Abstract
Plant oxylipins are involved in defense responses against pathogens or herbivores. Each oxylipin compound exerts its distinctive physiological function. The ability of many legumes to fix nitrogen with rhizobia gives them special importance in natural environments and agriculture. It has been postulated that oxylipins are somehow related to symbiosis; however, this is still a controversial issue. In this study, we isolated five genes at the branching point of the oxylipin pathway in Lotus japonicus, and their biochemical functions were identified as allene oxide synthases (AOSs), 13-hydroperoxide lyase (13HPL), and 9/13-HPLs. When the leaves were mechanically wounded, AOS and 9/13HPL were upregulated in leaves and roots, respectively, from which their implications in wound response were suggested. When the plants were inoculated with rhizobia, no big change in the expression levels of genes was found. When high N was supplied to the nodulated plants, the number of nodules decreased, and simultaneously, AOS in the leaves was downregulated. Significance of AOS in response to the N status in the plants was suggested.
Introduction
Fatty acid hydroperoxides produced by lipoxygenases, either 9- or 13-hydroperoxides of C18 fatty acids (linoleic and linolenic acid), are intermediates in the oxylipin pathway in plants (Stumpe and Feussner Citation2006). 13-Hydroperoxides of fatty acids are the substrates for the oxylipin-specific P450s, such as allene oxide synthase (13AOS) or hydroperoxide lyase (13HPL). 13AOS and 13HPL comprises an unusual class of P450s and demonstrate low affinity for carbon monoxide and do not require molecular oxygen or NADPH-dependent P450 reductase, therefore, classified as subfamily CYP74A and CYP74B, respectively. 13AOS converts the hydroperoxide into allene oxide that is further converted into oxophytodienoic acid (OPDA) or jasmonates (JAs). They are signaling molecules regulating defense and development in plants (Koo and Howe Citation2009). 13HPL cleaves the hydroperoxides to yield C6 volatile aldehydes and the corresponding C12 oxo acids (Matsui Citation2006).
The other types of CYP74s act on 9-hydroperoxides of C18 fatty acids (Stumpe and Feussner Citation2006; Matsui Citation2006). CYP74C cleaves both 9- and 13-hydroperoxides and named 9/13HPL. AOS specific to 9-hydroperoxide (9AOS) is also known. Divinyl ether synthase (DES; CYP74D) catalyzes conversion of either 9- or 13-hydroperoxide into the corresponding divinyl ether. Most plants have several genes for CYP74s. The end product of each branch of oxylipin pathway takes a distinctive role in defense responses against biotic and abiotic attack, and structural diversity of each oxylipin compound permits functional specificity. The function of JAs has been extensively studied, and their roles as plant hormones related to stress responses and development have been established (Koo and Howe Citation2009). The products of HPL branch function as molecules exerting direct and indirect defense against pathogens or herbivores (Matsui Citation2006). In this context, it should be plausible that the metabolic flow of each branch of the oxylipin pathway is tightly controlled, most probably through regulation at the branching point catalyzed by each CYP74.
The ability of many legumes to fix nitrogen in association with rhizobia gives them special importance in natural environments and agriculture (Doyle and Luckow Citation2003). The symbiosis between legumes and rhizobium needs mutual exchange of signal molecules, such as flavonoids and Nodulation (Nod) factors (Ferguson et al. Citation2010). Several CYP74 genes in leguminous plants have been isolated so far, and their biochemical nature has been studied (Noordermeer et al. Citation2000; Stumpe et al. Citation2005; Hughes et al. Citation2006; De Domenico et al. Citation2007; Kongrit et al. Citation2007). However, their expression has been seldom reported. Leguminous plants accept rhizobia and arbuscular mycorrhizal fungi, and establish symbiotic interactions with them, while they start defense responses against the pathogenic microorganisms, insects, or parasitic plants in order to deter them. Thus, there should be a device that can distinguish the foes and friends. Oxylipins are generally involved in defense responses against biotic invaders (Koo and Howe Citation2009; Matsui Citation2006), hence, the oxylipin pathway should be coordinately regulated with the distinguishing device.
In order to get insight into the coordination, we report here the isolation of genes encoding CYP74 enzymes in Lotus japonicus. L. japonicus is one of the model legumes, and its genome is almost completely sequenced (Sato et al. Citation2008). The aim of the present work was to gain an understanding of the molecular basis of CYP74-dependent metabolism of fatty acid hydroperoxides in L. japonicus. Owing to the wealth of knowledge of plant–symbiont interactions in L. japonicus, this system is likely to provide a good model for assessing the role of oxylipins in plant symbiosis with rhizobia.
Materials and methods
Plant materials
L. japonicus (Miyakojima, MG-20) seeds were damaged using a sheet of sandpaper in a mortar and sown on Kimwipe moistened with water. The seeds were grown at 22°C under a 14/10 h light/dark cycle. After three days, the seedlings were placed between ‘pillows’ (11×11×4 cm) made of a nylon mesh bag filled with vermiculite and perlite (6/1, v/v). The plants were grown with B & D medium containing 0.05 or 5 mM KNO3 under the same condition. Rhizobium (Mesorhizobium loti, MAFF 303099) was cultured in fresh TY medium for 72 h at 28°C. The culture (1 µl) containing 1×106?7cells was diluted with 200 ml of fresh B & D medium. Inoculation was carried out by drenching the diluted suspension of the bacteria to the pillows.
Cloning and expression of CYP74s
Expressed sequence tag (EST) clones were obtained from Frontier Science Research Center, University of Miyazaki, Japan. The list of the clones is shown in Supplemental Table S1 (All supplementary material be found by clicking on the Supplementary Content tab at http://dx.doi.org/10.1080/1729145.2011.562985). For AOS2, PCR cloning from the genomic DNA was carried out. The primers used in this study are listed in Supplemental Table S2. For expression of each gene in E. coli, restriction enzyme sites were generated using PCR with the primers listed in Table S2, then, the open reading frame was inserted into the pQE vector (QIAGEN, Valencia, CA). E. coli M15 (QIAGEN) was used for expression. The recombinant enzymes were prepared as described previously (Matsui et al. Citation2000).
Substrate and product specificities of recombinant CYP74s
Fatty acid hydroperoxides were prepared by using soybean lipoxygenase-1 or tomato fruit lipoxygenase as described previously (Matsui et al. Citation2000). Hydroperoxide decomposing activity was determined by recording the absorption at 234 nm for conversion of the hydroperoxides at 25°C. AOS activity was determined by reacting the enzyme with 10 mM 13-hydroperoxy-(Z,E,Z)-9,11,15-octadedatrienoic acid (13HPOT) suspended with 50 mM Tris-HCl, pH 7.5 for 10 min at 25°C. The AOS products were analyzed with high performance liquid chromatography (HPLC) by comparing those formed from 13HPOT by E. coli-expressed soybean AOS (Kongrit et al. Citation2007). HPL activity was determined by reacting the enzyme with 10 mM 13HPOT or 9-hydroperoxy-(Z,E)-10,12-octadecadienoic acid (9HPOD) suspended with 50 mM MES-KOH, pH 5.5 for 5 min at 25°C The products were analyzed as their hydrozaone derivatives with HPLC (Matsui et al. Citation2000).
Determination of heme b and volatiles
For heme b determination, the roots were frozen in liquid N2 and homogenized in 50 mM phosphate buffer (pH 7.4) by using a homogenizer (MicroSmash, TOMY, Tokyo, Japan). After centrifugation, the supernatant was filled up to 1.5 ml with cold acetone. The mixture was incubated for 10 min on ice, then, for 10 min at 25°C, and centrifuged. The supernatant was dried up and solubilized with 0.5 ml each of pyridine and 1N NaOH, and 1.5 ml of H2O. The mixture (pyridine ferro-hemochrome) was divided into two parts, and the one part was reduced with sodium hydrosulfite. The amount of heme b was determined from reduced minus oxidized difference spectra, through determining the absorbance at 555 and 535 nm.
For volatile analysis, the leaves or roots (ca. 40 mg fr wt) of L. japonicus (21-day old, without inoculation) was collected and homogenized with 1.5 ml of 50 mM Na-phosphate (pH 6.3) containing 0.1 mM diethylenetriamine pentaacetic acid (DETAPAC) on ice. The homogenate was put into a glass vial and incubated at 26°C for 10 min. After addition of 1.5 ml of saturated CaCl2 solution in order to kill the enzymes, an internal standard (nonyl acetate, 100 ng) was added. ll the enzymes, an internal standard solid-phase microextraction (SPME) fiber (50/30 µm DVB/Carboxen/PDMS, Supelco, Bellefonte, PA, USA) was exposed to the headspace of the vial for 30 min at 80°C. The volatiles were analyzed with gas chromatography-mass spectrometer (GC-MS) (QP-2010Plus, Shimadzu) equipped with 0.25 µm×30 m DB-WAX column (Agilent Technologies, Santa Clara, CA). The column temperature was programed as 40°C for 5 min to 200°C for 10 min at 5°C min-1 with a carrier gas (He) at 5 ml min-1. The temperature of the injector and interface was 200 and 230°C, respectively. The mass detector was operated in an electron impact mode (70 eV). In order to assign each compound, retention indices and MS profiles of corresponding authentic specimens were used.
RNA extraction and real time RT-PCR
RNA was extracted with RNeasy Plant Mini Kit (QIAGEN). cDNA was synthesized with ThermoScript RT-PCR System (Invitrogen). RT-PCR was performed with ABI PRISM 7300 (Life Technologies Co., Carlsbad, CA, USA) using an Express SYBR GreenER qPCR Super mix (Invitrogen, Life Technologies Co.). The ubiquitin gene (acc. no. DQ249171) was used as an internal standard. Primers used in this study are listed in Supplemental Table S2.
Results
Assigning five CYP74s in L. japonicus
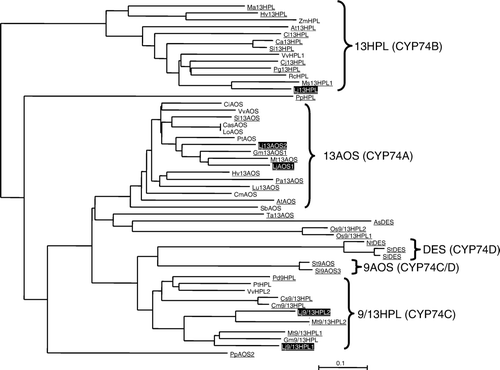
We performed BLAST search with the EST database of L. japonicus (Lotus japonicus EST index, http://est.kazusa.or.jp/en/plant/lotus/EST/) (Asamizu et al. Citation2004). We found 41 entries derived from four genes that showed high similarity to CYP74 genes (see Supplementary Table S1). The longest EST clone of each was sequenced, and from the sequences we validated that they were mostly full-length. Based on their sequences, they were tentatively identified as LjAOS1 (LjCYP74A1, DDBJ acc. no., AB600747), Lj13HPL (LjCYP74B, AB600748), Lj9/13HPL1 (LjCYP74C1, AB600749), or Lj9/13HPL2 (LjCYP74C2, AB600750). We also performed BLAST search with the genome database of L. japonicus (miyakogusa.jp, http://www.kazusa.or.jp/lotus/index.html) (Sato et al. Citation2008). We found another CYP74-related sequence within a BAC clone (CM0314) other than the four genes. The sequence showed high similarity to LjAOS1, and it was tentatively named LjAOS2 (LjCYP74A2). The genes for DES (CYP74D) and 9AOS were not found. When the phylogenetic tree was constructed, the CYP74s in L. japonicus were located in the clade as expected from their tentative assignments (). In the clade, LjCYP74s were located nearby the counterparts of the other leguminous plant, Medicago trancatula, except for LjAOS2, whose counterpart was absent from M. trancatula. Among the five LjCYP74s, two (Lj9/13HPL2 in chromosome III and LjAOS1 in choromosome V) occur in the synteny blocks found between chromosomes of L. japonicus and M. trancatula with their counterparts in M. trancatula (Cannon et al. Citation2006) (Supplemental Figure S1). Prediction of subcellular localization with TargetP analysis (http://www.cbs.dtu.dk/services/TargetP/) suggested that LjAOS1, LjAOS2, and Lj13HPL were localized in the chloroplasts with 37, 57, and 85 amino acid stretch of transit peptide, respectively. The TargetP analysis showed that Lj9/13HPL1 and 2 did not have any sequences directing them to a specific organelle.
Figure 1. Phylogenetic tree analysis of the CYP74 family. Amino acid sequences were aligned using ClustalW, then, the tree was constructed with Njplot. LjCYP74s are highlighted, and the entries whose enzymatic activities were confirmed are underlined. At13AOS; Arabidopsis thaliana AOS (CAA63266), At13HPL; A. thaliana 13HPL (AAC69871), AsDES; Allium sativum DES (CAI30435), Ca13HPL; Capsicum annuum 13HPL (AAK27266), CasAOS; Camelia sinensis AOS (ACU30142), CiAOS; Citrus cinensis AOS (AAO72741), Cj13HPL; Citrus jambhiri 13HPL (BAC55161), Cl13HPL; Citrullus lanatus 13HPL (AAU12570), CmAOS; Cucumis melo AOS (AAM66138), Cm9/13HPL; C. melo 9/13HPL (AAK54282), Cs9/13HPL; Cucumis sativum 9/13HPL (AAF64041), Gm13AOS1; Glycine max 13AOS1 (ABB91776), Gm9/13HPL; G. max 9/13HPL (ABC68416), HvAOS1; Hordeum vulgare AOS1 (CAB86384), Hv13HPL; H. vulgare 13HPL (CAC82980), LoAOS; Lonicera japonica AOS (ABC17856), Lu13AOS; Linum usitatissimum 13AOS (P48417), Ma13HPL; Musa acuminata 13HPL (CAB39331), MtAOS; Medicago trancatula 13AOS (CAC86897), Mt9/13HPL1; M. trancatula 9/13HPL1 (CAC86899), Mt9/13HPL2; M. trancatula 9/13HPL2 (CAC86898), Ms13HPL1; Medicago sativa 13HPL1 (CAB54847), NtDES; Nicotiana tabucum DES (AAL40900), Os9/13HPL1; Oryza sativa 9/13HPL1 (AK105964), Os9/13HPL2; O. sativa 9/13HPL2 (AK107161), PaAOS; Parthenium argentatum 13AOS (CAA55025), PdHPL; Prunus dulcis HPL (CAE18065), Pg13HPL; Psidium guajava 13HPL (AAK15070), PtAOS; Populus trichocarpa AOS (B9GP05), PpAOS2; Physcomitrella patens AOS (CAC86919), PpHPL; P. patens HPL (CAC86920), PtHPL; Populas trichocarpa HPL (XP 002305404), RcHPL; Ricinus communis (XP 002529334), SbAOS; Sorghum bicolor AOS (XP 002463829), Sl13AOS; Solanum lycopersicon 13AOS (CAB88032), Sl9AOS3; S. lycopersicon 9AOS (AAN7687), SlDES; S. lycopersicon DES (AAG42261), Sl13HPL; S. lycopersicon 13HPL (CAB43022), St9AOS; S. tuberosum 9AOS (CAI30876), StDES; S. tuberosum DES (CAC28152), TaAOS; Triticum aestivum 13AOS (CAC28152), VvAOS; Vitis vinifera AOS (XP_002283780), VvHPL1; V. vinifera HPL1 (ACZ17394), VvHPL2; V. vinifera HPL2 (XP_002281213), ZmHPL; Zea maize HPL (AAS47027).
Functional expression of recombinant proteins
Active recombinant enzymes of LjAOS1, LjAOS2, Lj13HPL, and Lj9/13HPL2 were expressed with E. coli. Lj9/13HPL1 could not be functionally expressed in E. coli with unknown reasons. 13HPOT was the best substrate for the recombinant LjAOS1, and 13-hydroperoxy-(Z,E)-9,11-octadedadienoic acid (13HPOD) followed (A). Recombinant LjAOS1 showed only 20% activity with 9HPOD, and 9-hydroperoxy-(E,Z,Z)-10,12,15-octadecatrienoic acid (9HPOT) was not essentially the substrate. When products from 13HPOT formed by recombinant LjAOS1 was analyzed, α- and γ-ketols, and OPDA were detected (B), but HPL products could not be detected. Collectively, it was concluded that LjAOS1 encoded 13-hydroperoxide-specific AOS. Unlike most AOSs examined so far (Stumpe et al. 2005; Kongrit et al. Citation2007; Lee et al. Citation2008), the activity of recombinant LjAOS2 with 13HPOD was significantly higher than that with 13HPOT. Products formed by recombinant LjAOS2 from 13HPOT were also α- and γ -ketols, and OPDA. These analyses confirmed that LjAOS2 also encoded 13-hydroperoxide-specific AOS but with a preference to the hydroperoxides of dienoic acids rather than those of trienoic acids as substrates.
Figure 2. Substrate specificities (A) and product specificities (B, C) of recombinant CYP74s derived from L. japonicus. The hydroperoxide-decomposing activity was determined spectrophotometrically with 13-hydroperoxy-(Z,E,Z)-9,11,15-octadecatrienoic acid (13HPOT), 13-hydroperoxy-(Z,E)-9,11-octadecadienoic acid (13HPOD), 9-hydroperoxy-(E,Z,Z)-10,12,15-octadecatrienoic acid (9HPOT), or 9-hydroperoxy-(E,Z)-10,12-octadecadienoic acid (9HPOD). The highest activity for each enzymatic activity, 1.66 (LjAOS1), 3.62 (LjAOS2), 2.16 (Lj13HPL), or 11.8 (Lj9/13HPL2) µkat ml−1, is set as 100%, respectively. For analysis of products, HPLC for detection of AOS products (B) or of HPL products (C) was carried out as in Method section, and the representative chromatograms are shown. The profiles of products formed from Lj13HPL and Lj9/13HPL2 from 13HPOT were essentially same, then, only one chromatogram is shown. Peaks 1; γ-ketol, 2; α-ketol, 3; OPDA, 4; 12-oxo-(Z)-9-dodecenoic acid, 5; (Z)-3-hexenal, 6; (Z)-3-nonenal.
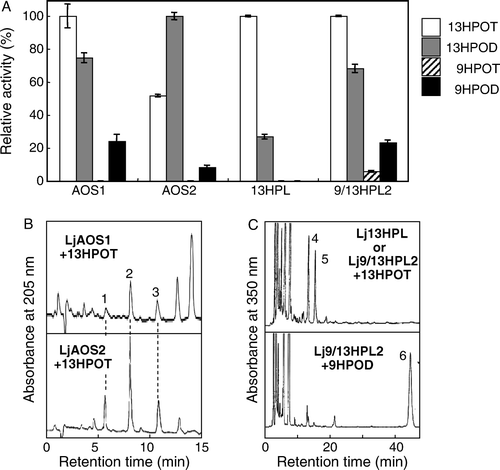
Recombinant Lj13HPL showed highest activity against 13HPOT, and 13HPOD followed. 9HPOs were hardly catalyzed by Lj13HPL as found with most 13HPLs of the other plant sources (Matsui Citation2006; Stumpe and Feussner Citation2006). (Z)-3-Hexenal and 12-oxo-(Z)-9-dodecenoic acid were the major products, and the AOS products could not be detected (C). Thus, it was clarified that Lj13HPL encoded 13-hydroperoxide-specific HPL. Recombinant Lj9/13HPL2 showed high activity with 13HPOs, but also showed a substantial activity against 9HPOD, which is a distinguishing feature of most 9/13HPLs. The recombinant Lj9/13HPL2 yielded a typical HPL product, (Z)-3-nonenal from 9HPOD (C); thus, it is reasonable to assign the gene as the one encoding 9/13HPL.
Expression of CYP74 genes
Expression levels of LjAOS1 and 2, and Lj9/13HPL2 were largely equivalent among leaves, stems, and roots of 21 days old seedlings grown without inoculation of rhizobia but with B & D medium containing 5 mM KNO3 (A). The levels of the other two LjCYP74 genes in these organs varied. Lj13HPL transcript level was higher in leaves, and lower in stems and roots. Lj9/13HPL1 transcript level was higher in roots than in leaves or stems. The same tendency was observed with the rhizobia-inoculated plants (cf. ); therefore, organ-specificity of expression of each CYP74 in L. japonicus was little affected by symbiosis. The organ-specific expression of Lj13HPL and Lj9/13HPL genes was further confirmed by the analysis of volatile products of these two types of HPLs (B). In leaves the amounts of C6 volatiles, such as (E)-2-hexenal, (Z)-3-hexenal, and the alcohols and acetates derived from them, were significantly higher than those in roots. On the contrary, C9 volatiles, such as (E,Z)-2,6-nonadienal and (E)-2-nonenal were more abundant in roots than in leaves.
Figure 3. Expression of each LjCYP74 in leaves, stems, and roots of L. japonicus (21 days old, uninoculated) and the effect of mechanical wounding (A). The transcript level was first normalized to the level of ubiquitin in each sample, and the expression level in the roots before wounding was then set as 1. The leaves were mechanically wounded, and the expression levels after 6 h was determined. The means±SE (n=3) are shown. The values followed by the same letter indicate means that are not significantly different (P<0.05, Tukey's test). Volatiles formed after disruption of leaves or roots of the seedlings (B). The relative area ratio normalized with the area of internal standard calculated with total ion chromatogram is shown (means±SE, n=3). The values followed with asterisks indicate means significantly different between leaves and roots (P<0.05, t-test). Compounds 1; n-hexanal, 2; (Z)-3-hexenal, 3; (E)-2-hexenal, 4; 2-octanone, 5; (Z)-3-hexenyl acetate, 6; (Z)-2-penten-1-ol, 7; n-hexan-1-ol, 8; (Z)-3-hexen-1-ol, 9; n-nonanal, 10; (E)-2-hexen-1-ol, 11; 1-octen-3-ol, 12; (E)-2-nonenal, 13; 2,6-(E,Z)-nonadienal.
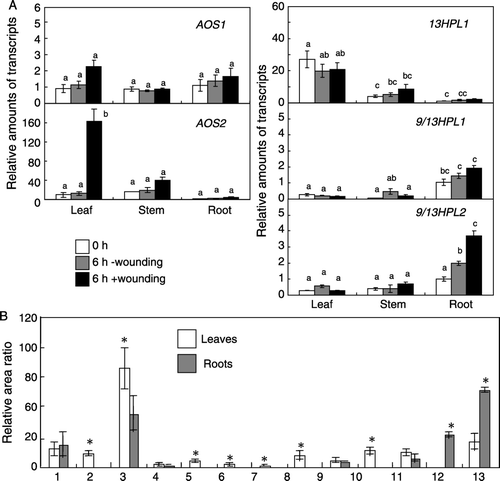
Oxylipin metabolism is known to be involved in wound-inducible defense gene expression in various plant species (Koo and Howe Citation2009; Matsui Citation2006). When the leaves of 21 days old non-inoculated seedlings of L. japonicus were mechanically wounded, LjAOS2 was upregulated in leaves (A). LjAOS1 was also slightly upregulated. The transcript levels of Lj13HPL showed no change. Lj9/13HPL2 was upregulated in roots when the leaves were wounded even though upregulation of the gene in the leaves was not observed. Lj9/13HPL1 was also slightly upregulated in roots after mechanical wounding on leaves.
Effect of nodulation and autoregulation
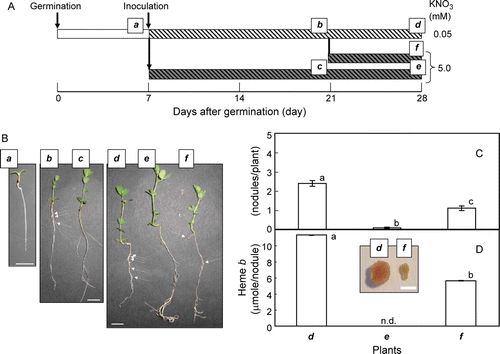
In order to examine the effects of nodulation and autoregulation of nodulation (AON) caused by high supply of N (Ferguson et al. Citation2010; Oka-Kira and Kawaguchi Citation2006) to the nodulated L. japonicus plants on expression of LjCYP genes, the seedlings (7 days old) were inoculated with Mesorhizobium loti, then, grown with low (0.05 mM) or high (5.0 mM) N supply (A). At 14 days after inoculation, formation of nodules was evident with the plants grown with low N supply while few nodules were observed with the ones with high N supply (B). At 14 days after inoculation (i.e. at 21 days after germination), the nodulated plants grown with low N were divided into two groups; then, one group was further grown with low N supply while the other group was grown with high N supply in order to onset AON (Ferguson et al. Citation2010; Oka-Kira and Kawaguchi Citation2006). Seven days after the supply of high N (i.e. 21 days after inoculation and 28 days after germination), the number of nodules was reduced to less than half of that with low N because of AON (B, C). The AON was also evident when the amount of heme b, which was an indicator of active nodules containing leghemoglobin, was quantified (D).
Figure 4. The growth schedule of L. japonicus seedlings (A), photos of plants used in this study (B), number of nodules (C) and the amount of heme b derived from leghemoglobin in roots (D). The seeds were germinated and grown with 0.05 mM KNO3 for 7 days. Then, the seedlings were inoculated with M. loti, and grown with 0.05 or 5.0 mM KNO3. At 14 days after inoculation, the ones grown with 0.05 mM KNO3 were divided into two halves, and one of them were grown with 0.05 mM KNO3 while the others were grown with 5.0 mM KNO3 for additional 7 days. The plants used for the analyses were named as a to f. On the photos (B), the nodules are shown with white arrows. For nodule number (C) and the amount of heme b (D), the means±SE (n=3) are shown. The values followed by the different letter indicate means that are significantly different (P<0.05, Tukey's test). In panel D, the shape and color of nodules isolated from plant d and f is shown in the inset.
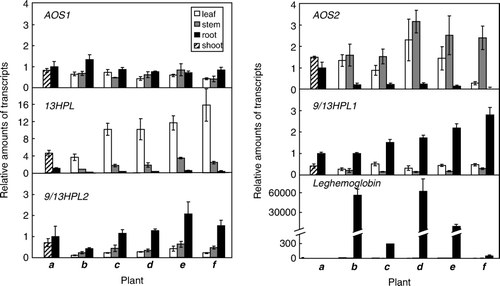
The transcript level of leghemoglobin extensively increased after inoculation when the plants were grown with low N supplies (). When the inoculated L. japonicus plants were grown with high N supply, the transcript level of leghemoglobin was effectively reduced. Such effect of high N supply on the transcript level of leghemoglobin was evident even with the nodulated L. japonicus plants, and a shift of N supply from 0.05 to 5 mM from 14 days after inoculation extensively reduced the expression of leghemoglobin, from which it was confirmed that AON proceeded by high N supply under the experimental condition employed here. When the transcript levels of CYP74s were examined, they were mostly constant irrespective of the presence or absence of rhizobium, or to the N availability. The only exception being LjAOS2 in leaves. The amount of LjAOS2 transcript in the leaves of inoculated L. japonicus plants (14 days post inoculation) grown with high N was lower than that found with the inoculated plants grown with low N (). This is also the case with the plants at 21 days post inoculation. The amount of LjAOS2 transcript was extensively suppressed at 7 days after changing the N supply from 0.05 mM to 5 mM when suppression of nodulation caused by AON was obvious.
Figure 5. Expression of each CYP74 in nodulated L. japonicus grown with different N availability. The plants were grown as shown in Results section, then, the expression level of each gene with the plant a to f was determined. For the details of the plants, refer A. The transcript level was first normalized to the level of ubiquitin in each sample, and the expression level in the roots of 7 days old seedling was then set as 1. The means±SE (n=3) are shown.
Discussion
Comprehensive analysis showed that L. japonicus had two AOSs, one 13HPL, and two 9/13HPLs. The phylogenetic analysis based on their amino acid sequences indicated that each CYP74 in L. japonicus was placed in the appropriate clade, such as CYP74A, B, or C, which was consistent with its enzymatic activity identified in this study. When BLASTP search was performed with genome sequences of M. trancatula, Glycine max, Cucumis sativus, and Prunus persica with phytozome (http://www.phytozome.net/) by using either of CYP74s in L. japonicus as a query, five hypothetical gene families, namely, #26459994, #26457455, #26477720, #26478987, and #26455604, were identified. They were corresponding to LjAOS1, LjAOS2, Lj9/13HPL2, Lj9/13HPL1, and 13HPL, respectively. Among them, the gene corresponding to Lj9/13HPL1 was found only with leguminous plants but not found with the Cucurbitales and Fagales. Thus, it was assumed that the five genes except for that corresponding to Lj9/13HPL1 were acquired before segregation of the clade consisting of Fabales, Rosales, Cucurbitales, and Fagales in the eudicots, but duplication of 9/13HPL genes occurred after establishing leguminous plants (Stevens Citation2008). Every gene for 13HPL, including that in monocots and eudicots, was found at the same clade in the phylogenic tree, which suggested that the gene was acquired before segregation of monocots and dicots (Lee et al. Citation2008). Among dicots, the genes corresponding to 9/13HPL have been found only with rosids. Some monocots also have 9/13HPLs; however, they were placed apart from those of rosids on the phylogenetic tree. Therefore, it is assumed that 9/13HPL genes in rosids and monocots acquired their catalytic function independently after divergence of eudicots and monocots.
With L. japonicus seedlings, 13HPL highly expressed in leaves, while both the 9/13HPLs expressed profoundly in roots. Volatile analysis indicated that C6 volatiles were more abundant in leaves but C9 volatiles were more abundant in roots, which coincided with the expression profiles of Lj13HPL, and Lj9/13HPL1 and 2. C6 volatiles thus formed in leaves are thought to be involved in direct or indirect defense against pests as reported with Arabidopsis (Shiojiri et al. Citation2006); however, the physiological significance of C9 volatiles formed in roots is scarcely known. In cucumber, it was expected that one of the products of 9/13HPL, C9 aldehyde, functioned as toxic agents against pathogens (Matsui et al. Citation2006). Azelaic acid, which might be formed from the counterpart of 9/13HPL products, 9-oxononanoic acid, is a mobile metabolite accountable for plant systemic immunity involved in priming defenses (Jung et al. Citation2009). Taken together, root expression of 9/13HPL might be involved in plant defense against pathogens in the rhizosphere. We found that mechanical wounding on leaves resulted in upregulation of Lj9/13HPLs. It was reported that wounding of pepper leaves by infestation of whitefly (Bemisia tabaci) resulted in induced resistance at the below-ground organs against a soil-borne pathogen, Ralstonia solanacearum, and also in modifications of the rhizosphere microflora (Yang et al. Citation2011). The systemic induction of Lj9/13HPL expression in roots observed in this study was in line with this orchestrated defense responses between the above-ground and below-ground organs, which supported the hypothesis for the role of Lj9/13HPLs in defense responses against pathogens in rhizosphere. Because LjAOS2 was upregulated in leaves after mechanical wounding, the products of the corresponding enzymes, for examples, OPDA or JAs, might be involved in the systemic signaling from the above-ground to the below-ground. JA as a long-distance signaling molecule that is transported in the phloem system has been reported in various plant species (Schilmiller and Howe Citation2005).
Oxylipin pathway is induced under biotic stresses in leguminous plants. When M. trancatula was infected by pathogenic fungi or by a parasitic plant, the genes for JA synthesis were upregulated (Hiraoka et al. Citation2009; Uppalapati et al. Citation2009; Anderson et al. Citation2010). Aphid infestation resulted in induction of oxylipin pathway through upregulation of LOX and several CYP74 genes in M. trancatula (Gao et al. Citation2007). Damage by herbivores enhanced emission of C6 compounds (Shimoda Citation2010). On the contrary, our study showed that inoculation of rhizobia did not result in upregulation or downregulation of the LjCYP74 genes. Apparently, legume plants have a mechanism to distinguish the foes and friends. In fact, β-glucan and chitin tetramer, which are pathogen-related microbial elicitors for plants, induced accumulation of JA while no accumulation of JA could be observed after treating M. trancatula with Nod-factors, which are also oligosaccharides (Leitner et al. Citation2008). Thus, leguminous plants have a machinery to distinguish the foes and friends, and because of it, leguminous plants induce expression of CYP74s only when they were attacked by destructive invaders but do not modulate expressions of CYP74s when they recognize their symbionts.
Leguminous plants control the number of nodules through a mechanism known as the AON pathway involving long-distance root-shoot signaling (Ferguson et al. Citation2010; Oka-Kira and Kawaguchi Citation2006). Several reports pointed out a possible involvement of JAs in AON. Spraying methyl JA on shoots strongly suppressed nodulation in L. japonicus (Nakagawa and Kawaguchi Citation2006). Involvement of JA and AOS in controlling the degree of nodulation has also been reported with soybean (Kinkema and Gresshoff Citation2008). However, it was recently reported that JA might not be involved in a typical AON that needed a perception of shoot-derived signal by a product of gene named Too Much Love (TML) because the suppression of nodulation by JA was also apparent with a mutant tml that showed a hypernodulating phenotype (Magori and Kawaguchi Citation2010). The result shown in this study indicated that expression of CYP74 genes, except for LjAOS2, in L. japonicus showed no big change when the plants were forced to launch AON by supplying high N. In contrast, LjAOS2 was highly suppressed in the leaves when the nodulated plants were grown with high N supply. From this, it was suggested that any CYP74 genes including the two AOSs were not involved in launching or maintaining AON. However, at this moment, it is unknown why LjAOS2 in leaves was downregulated after onset of AON. Nitrogen status affects nodulation because the symbiosis would be no longer necessary when N availability is high. JA-treatment on leaves of tomato enhanced allocation of N to roots (Gómez et al. Citation2010). It was also reported that surplus N supply induced expression of JA-responsive genes in soybean (Staswick et al. Citation1991). JA was also involved in partitioning of nitrogen among organs of alfalfa (Meuriot et al. Citation2004). Thus, it was assumed that the downregulation of LjAOS2 under high N status might be related to the nitrogen dynamics in L. japonicus that was coordinated with AON.
tjpi_a_562985_sup_18319880.ppt
Download MS Power Point (362.5 KB)Acknowledgements
We thank Dr Jisaka (Shimane University, Japan) for kind supply of soybean AOS gene, Prof. Kawaguchi (National Institute for Basic Biology, Japan) for valuable suggestions for this study, Dr Sato (Kazusa DNA Institute, Japan) for providing us sequence information of L. japonicus, and Ms Mugo (Yamaguchi University, Japan) for critical reading of the manuscript. This study was partially supported by the Japan Society of the Promotion of Science Grants (19101009).
References
- Anderson , JP , Lichtenzveig , J , Gleason , C , Oliver , RP and Singh , KB. 2010 . The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia . Plant Physiol. , 154 : 861 – 873 .
- Asamizu , E , Nakamura , Y , Sato , S and Tabata , S. 2004 . Characterization of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis. Plant Mol . Biol. , 54 : 405 – 410 .
- Cannon , SB , Sterck , L , Rombauts , S , Sato , S , Cheung , F , Gouzy , J , Wnag , X , Mudge , J , Vasdewani , J Schiex , T . 2006 . Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes, Proc. Natl. Acad. Sci . USA. , 103 : 14959 – 14964 .
- De Domenico S , Tsesmetzis N , Di Sansebatiano GP , Hughes RK , Casey R , Santino A. 2007 . Subcellular localization of Medicago truncatula 9/13-hydroperoxide lyase reveals a new localization pattern and activation mechanism for CYP74C enzymes BMC Plant Biol . 7 : doi: 10.1186/1471-2229-7-58 .
- Doyle , JJ and Luckow , MA. 2003 . The rest of the iceberg. Legume diversity and evolution in a phylogenetic context . Plant Physiol. , 131 : 900 – 910 .
- Ferguson , BJ , Indrasumunar , A , Hayashi , S , Lin , MH , Lin , YH , Reid , DE and Gresshoff , PM. 2010 . Molecular analysis of legume nodule development and autoregulation. J. Integr . Plant Biol. , 52 : 61 – 76 .
- Gao , LL , Anderson , JP , Klingler , JP , Nair , RM , Edwards , OR and Singh , KB. 2007 . Involvement of the octadecanoid pathway in bluegreen aphid resistance in Medicago trancatula . Mol. Plant-Microbe Interact. , 20 : 82 – 93 .
- Gómez , S , Ferrieri , RA , Schueller , M and Orians , CM. 2010 . Methyl jasmonate elicits rapid changes in carbon and nitrogen dynamics in tomato . New Phytol. , 188 : 835 – 844 .
- Hiraoka , Y , Ueda , H and Sugimoto , Y. 2009 . Molecular responses of Lotus japonicus to parasitism by the compatible species Orobanche aegyptiaca and the incompatible species Striga hermonthica . J. Exp. Bot. , 60 : 641 – 650 .
- Hughes , RK , Belfield , EJ , Muthusamay , M , Khan , A , Rowe , A , Harding , SE , Fairhurst , SA , Bornemann , S , Ashton , R Thorneley , RNF . 2006 . Characterization of Medicago truncatula (barrel medic) hydroperoxide lyase (CYP74C3), a water-soluble detergent-free cytochrome P450 monomer whose biological activity is defined by monomer-micelle association . Biochem. J. , 395 : 641 – 652 .
- Jung , HW , Tschaplinski , TJ , Wang , L , Glazebrook , J and Greenberg , JT. 2009 . Priming in systemic plant immunity . Science , 324 : 89 – 91 .
- Kinkema , M and Gresshoff , PM. 2008 . Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Mol . Plant-Microbe Interact. , 21 : 1337 – 1348 .
- Kongrit , D , Jisaka , M , Iwanaga , C , Yokomichi , H , Katsube , T , Nishimura , K , Nagaya , T and Yokota , K. 2007 . Molecular cloning and functional expression of soybean allene oxide synthases. Biosci. Biotechnol . Biochem. , 71 : 491 – 498 .
- Koo , AJK and Howe , GA. 2009 . The wound hormone jasmonate . Phytochemistry , 70 : 1571 – 1580 .
- Lee , DS , Nioche , P , Hamberg , M and Raman , CS. 2008 . Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes . Nature , 455 : 363 – 368 .
- Leitner , L , Kaiser , R , Rasmussen , RO , Driguez , H , Boland , W and Mithöfer , A. 2008 . Microbial oligosaccharides differentially induce volatiles and signaling components in Medicago truncatula . Phytochemistry , 69 : 2029 – 2040 .
- Magori , S and Kawaguchi , M. 2010 . Analysis of two potential long-distance signaling molecules, LjCLE-RS1/2 and jasmonic acid, in a hypernodulating mutant too much love. Plant Signal . Behav. , 5 : 403 – 405 .
- Matsui , K. 2006 . Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr. Opin . Plant Biol. , 9 : 274 – 280 .
- Matsui , K , Minami , A , Hornung , E , Shibata , H , Kishimoto , K , Ahnert , V , Kindl , H , Kajiwara , T and Feussner , I. 2006 . Biosynthesis of fatty acid derived aldehydes in induced upon mechanical wounding and its products show fungicidal activities in cucumber . Phytochemistry , 67 : 649 – 657 .
- Matsui , K , Miyahara , C , Wilkinson , J , Hiatt , B , Knauf , V and Kajiwara , T. 2000 . Fatty acid hydroperoxide lyase in tomato fruits: Cloning and properties of a recombinant enzyme expressed in Escherichia coli. Biosci. Biotechnol . Biochem. , 64 : 1189 – 1196 .
- Meuriot , F , Noquet , C , Avice , JC , Volenec , JJ , Cunningham , SM , Sors , TG , Caillot , S and Ourry , A. 2004 . Methyl jasmonate alters N partitioning, N reserves accumulation and induces gene expression of a 32-kDa vegetative storage protein that possesses chitinase activity in Medicago sativa taproots. Physiol . Plant. , 120 : 113 – 123 .
- Nakagawa , T and Kawaguchi , M. 2006 . Shoot-applied MeJA suppresses root nodulation in Lotus japonicus . Plant Cell Physiol. , 47 : 176 – 180 .
- Noordermeer , MA , Van Dijken , AJH , Smeekens , SCM , Veldink , GA and Vliegenthart , JFG. 2000 . Characterization of three cloned and expressed 13-hydroperoxide lyase isoenzymes from alfalfa with unusual N-terminal sequences and different enzyme kinetics. Eur. J . Biochem. , 267 : 2000 – 2007 .
- Oka-Kira , E and Kawaguchi , M. 2006 . Long-distance signaling to control root nodule number. Curr. Opin . Plant Biol. , 9 : 496 – 502 .
- Sato , S , Nakamura , Y , Kaneko , T , Asamizu , E , Kato , T , Nakao , M , Sasamoto , S , Watanabe , A , Ono , A Kawashima , K . 2008 . Genome structure of the legume, Lotus japonicus . DNA Res. , 15 : 227 – 239 .
- Schilmiller , AL and Howe , GA. 2005 . Systemic signaling in the wound response. Curr. Opin . Plant Biol. , 8 : 369 – 377 .
- Shimoda , T. 2010 . A key volatile infochemical that elicits a strong olfactory response of the predatory mite Neoseiulus californicus, an important natural enemy of the tow-spotted spider mite Tetranychus urticae. Exp. Appl . Acarol. , 50 : 9 – 22 .
- Shiojiri , K , Kishimoto , K , Ozawa , R , Kugimiya , S , Urashimo , S , Arimura , G , Horiuchi , J , Nishioka , T , Matsui , K and Takabayashi , J. 2006 . Changing green leaf volatile biosynthesis in plants: an approach for improving plant resistance against both herbivores and pathogens . Proc Natl Acad Sci USA. , 103 : 16672 – 16676 .
- Staswick , PE , Huang , JF and Rhee , Y. 1991 . Nitrogen and methyl jasmonate induction of soybean vegetative storage protein genes . Plant Physiol. , 96 : 130 – 136 .
- Stevens PF . 2008 Angiosperm Phylogeny Website. Version 9
- Stumpe , M , Carsjens , JG , Stenzel , I , Göbel , C , Lang , I , Pawlowski , K , Hause , B and Feussner , I. 2005 . Lipid metabolism in arbuscular mycorrhizal roots of Medicago truncatula . Phytochemistry , 66 : 781 – 791 .
- Stumpe , M and Feussner , I. 2006 . Formation of oxylipins by CYP74 enzymes. Phytochem . Rev. , 5 : 347 – 357 .
- Uppalapati , SR , Marek , SM , Lee , HK , Nakashima , J , Tang , Y , Sledge , MK , Dixon , RA and Sysore , KS. 2009 . Global gene expression profiling during Medicago truncatula–Phymatotrichopsis omnivora interaction reveals a role for jasmonic acid, ethylene, and the flavonoid pathway in disease development . Mol. Plant-Microbe Interact. , 22 : 7 – 17 .
- Yang , JW , Yi , HS , Kim , H , Lee , B , Lee , S , Ghim , SY and Ryu , CM. 2011 . Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora . J. Ecol. , 99 : 46 – 56 .