ABSTRACT
In this study, BcHHP3 was isolated from Pak-choi; it has an open-reading frame (ORF) of 1044 base pairs, encoding 347 amino acids, a molecular weight of 39.35 kDa, isoelectric point (pI) of 9.08, an instability index of 48.35, and grand average of hydropathicity of 0.382. Multi-alignment and phylogenetic analysis showed that BcHHP3 bears a high similarity to AtHHP2. As predicted by SOMPA and SWISS-MODEL databases, the structure of the BcHHP3 protein is relatively stable and highly conservative. Real-time quantitative polymerase chain reaction (qRT-PCR) analysis showed that BcHHP3 was induced to co-express under cold and abscisic acid (ABA) stresses. The BcHHP3-GFP fusion protein was localized on the cell membrane and nuclear membrane. This work might be useful for future analysis of other HHP-like genes in Pak-choi.
1. Introduction
Low temperature is an important environmental stress that plants have to cope with during their life cycles. Many plants increase their cold tolerance in response to low temperature. This process is called cold acclimation (CA) (Thomashow Citation1999). CA is a complex process involving a number of biochemical and physiological changes during gene expressions and protein metabolisms (Chinnusamy et al. Citation2007). So plants have to use a signaling network to respond to low or even freezing temperature (Chinnusamy et al. Citation2005; Yamaguchi-Shinozaki and Shinozaki Citation2006).
Due to the complex environments that plants have to face, a number of signaling components are supposed to be involved in plant respond to abiotic stresses. Heptahelical protein 1 (HHP1) has been shown to function as a negative regulator in ABA and osmotic signals (Chen et al. Citation2009). HHP1 is an important member of the HHP gene family that consists of at least five members HHP1, HHP2, HHP3, HHP4, and HHP5 in Arabidopsis thaliana (Hsieh and Goodman Citation2005). Of the five HHP proteins in A. thaliana, HHP1 has been characterized as a reliable link for ABA regulation of cold signal transduction components (Chen et al. Citation2009, Citation2010). However, the function of HHP3 protein needs to be studied. The five HEPTAHELICAL PROTEIN (HHP) genes in the A. thaliana genome are homologous to human adiponectin receptors and membrane progestin receptors (Hsieh and Goodman Citation2005). Under cold stress, the expression of CBF3 is activated by ICE1 (Chinnusamy et al. Citation2007). In addition to the induction of CBF3 expression, ICE1 appears to regulate MYB15 negatively, an upstream negative regulator of the CBF genes, such as CBF1 and CBF3 (Agarwal et al. Citation2006). The mRNA expression patterns of the five HHP genes in response to low temperature, NaCl and phytohormones stresses, have been described. However, further experiments are needed to elucidate the functions of the HHP gene family (Hsieh and Goodman Citation2005). HHP proteins are identified from the primary structural analysis by the seven predicted transmembrane domains and the lack of similarity to G protein-coupled receptors. Among them, AtHHP2 and AtHHP3 form one group with 77.3% similarity, while AtHHP4 and AtHHP5 form another group with 96.1% similarity (Hsieh and Goodman Citation2005). The sequence similarities between AtHHP1 and the other two groups are 52.2% and 44.0%, respectively (Hsieh and Goodman Citation2005). Osmotic and low-temperature stresses both lead to the expressions of several kinds of genes including the ABA signaling genes (ABI1, ABI2, ABI3, and ABI5), ABA biosynthetic genes (NCED3, AAO3, ABA1, and ABA3), and common downstream stress-responsive genes (RD29A, RD29B, ADH1, KIN1, COR15A, and COR47) (Kurkela and Franck Citation1990; Jarillo et al. Citation1993; Yamaguchi-Shinozaki and Shinozaki Citation1993; Stockinger et al. Citation1997; Ishitani et al. Citation1998).
Pak-choi (Brassica rapa ssp. chinensis), also known as non-heading Chinese cabbage, has a long history of cultivation and is widely cultivated in the middle and lower reaches of the Yangtze River China. Pak-choi is a subspecies of Brassica species, which is divided into six varieties: (1) var.communis Tsen et Lee (var.erecta Mao); (2) var.rosularis Tsen et Lee (var.atrovirens Mao); (3) var.tsaitai Hort et Lee (var.purpurea Mao); (4) var.tai-tsai Hort et Lee; (5) var.multiceps Hort et Lee (var.nipponsinica Hort) and (6) var.utilis Tsen et Lee (var.oleifera Makino). Pak-choi is related to A. thaliana and has a cold acclimation character. Therefore, we speculate that there is an HHP cold signaling pathway similar to the A. thaliana HHP cold signaling pathway in Pak-choi. Some cold-related genes, such as BcICE1, BcCBF3 and BcWRKY46, were successfully cloned and analyzed and their functions were analyzed and identified. However, the BcHHP3 gene remains to be studied. In this study, the HHP3 gene from Pak-choi was cloned and named as BcHHP3 (CabbageG_a_f_g047327). We analyzed the sequence structure and studied evolutionary position through phylogenetic trees, SOMPA database and SWISS-MODEL database. Furthermore, we found that the homologous relationship between BcHHP3 and AtHHP2 is very close and highly conservative. We analyzed the phylogenetic relationships, conserved motifs and subcellular localization of BcHHP3, to demonstrate whether BcHHP3 and AtHHP2 have similar functions in response to cold stress. In addition, the expression patterns of BcHHP3 under multiple abiotic stresses were examined through real-time quantitative PCR analysis. This work might help to reveal the biological functions of BcHHP3, which could be a candidate gene for Pak-choi genetic improvement.
2. Materials and methods
2.1. Plant material, growth conditions, and stress treatments
As for plant material, Pak-choi (B. rapa ssp. chinensis cv. Suzhouqing), obtained from the non-heading Chinese cabbage project team in Nanjing Agricultural University (Nanjing, China), was used in the following experiments. As for growth conditions, first, healthy seeds were harvested from the same batch, which were grown in an artificial climate chamber at 24°C (Gilmour et al. Citation1998). Then, those healthy seeds were soaked for germination and sowed in a soil: sand mixture (3:1). Finally, those healthy seeds were placed in a growth chamber (16 h 22°C, light; 8 h 20°C, dark). As for stress treatments, seedlings at their five-leaf stage were collected and treated by various stresses (cold, NaCl, ABA and SA) under hydroponic conditions. In order to provide a large number of elements and trace elements needed for plant growth, the Murashige and Skoog (MS) culture medium was used. For all stress treatments, light and nutrition supplying conditions remained unchanged. Those seedlings were grown in hydroponic containers. For cold stress treatment, plants were incubated at 4 °C under continuous cool white fluorescent illumination at approximately 100 μmol m−2 s−1 light intensity. For NaCl, ABA and SA treatments, the plants were watered with 100 mM NaCl, 100 μM ABA and 0.1 mM SA, respectively (Tang et al. Citation2013). Samples were harvested at 0, 0.5, 1, 2, 4, 8 and 24 h, and then were frozen in liquid nitrogen and stored at −80°C for further use.
2.2. Identification of cDNA encoding BcHHP3 protein
Taking into account the feature that the nucleotide sequences are relatively conservative evolutionarily in different plant species, we extracted the A. thaliana gene sequence of HHP3 (AT2G24150) from the NCBI GeneBank (http://www.ncbi.nlm.nih.gov/genbank/). Using this protein sequence as the probe to search the Brassica EST database (http://www.Brassica.nbi.ac.uk), we successfully obtained several homologous fragments, using the encoding products’ sequence of the AtHHP3 gene as the control. Then we inspected the integrity of the open-reading frame (ORF) of the obtained sequences through BLAST (http://brassicadb.org/brad/). Subsequently, the corresponding forward primer and reverse primer were designed by the Primer 5.0 software with the sequence of the homologous gene ().
Table 1. Primers used in present study.
The cDNA of BcHHP3 was amplified from a cDNA library of Pak-choi, treated with abiotic stresses, and tested by real-time quantitative PCR (qRT-PCR). First, total RNA was extracted from leaves with the RNAeasy mini kit (Tiangen, Beijing, China) following the manufacturer’s instructions. The first-strand cDNA was synthesized with 1 μg of mixed total RNA using a Superscript II kit (Takara, Dalian, China) to construct a stress-induced cDNA library of Pak-choi. Conserved regions were identified based on sequence information from the A. thaliana HHP gene family (http://arabidopsis.org/index.jsp) and the Chinese cabbage chiifu genome (http://brassicadb.org/brad/). We designed the primers of qBcHHP3 and qBcACTIN according to the sequences of the conserved regions to amplify the core fragment by qRT-PCR (). For over-expression of the BcHHP3 gene, we designed the primers of gBcHHP3 (). For molecular cloning of BcHHP3 gene, we designed primers of BcHHP3 (). The PCR program was set as follows: 5 min at 94°C, 35 cycles for 30 s at 94°C, 30 s at 61°C and 1 min at 72°C and extension for 10 min at 72°C, and then the PCR product was cloned into the pMD19-T vector (Takara, Dalian, China).
2.3. Cloning the cDNA of BcHHP3 gene
The leaves of Pak-choi (B. rapa ssp. chinensis cv. Suzhouqing) were selected at their five-leaf stage, and then the reverse transcription and synthesis of the first-strand cDNA was carried out. The cDNA, as a template, and the primer of BcHHP3 () were used to perform PCR amplification. The amplification program was set the same as above. One percent agarose gel electrophoresis was used to detect the PCR product. The target fragment was recovered and ligated with the cloning vector pMD19-T in a 16°C water bath for 12 h and then was transformed into DH5α competent cells, where blue and white spots were screened, and white colonies were selected to activate the extraction of plasmids from the culture. The plasmid, named as BcHHP3-pMD19-T, was extracted and sent to invitrogen sequenced, and PCR amplification was performed. Then the PCR product was assayed with 1% agarose gel electrophoresis.
2.4. Sequence alignment and phylogenetic analysis
Comparing NCBI homologous sequences online (https://www.ncbi.nlm.nih.gov/pubmed/), we obtained HHP protein sequences from some different crops, as listed in supplementary Table S1. We used DNAMAN software (Lynnon Biosoft) to analyze the deduced amino acid sequence of BcHHP3. Sequence analysis was conducted by MEGA 5.0 software: the multiple sequence alignment of BcHHP3 was performed with default parameters and manual correction (Tamura et al. Citation2014) and phylogenetic tree was drawn with bootstrap values estimated as 1000 replicates. The conserved motifs were analyzed using the MEME Suite (http://meme.nbcr.net/meme/) with the default settings except that the maximum width which was set to 200, and the minimum and maximum numbers of motifs were defined as 2 and 10, respectively.
2.5. BcHHP3 protein sequence analysis and structural analysis in Pak-choi
The ORF sizes of the BcHHP3 gene were confirmed by sequencing. The molecular weight (kDa), isoelectric point (pI), half life (h) and instability index were predicted (in press) through the online ExPasy program (http://www.expasy.org/tools/) (). The structure of the BcHHP3-encoded protein was predicted through SOMPA (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page = npsa_sopma.html) and SWISS-MODEL (http://sissmodel.expasy.org/) databases.
Table 2. The overall information on HHP proteins from different crops.
2.6. Real-time quantitative PCR expression analysis of BcHHP3 under stress treatments
To measure the changes of BcHHP3 expression under diverse stress treatments, we performed real-time quantitative PCR (qRT-PCR). The qRT-PCR primers were designed by the online primer design tool from the Genscript Biotechnology Company (https://www.genscript.com/ssl-bin/app/primer) and are listed in . From the five-leaf stage seedlings of Pak-choi (B. rapa ssp. chinensis cv. Suzhouqing), we collected plant leaves and tried to avoid all leaf veins, and these were utilized for qRT-PCR analysis. Plants undergoing different treatments were prepared separately. For cold stress treatment, seedlings were transferred to 4 °C for varying lengths of time (0, 0.5, 1, 2, 4, 8 and 24 h). For NaCl stress and hormone stress treatments, seedlings were raised through hydroponics in the artificial climate chamber at 22°C. NaCl stress treatment was carried out by 100 mM NaCl for 0, 0.5, 1, 2, 4, 8 and 24 h, while hormone stress treatments were carried out by 100 μM ABA and 0.1 mM SA for 0, 0.5, 1, 2, 4, 8 and 24 h, respectively. Plant leaves were collected. Total RNA was extracted from the previously frozen plant tissues with an RNA extraction kit (RNA simply total RNA Kit, Tiangen, Beijing, China). Genomic DNA contamination was removed using DNase I (Takara, Dalian, China). The qRT-PCR programs were executed as follows: denaturating at 95°C for 4 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s and polymerizing at 72°C for 30 s. In order to confirm that the amplified and detected product was specific, a melting curve was generated after each qRT-PCR. The relative expression of BcHHP3 gene was calculated by the 2−ΔΔCT method. The Pak-choi BcACTIN gene was used as a standard (Livak and Schmitt Citation2001). The experiment was repeated three times.
2.7. Statistical analysis
The data from the qRT-PCR experiments were tested using one-way analysis of variance (ANOVA) by SPSS v.19.0 software. Statistical analysis was performed to find the differences between experimental data and the control, while the treated plants were analyzed by two-way ANOVA, according to Duncan’s test. Three biological replicates and three trial replicates were used.
2.8. Subcellular localization analysis of BcHHP3
We used WOLF PSORT and Nuc-PLoc to predict the subcellular localization of the BcHHP3 protein. In order to determine the subcellular localization of the BcHHP3 protein, we constructed two vectors. The full-length coding region of BcHHP3 was amplified with Gateway-specific primers gBcHHP3 (). The coding region was cloned into an entry vector and further introduced into the pEarleyGate101 vector using the Gateway system (Invitrogen, Carlsbad, CA, USA) to generate a 35S: BcHHP3-GFP fusion construct. About 20 base pairs from each end of the ORFs were added to attB joint sequences to obtain Gateway primers (). The PCR was performed as described above. The pDONR221 and pEarleyGate101 vectors were provided by our laboratory, and the BP and LR enzyme mixes were purchased from Invitrogen Company and used in accordance with their instructions (Shanghai, China). To transiently express BcHHP3, 35S: BcHHP3-GFP was bombarded into tobacco cells with a PDS-100/He particle delivery system (Bio-Rad, Hercules, CA, USA). The infected plants were incubated in the dark at 22°C for 3 days and then subjected to confocal microscopy (Leica, TCS SP2, Wetzlar, Germany).
3. Results
3.1. Cloning and identification of the full-length BcHHP3 in Pak-choi
The full-length BcHHP3 gene was isolated. This gene had an open-reading frame (ORF) of 1044 base pairs encoding 347 amino acids, an estimated molecular weight of 39.35 kDa, a theoretical isoelectric point (pI) of 9.08, an instability index of 48.35, and grand average of hydropathicity (GRAVY) of 0.382 (in press). The ORF was confirmed by sequencing three times. The predicted amino acid sequence lacked secretory and nuclear localization signal sequences.
3.2. Sequence alignment and phylogenetic analysis of HHP proteins
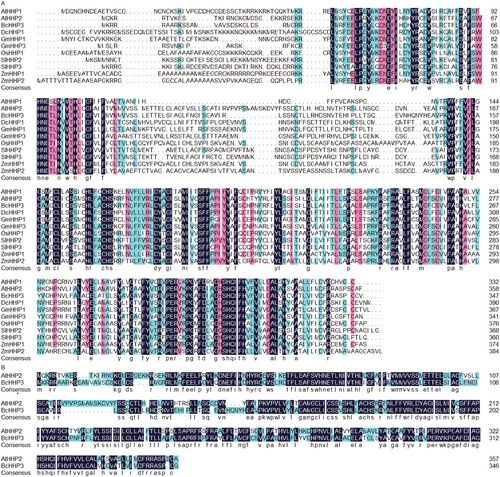
The deduced protein sequence of BcHHP3 was aligned with HHP protein sequences from several representative monocots and dicots by using DNAMAN software (Lynnon Biosoft). As shown in , BcHHP3 is similar to other plant HHPs in sequence with higher sequence similarities in the N-terminus. The sequence alignment revealed that BcHHP3 and AtHHP2 proteins have approximately 67.49% amino acid sequence homology. Thus, we speculated that BcHHP3 might have a similar function to AtHHP2. The results revealed that the BcHHP3 sequence is more homologous to the Arabidopsis HHP2 sequence with 67.49% similarity than Oryza sativa, Zea mays and Glycine max HHP sequences (), suggesting that the HHP proteins between Pak-choi and Arabidopsis are highly conservative.
Figure 1. Amino acid sequence alignments of heptahelical proteins (HHPs) from Pak-choi and other crops. BcHHP3 was isolated in the study and some other crop proteins were selected from GenBank (A). Less conserved residues, highly conserved residues and perfectly matched residues are represented by a green box, a pink box and a black box, respectively (B).
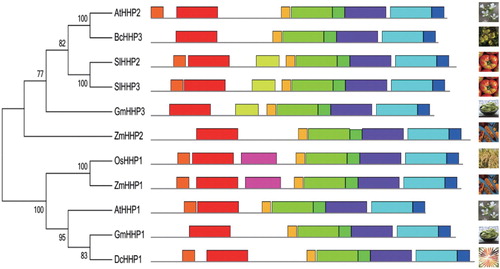
To examine the evolutionary relationship between BcHHP3 and HHPs from other plant species, an unrooted phylogenetic tree was constructed by using the amino acid sequence alignment by the maximum likelihood method with 1000 bootstrap replicates (). The results showed that BcHHP3 in Pak-choi is the most homologous to AtHHP2 in A. thaliana. For further analysis, the schematic structures of the deduced BcHHP3 protein and other plant HHP proteins were investigated (). BcHHP3 and AtHHP2 displayed similar schematic structures, especially in the types and numbers of motifs. This was consistent with the above results.
Figure 2. Phylogenetic and structural analyses of the putative BcHHP3 protein and other HHPs available from GenBank and other databases were carried out. The unrooted tree on the left of the figure is based on an alignment of the full-length protein sequences and was constructed by the maximum likelihood method. The schematic structures of HHP proteins in the selected plants are shown on the right side of the figure (the left panel). The schematic structure of some of the HHP proteins in the selected plants is shown. Different types of patterns are represented by boxes of different color types. The same box type in different proteins indicates the same motif (the right panel). Species are as follows: At, Arabidopsis thaliana; Bc, Brassica campestris; Dc, Daucus carota; Gm, G. max; Os, O. sativa; Sl, Solanum lycopersicum; Zm, Z. mays.
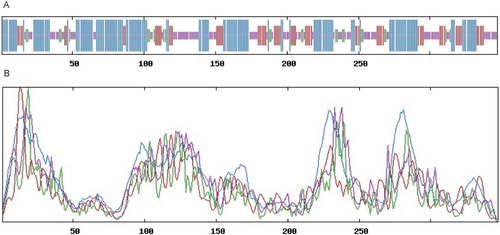
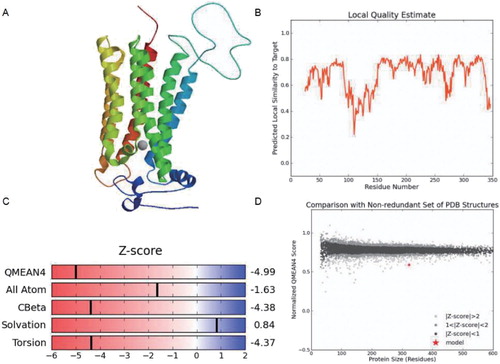
3.3. Sequence analysis and structural analysis of BcHHP3 protein
The secondary structure of the BcHHP3 protein was analyzed and predicted through SOPMA database in Pak-choi. The BcHHP3 protein was mainly composed of free-curl, α-helix and β-sheet, which was consistent with previous studies on BcHHP3 protein structure. SOPMA: with the Alpha helix (Hh): 154, 44.38%; 310 helix (Gg): 0, 0.00%; Pi helix (Ii): 0, 0.00%; Beta bridge (Bb): 0, 0.00%; Extended strand (Ee): 67, 19.31%; Beta turn (Tt): 24, 6.92%; Bend region (Ss): 0, 0.00%; Random coil (Cc): 102, 29.39%; Ambiguous states (?): 0, 0.00%; Other states: 0, 0.00%. Window width: 17, Similarity threshold: 8 and Number of states: 4 (). The secondary structure might function as protection of the BcHHP3 protein from being destroyed by some endonucleases. In addition, the three-dimensional structure of the BcHHP3 protein was predicted by the SWISS-MODEL database. The Seq Identity: 34.89; Oligo-state: monomer; Method: X-ray; Resolution: 2.40 Å; Seq Similarity: 0.37; Range: 25–346; Coverage: 0.80; and Description: Adiponectin receptor protein 2 (). It is noteworthy that the α-helix occurs predominantly in the structure of the BcHHP3 protein and there are amino acid residues as predicted to be exposed. This revealed that the BcHHP3 protein might not be globular. The results showed that the structure of the BcHHP3 protein is relatively stable and highly conservative.
3.4. BcHHP3 gene expression patterns of Pak-choi under different abiotic stresses
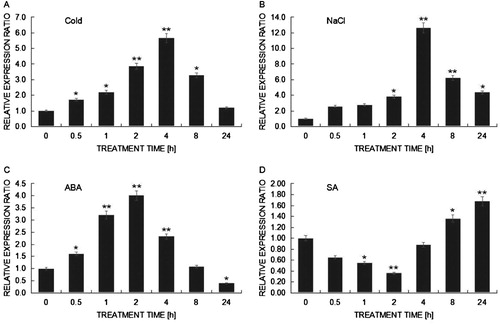
To investigate expression patterns of the BcHHP3 gene under various abiotic stresses, we performed qRT-PCR analysis of the BcHHP3 gene. As shown in , the BcHHP3 gene showed various expression patterns under different abiotic stresses. Under cold stress treatment, the induction dynamics of BcHHP3 were that the BcHHP3 transcript level was rapidly increased and reached a maximum rate at 4 h after treatment, then followed by a slow descent ((A)). BcHHP3 expression increased slowly and reached a maximum rate at 4 h after NaCl stress treatment, then followed by a rapid descent ((B)). The relative expression of BcHHP3 was marginally up-regulated after 2 h and then markedly up-regulated at 2 h after ABA stress treatment, reaching a maximum level of 4-fold (P > .05) compared with the control ((C)). However, under SA stress, the transcription level of BcHHP3 slightly decreased compared with the control, and then increased ((D)). These data suggested that BcHHP3 expression is influenced by cold, NaCl, ABA, and SA stress treatments. More importantly, these data also showed that in the cold stress treatment, the induction dynamics of BcHHP3 was similar to that under ABA stress treatment. After the respective cold and ABA treatments, the BcHHP3 expression in leaves increased significantly (0.01 < P < .05) at 4 h, indicating that BcHHP3 is cold-inducible and involved in co-responding to cold and ABA treatments. Besides, because the expression level of BcHHP3 did not increase rapidly after the drop in temperature, but was induced after 24 h of cold treatment, we speculated that BcHHP3 was related to recovery from translational arrest under low temperature stress.
Figure 5. qRT-PCR analysis of expression patterns on BcHHP3 in Pak-choi under abiotic stress. BcHHP3 expression under cold treatment (A). BcHHP3 expression under salt treatment (B). BcHHP3 expression under ABA treatment (C). BcHHP3 expression under SA treatment (D). The transcript levels of BcHHP3 relative to those of BcACTIN were quantified using 2−ΔΔCT ; ΔΔCT = ΔCT (treated sample) − ΔCT (untreated sample), ΔCT = CTtarget − CTBcACTIN. Each column represents the average and standard deviation of three replicates. *.01 < P < .05, **P < .01, significant differences between treatments, respectively (ANOVA calculated using SPSS). Each bar value represents the Means ± SEs of the three replicates.
3.5. Subcellular localization analysis of BcHHP3
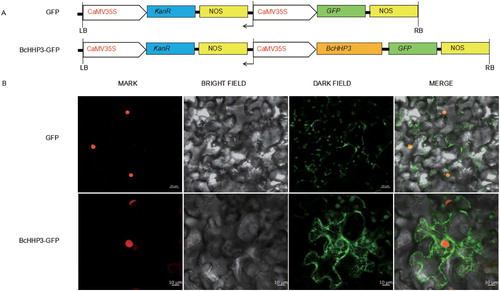
According to WOLF PSORT, the nuclear localization score of BcHHP3 was 8.0 (KNN = 14); and according to Nuc-PLoc, the subcellular location predicted was the nuclear membrane. These results suggested that BcHHP3 might be located in the cell membrane and nuclear membrane. In the experiment, transient expression of the BcHHP3-GFP fusion construct in tobacco cells was tested for BcHHP3 subcellular localization (). GFP alone was used as a control and was distributed on the cell membrane and nuclear membrane ((A)). Confocal images showed that BcHHP3-GFP was localized on the cell membrane and nuclear membrane ((B)). These results suggested that BcHHP3 localized on both the cell membrane and nuclear membrane, which was consistent with its predicted function in destabilizing RNA secondary structures as an RNA chaperone.
Figure 6. Subcellular localization of BcHHP3 in tobacco cells. Constructs were used in the experiments (A). BcHHP3-GFP were transferred to tobacco cells through injecting leaves to test and verify the subcellular localization of BcHHP3 (B). The upper panel shows mark, bright field, dark field and merge images of the GFP control; the lower panel shows mark, bright field, dark field and merge images of BcHHP3-GFP.
4. Discussion
4.1. HHP genes in Arabidopsis, O. sativa, Z. mays, and G. max
To analyze the phylogenetic relationships between HHP genes in various plant species, a combined maximum likelihood tree was generated, which showed that BcHHP3, AtHHP2 and BcHHP2 are orthologous genes and have similar schematic structures. Through our research, we found at least five protein homologues HHP1, HHP2, HHP3, HHP4 and HHP5 in Arabidopsis (Hsieh and Goodman Citation2005). In this study, the BcHHP3 gene was isolated from Pak-choi based on amino acid sequence alignment. The results revealed that BcHHP3 protein sequence is highly similar to HHP protein sequences of A. thaliana and was homologous to HHP protein sequences of O. sativa, Z. mays and G. max, indicating that HHP proteins in the dicotyledonous plants are conservative ((A)). Phylogenetic and structural analyses of the putative BcHHP3 protein and other HHP proteins available from GenBank and other databases was carried out. The results suggested that these orthologous genes might be derived from a common ancestor, and tend to be conserved during evolution.
4.2. Secondary structure and three-dimensional structure analysis of BcHHP3 protein
HHP1 in Arabidopsis is the most closely related to the adiponectin receptor-related PAQR subgroup (Chen et al. Citation2009). Whether HHPs and other PAQR members have the structure and function similar to GPCRs is intriguing. Seatrout mPR has been predicted to have an N-terminal outside topology as in the GPCRs (Zhu et al. Citation2003), whereas the adiponectin receptor has been shown to adopt an N-terminal inside topology, as revealed by epitope tagging experiments (Tamura et al. Citation2014).
The HHP proteins family is homologous to the PAQR family of proteins, which include the membrane progestin receptor (mPR), adiponectin receptor (AdipoR), and YOL002c (Lyons et al. Citation2004; Tang et al. Citation2005; Tamura et al. Citation2014). First, the promoter region of HHP1 was found to contain an ABRE-related element and DPBF, MYB, and MYC recognition sequences, which are the downstream targets of ABA-dependent osmotic stress signaling pathways (Chinnusamy et al. Citation2004). HHP1 is likely to be an upstream positive regulator of cold, drought, and other abiotic stress signaling pathways, as we speculated. Interestingly, it plays a negative regulatory role in seed germination and early growth (Chen et al. Citation2009). As one of the most popular vegetables, Pak-choi is often subject to a variety of biological (especially Aphids and Spodoptera litura, etc.) and abiotic (cold, salt and drought, etc.) stresses throughout all the growth and development stages (Wei et al. Citation2016). Low temperature limits normal growth development of plants directly through the inhibition of metabolic reactions, or indirectly through cold-induced osmotic and oxidative stresses (Chinnusamy et al. Citation2007). The mRNA expression patterns of the five HHP genes in response to phytohormones, low temperature and salt stresses (Hsieh and Goodman Citation2005). However, further experiments are needed to elucidate the functions of the HHP gene family. In this study, BcHHP3 and AtHHP2 were shown to have a high degree of homology and to be conservative, therefore, we suspect that this extra area might be the reason why BcHHP3 and AtHHP2 differ in some functions. In Arabidopsis, at least one gene, AtHHP2 gene, has been found to respond to low-temperature stress, while BcHHP3 has a high degree of homology to AtHHP2 ((B)). Thus, we speculated that BcHHP3 might also be involved in the regulation of low-temperature response.
4.3. BcHHP3 transcription factor responses to abiotic stress in Pak-choi
HHP1 has been shown to be involved in ABA-mediated osmotic stress signaling and to act as a negative regulator (Chen et al. Citation2009). Work was carried out to examine the role of HHP1 in the signaling network for abiotic stresses, including drought, salinity and low temperature, phenotypic analysis related to cold signaling. Sensitivity to abiotic stresses is a very complex phenomenon and there are intricate signaling pathways which enable plants to tolerate and adapt in response to exogenous osmotic stress. The elucidation of the mechanism of osmotic stress signaling may be beneficial in breeding by improving stress tolerance in crops (Xiong et al. Citation2002; Fujita et al. Citation2006). In this study, real-time quantitative PCR analysis revealed that BcHHP3 played an important regulatory role in response to cold, NaCl, ABA, and SA stress treatments in Pak-choi. On exposure to low but not-freezing temperature stresses, plants can acquire cold tolerance, this process is called cold acclimation (Chinnusamy et al. Citation2007). Relative expression of HHP1 increased, in response to low temperature, NaCl, and abscisic acid (ABA) stress treatments as shown by real-time quantitative PCR (qRT-PCR) (Chen et al. Citation2009). The expression patterns of BcHHP3 under continuous low temperature and ABA stress treatments showed similar induction dynamics (). The expression and regulation of the A.thaliana HHP gene family have been studied. In addition to that low temperature and NaCl stress treatments also differently affect the expression patterns of HHP genes (Hsieh and Goodman Citation2005). These results suggested that the physiological functions of BcHHP3 might differ from those of AtHHP2 in response to multiple stresses. BcHHP3 might cross-respond to multiple stresses and produce an overlapping effect or cross-protection against environmental stress factors.
4.4. Localization of BcHHP3 in tobacco cells
Significant HHP1 expression has been observed in the reproductive organs (Hsieh and Goodman Citation2005). In the experiment, transient expression of the BcHHP3-GFP fusion protein showed that BcHHP3 was localized on the cell membrane and nuclear membrane in tobacco cells (). In the reproductive organs, we could see a significant expression of HHP1 (Hsieh and Goodman Citation2005). It is possible that HHP1 plays different roles in different organs or cells and at different growth stages under low-temperature stress in A. thaliana (Chen et al. Citation2009). Considering the conservative structures of BcHHP3 and AtHHP2, similar expression patterns under low-temperature stress, and subcellular localization of BcHHP3 on the cell membrane and nuclear membrane, we hypothesized that BcHHP3 might act as an RNA chaperone during the cold adaptation, playing an important role in RNA processing.
5. Conclusions
In this study, the HHP3 gene was cloned from Pak-choi and named as BcHHP3. We analyzed the sequence structure and studied the evolutionary position through phylogenetic trees. The phylogenetic relationship analysis of HHP transcription factors showed that these HHP genes have a certain homology more or less and are relatively conservative in Pak-choi. More importantly, we found that the homologous relationship of BcHHP3 and AtHHP2 is very close and highly conservative, which reveal that these genes might have some similar functions in some aspects. The BcHHP3 gene was selected and induced by four abiotic stress (cold, salt, ABA, and SA) treatments. Besides, this study also demonstrated that BcHHP3 was localized on the cell membrane and nuclear membrane. In addition, the comparative study of between BcHHP3 in Pak-choi and HHPs in other plant species, provided valuable information for further functional studies of other HHP-like factors in Pak-choi. Our study might support recourses for understanding the expression patterns of BcHHP3 protein under various abiotic stresses. Finally, it should be noted that besides BcHHP3 other cold-responsive HHP-like genes may also exist in Pak-choi. Their functions need to be explored.
TJPI-2018-0103-File010.doc
Download MS Word (29 KB)Acknowledgements
Jin Wang was responsible for the experimental design, experimental work, data analysis and manuscript writing. Feiyi Huang and Xiong You also contributed to the experimental work. Xilin Hou and Xiong You contributed to the interpretation of results and coordinated the study. All authors have read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Agarwal M, Hao Y, Kapoor A, Dong C, Fujii H, Zheng X, Zhu J. 2006. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 281:37636–37645. doi: 10.1074/jbc.M605895200
- Chen CC, Liang CS, Kao AL, Yang CC. 2009. HHP1 is involved in osmotic stress sensitivity in Arabidopsis. J Exp Bot. 60:1589–1604. doi: 10.1093/jxb/erp039
- Chen CC, Liang CS, Kao AL, Yang CC. 2010. HHP1, a novel signalling component in the cross-talk between the cold and osmotic signalling pathways in Arabidopsis. J Exp Bot. 61:3305–3320. doi: 10.1093/jxb/erq162
- Chinnusamy V, Jagendorf A, Zhu JK. 2005. Understanding and improving salt tolerance in plants. Crop Sci. 45:437–448. doi: 10.2135/cropsci2005.0437
- Chinnusamy V, Schumaker K, Zhu JK. 2004. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 55:225–236. doi: 10.1093/jxb/erh005
- Chinnusamy V, Zhu J, Zhu JK. 2007. Cold stress regulation of gene expression in plants. Trends Plant Sci. 12:444–451. doi: 10.1016/j.tplants.2007.07.002
- Fujita M, Fujita AY, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. 2006. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 9:436–442. doi: 10.1016/j.pbi.2006.05.014
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. 1998. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J Cell & Mol Biol. 16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x
- Hsieh MH, Goodman HM. 2005. A novel gene family in Arabidopsis encoding putative heptahelical transmembrane proteins homologous to human adiponectin receptors and progestin receptors. J Exp Bot. 56:3137–3147. doi: 10.1093/jxb/eri311
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. 1998. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell. 10:1151–1162. doi: 10.1105/tpc.10.7.1151
- Jarillo JA, Leyva A, Salinas J, Martinezzapater JM. 1993. Low temperature induces the accumulation of alcohol dehydrogenase mRNA in Arabidopsis thaliana, a chilling-tolerant plant. Plant Physiol. 101:833–837. doi: 10.1104/pp.101.3.833
- Kurkela S, Franck M. 1990. Cloning and characterization of a cold- and ABA-inducible Arabidopsis gene. Plant Mol Biol. 15:137–144. doi: 10.1007/BF00017731
- Livak KJ, Schmitt TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ C T method. Methods. 25:402–408. doi: 10.1006/meth.2001.1262
- Lyons TJ, Villa NY, Regalla LM, Kupchak BR, Vagstad A, Eide DJ, Palmiter RD. 2004. Metalloregulation of yeast membrane steroid receptor homologs. Proc Nat Acad Sci USA. 101:5506–5511. doi: 10.1073/pnas.0306324101
- Stockinger EJ, Gilmour SJ, Thomashow MF. 1997. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Nat Acad Sci USA. 94:1035–1040. doi: 10.1073/pnas.94.3.1035
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2014. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. doi: 10.1093/molbev/msr121
- Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Emtage PC, Funk WD. 2005. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol. 61:372–380. doi: 10.1007/s00239-004-0375-2
- Tang J, Wang F, Wang Z, Huang Z, Xiong A, Hou X. 2013. Characterization and co-expression analysis of WRKY orthologs involved in responses to multiple abiotic stresses in Pak-choi (Brassica campestris ssp. chinensis). BMC Plant Biol. 13:1–13. doi: 10.1186/1471-2229-13-188
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Ann Rev Plant Biol. 50:571–599. doi: 10.1146/annurev.arplant.50.1.571
- Wei Y, Hu W, Xia F, Zeng H, Li X, Yan Y, He C, Shi H. 2016. Heat shock transcription factors in banana: genome-wide characterization and expression profile analysis during development and stress response. Sci Rep. 6:36864. doi: 10.1038/srep36864
- Xiong L, Schumaker KS, Zhu JK. 2002. Cell signaling during cold, drought, and salt stress. Plant Cell. 14:S165–S183.
- Yamaguchi-Shinozaki K, Shinozaki K. 1993. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gener Genet Mgg. 236:331–340. doi: 10.1007/BF00277130
- Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444
- Zhu Y, Rice CD, Pang Y, Pace M, Thomas P. 2003. Cloning, expression, and characterization of a membrane progestin receptor and evidence it is an intermediary in meiotic maturation of fish oocytes. Proc Nat Acad Sci USA. 100:2231–2236. doi: 10.1073/pnas.0336132100