers by RT-PCR using tcf-DNA, and the results were 84.6% concordant with conventional analysis. We notified that 5 of 13 couples at risk with different mutations showed a negative paternal inherited mutation pattern, which suggested the mothers can choose not to undergo additional invasive procedures. The major aim of this study is to exclude the cases from noninvasive procedures where fetal inheritance is negative for paternally inherited beta-thalassemia mutations using maternal plasma. Currently, many techniques are being used for screening paternally inherited mutations including microarrays [Citation19], multiplex PCR, target capture, next-generation sequencing [Citation20–22], TaqMan genotyping assay [Citation23], droplet digital PCR [Citation22,Citation24], and allele-specific RT-PCR by f-DNA enhancement techniques [Citation25,Citation26]. This study shows the modified ARMS RT-PCR using allele-specific primers for common mutation in Pakistan could be used as cost-effective and simple for detecting paternally inherited beta-thalassemia mutations. This technique can be successfully applied for NIPT in pregnancies starting from 10th gestational weeks where couples are with different mutations. However, this method is unable to differentiate maternally inherited mutations in cf-DNA extracted from maternal plasma. Some of the methods such as droplet digital PCR [Citation27,Citation28] and next-generation sequencing will be available in near future for NIPT of thalassemia through multiplex PCR, target capture, and next-generation sequencing [Citation20] for clinical diagnosis in developed countries which are costly and required more expensive types of the equipment and expertise.
In summary, the present NIPT using cf-DNA by allele-specific ARMS RT-PCR was simple, accurate, and can be used to rule out pregnant women undergoing invasive procedures where a NIPT is negative for paternally inherited mutations. However, a low amount of fetal DNA in maternal plasma is a limiting factor and required further improvement to enrich fetal cf-DNA for complete concordance with conventional invasive prenatal testing. Furthermore, this method may reduce additional routine invasive prenatal procedures and chances of miscarriages.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Ahmed S. Prenatal diagnosis of beta-thalassemia: 12 years’ experience at a single laboratory in Pakistan. Prenat Diagn. 2007;27(13):1224–1227.
- Zaheer HA, Waheed U, Abdella YE, et al. Thalassemia in Pakistan: a forward-looking solution to a serious health issue. Glob J Transfus Med. 2020;5:108–110.
- Zafari M, Kosaryan M, Gill P, et al. Non-invasive prenatal diagnosis of β-thalassemia by detection of the cell-free fetal DNA in maternal circulation: a systematic review and meta-analysis. Ann Hematol. 2016;95(8):1341–1350.
- Swanson A, Sehnert AJ, Bhatt S. Non-invasive prenatal testing: technologies, clinical assays and implementation strategies for women's healthcare practitioners. Curr Genet Med Rep. 2013;1(2):113–121.
- Papageorgiou EA, Patsalis PC. Non-invasive prenatal diagnosis of aneuploidies: new technologies and clinical applications. Genome Med. 2012;4(5):46.
- Ahmed S, Saleem M, Rashid Y. The first prenatal diagnosis of thalassaemia in Pakistan: a case report. Pak J Pathol. 1994;5:69–71.
- Pan M, Chen P, Lu J, et al. The fragmentation patterns of maternal plasma cell-free DNA and its applications in non-invasive prenatal testing. Prenat Diagn. 2020;40(8):911–917.
- White HE, Dent CL, Hall VJ, et al. Evaluation of a novel assay for detection of the fetal marker RASSF1A: facilitating improved diagnostic reliability of noninvasive prenatal diagnosis. PLoS One. 2012;7(9):e45073.
- Chan KC, Ding C, Gerovassili A, et al. Hypermethylated RASSF1A in maternal plasma: A universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin Chem. 2006;52(12):2211–2218.
- Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487.
- Mahmoud ST, Aboalwafa H, Ali E, et al. Non-invasive prenatal diagnosis of β-thalassemia by detection of the cell-free fetal DNA in maternal circulation. Sohag Med J. 2019;23(3):156–167.
- Chen C, Li R, Sun J, et al. Noninvasive prenatal testing of α-thalassemia and β-thalassemia through population-based parental haplotyping. Genome Med. 2021;13(1):18.
- Chiu RW, Lau TK, Leung TN, et al. Prenatal exclusion of beta thalassaemia major by examination of maternal plasma. Lancet. 2002;360(9338):998–1000.
- Lo YM, Tein MSC, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62(4):768–775.
- Hu P, Liang D, Chen Y, et al. An enrichment method to increase cell-free fetal DNA fraction and significantly reduce false negatives and test failures for non-invasive prenatal screening: a feasibility study. J Transl Med. 2019;17(1):124.
- Akbariqomi M, Heidari R, Gargari SS, et al. Evaluation and statistical optimization of a method for methylated cell-free fetal DNA extraction from maternal plasma. J Assist Reprod Genet. 2019;36(5):1029–1038.
- Fan HC, Blumenfeld YJ, Chitkara U, et al. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem. 2010;56(8):1279–1286.
- Li Y, Di Naro E, Vitucci A, et al. Size fractionation of cell-free DNA in maternal plasma improves the detection of a paternally inherited beta-thalassemia point mutation by MALDI-TOF mass spectrometry. Fetal Diagn Ther. 2009;25(2):246–249.
- Galbiati S, Monguzzi A, Damin F, et al. COLD-PCR and microarray: two independent highly sensitive approaches allowing the identification of fetal paternally inherited mutations in maternal plasma. J Med Genet. 2016;53(7):481–487.
- Hudecova I, Chiu RW. Non-invasive prenatal diagnosis of thalassemias using maternal plasma cell free DNA. Best Pract Res Clin Obstet Gynaecol. 2017;39:63–73.
- Yang X, Ye Y, Fan D, et al. Non-invasive prenatal diagnosis of thalassemia through multiplex PCR, target capture and next-generation sequencing. Mol Med Rep. 2020;22(2):1547–1557.
- Pedini P, Graiet H, Laget L, et al. Qualitative and quantitative comparison of cell-free DNA and cell-free fetal DNA isolation by four (semi-)automated extraction methods: impact in two clinical applications: chimerism quantification and noninvasive prenatal diagnosis. J Transl Med. 2021;19(1):15.
- Breveglieri G, Travan A, D’Aversa E, et al. Postnatal and non-invasive prenatal detection of β-thalassemia mutations based on TaqMan genotyping assays. PLoS One. 2017;12(2):e0172756.
- Debrand E, Lykoudi A, Bradshaw E, et al. A non-invasive droplet digital PCR (ddPCR) assay to detect paternal CFTR mutations in the cell-free fetal DNA (cffDNA) of three pregnancies at risk of cystic fibrosis via compound heterozygosity. PLoS One. 2015;10(11):e0142729.
- Ramezanzadeh M, et al. Detection of paternally inherited fetal point mutations for β-thalassemia in maternal plasma using simple fetal DNA enrichment protocol with or without whole genome amplification: an accuracy assessment. J Matern Fetal Neonatal Med. 2016;29(16):2645–2649.
- Khordadpoor-Deilamani F, Akbari MT. The use of cell-free fetal DNA in maternal plasma for noninvasive prenatal linkage analysis in beta globin gene cluster. Bratisl Lek Listy. 2015;116(11):662–665.
- Chang MY, Ahn S, Kim MY, et al. One-step noninvasive prenatal testing (NIPT) for autosomal recessive homozygous point mutations using digital PCR. Sci Rep. 2018;8(1):2877.
- Perlado S, Bustamante-Aragonés A, Donas M, et al. Fetal genotyping in maternal blood by digital PCR: towards NIPD of monogenic disorders independently of parental origin. PLoS One. 2016;11(4):e0153258.
