ABSTRACT
Artificial light has been used to control plant growth and secondary metabolite production. Artemisia annua L. plants were illuminated with three light-emitting diode (LED) spectra to investigate proteomic and biochemical responses. After 7 days, proteomic data revealed different protein numbers in leaves, stems, and roots under particular treatments. Results demonstrated increased accumulation of several proteins, including secondary metabolite-related proteins. The red light (R) (660 nm) highly induced terpenoid proteins. Similar biochemical profiles were observed in white light (W) (445, 544 nm) and blue light (B) (445 nm) conditions, while profiles from R treatment were different. Functional proteins of W and B treatments were involved in the MVA and MEP pathways and sesquiterpene biosynthesis. By contrast, unique proteins in R treatment were mainly expressed in sesquiterpene and tetraterpene biosynthesis. Specific relationships between biosynthetic proteins and sesquiterpenes were observed, suggesting the indispensable role of the light spectrum in regulating terpenoid biosynthesis.
Introduction
Qinghao (Artemisia annua L.), also known as sweet wormwood, is an important Chinese herb used to treat many infectious diseases (Graziose et al. Citation2010; Chang Citation2016). The plant extract exhibited wide-ranging pharmacological properties including antioxidant, antimicrobial, antiparasitic, and antiviral activities (Efferth et al. Citation2001; Cai et al. Citation2004; Sharopov et al. Citation2020). Phytochemical analyses revealed that leaves and crude extracts of A. annua contained several useful compounds including terpenoids, flavonoids, coumarins, sterols, phenols, lipids, and other hydrocarbons (Bhakuni et al. Citation2001; Czechowski et al. Citation2018). Among these, terpenoids are the largest and most diverse group, consisting of more than 40,000 plant secondary compounds (Bohlmann and Keeling Citation2008). Artemisinin, a useful sesquiterpene compound found in leaves of A. annua, plays a central role in combating the malaria-causing parasite Plasmodium falciparum (Klayman Citation1985; Dondorp et al. Citation2009), while the triterpene squalene showed a synergistic effect with artemisinin to promote anti-falciparum activity (Karaket et al. Citation2014).
Terpenoids contain unit(s) of 5-carbon isoprene and can be classified as hemi- (C5: isoprene), mono- (C10), sesqui- (C15), di- (C20), sester- (C25), tri- (C30), and tetra- (C40: carotenoids) terpenoids (Yazaki et al. Citation2017). Biosynthesis of terpenoid compounds requires the common precursor isopentenyl diphosphate (IPP) that can be synthesized by two different pathways involving mevalonic acid (MVA) and 2-C-methyl-D-erythritol 4-phosphate (MEP). The MVA pathway operates in cytosol, while the MEP pathway is localized in plastids. Crosstalk and exchange of IPP between these two pathways have been investigated in many plant species (Lange and Croteau Citation1999; Hemmerlin et al. Citation2003; Laule et al. Citation2003; Opitz et al. Citation2014). Biosynthesis of mono-, di- and tetraterpenes occurs in plastids, while cytosol is mainly responsible for sesquiterpene and triterpene biosynthesis (Muhlemann et al. Citation2014; Abbas et al. Citation2017). Alteration of the terpenoid biosynthetic pathway affects the target and also other related compounds.
Plants grown in their natural habitats spontaneously encounter a variety of stresses including light stresses from sunlight radiation. To maintain cellular activity and survival, plants employ molecular mechanisms underlying plant responses involving the activity of metabolic enzyme networks associated with responsive proteins (Kong and Okajima Citation2016; Chi et al. Citation2019). Previous studies indicated the influence of light signals on biosynthesis of secondary metabolites in several plant species. High light emission enhanced accumulation of coniferin, syringin, and flavonoids in Arabidopsis roots (Hemm et al. Citation2004), while the presence of light increased levels of several terpenoids, indole alkaloids, and loganin in Catharanthus roseus L. plants (Yu et al. Citation2018). Cited reports suggested positive impacts of light on improving plant metabolite production; however, inappropriate light supply ultimately destroyed plant photosynthetic apparatus and induced the accumulation of reactive oxygen species (ROS), leading to alterations in plant growth and development or even death (Li et al. Citation2009; Xie et al. Citation2014; Huang et al. Citation2019). The role of light signals in regulating metabolite production was also demonstrated in A. annua. Levels of bioactive compounds such as artemisinin were promoted at more than 1.5 times the control when UV-B and UV-C were irradiated before transplantation (Rai et al. Citation2011). Improvement in bioactive compounds occurred, with significant induction of key genes in the biosynthetic pathway after UV irradiation. Blue and red spectra irradiation increased the contents of artemisinin and its derivatives via upregulation of biosynthesis genes and suppression of enzymes in the competitive pathway (Zhang et al. Citation2018).
The implication of light quality provided a method for significant improvement of bioactive and other useful compounds in plants. Effects of different light wavelengths on transcriptomic responses of biosynthetic genes have previously been reported but data on proteomic responses of plants to light signals remain limited. Previous studies mainly emphasized the role of light in regulating light sensory proteins (photoreceptor proteins) such as UV-A/blue light receptors (Fuglevand et al. Citation1996; Lin Citation2002), red/far-red photoreceptors (Sharrock Citation2008; Legris et al. Citation2019), and UV-B photoreceptors (Heijde and Ulm Citation2012; Jenkins Citation2014), while the impacts of light signals on biosynthetic proteins have not been investigated in A. annua. Hence, here, comprehensive analyses of proteomic expression and the phytochemical fingerprint of volatile terpenoid compounds in A. annua were conducted to examine the proteomic and metabolomic responses of the plant to different light signals. Relationships between unique terpenoid proteins and putative terpenoid compounds in each specific light spectrum were also investigated.
Materials and methods
Plant material, growth conditions, and LED spectra irradiation
Artemisia annua seeds have been kindly provided by Dr. Chalermpol Kirdmanee (BIOTEC, NSTDA) since the year 2000, under the cooperative research project between Mahidol University and the National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA), Thailand (Supaibulwatana et al. Citation2004). The seeds were germinated and subsequently multiplied in the tissue culture lab at the Faculty of Science, Mahidol University. Seedlings of A. annua L. were maintained in vitro in solidified Murashige and Skoog (Citation1962) medium supplied with 3% (w/v) sucrose and incubated at 25 ± 2°C and 60 ± 5 μmol m−2 s−1 light intensity with 16 h photoperiod. After 15 days, the plantlets were transferred to pots containing vermiculite, supplemented with sugar-free MS medium, and cultivated under 80 ± 5 μmol m−2 s−1 light intensity. Four biological replications of three-month-old A. annua plants were transferred to white light (W, 445, 544 nm), blue light (B, 445 nm), and red light (R, 660 nm) for 7 days. Environmental factors in the PFAL (plant factory with artificial lighting) chamber were controlled at 25 ± 2°C, 50 ± 5% relative humidity and 200 ± 10 μmol m−2 s−1 light intensity with 16 h photoperiod. Fresh leaves were harvested on day 7 and stored at −80°C until required for further analysis.
GeLC-MS/MS shotgun proteomics
Total protein was prepared using the modified protocol from Bryant et al. (Citation2016). Protein content was determined by Bradford assay (Citation1976) and 15 µg of total protein was used for gel packing with 12.5% polyacrylamide. The gel pieces were subjected to dehydration in 100% acetonitrile (ACN) and incubated with 10 mM dithiothreitol (DTT) in 10 mM ammonium bicarbonate (AmBic) at 56°C for 1 h. For the alkylation step, 100 mM iodoacetamide in 10 mM AmBic was added to the samples and incubated in the dark at room temperature (RT) for 45 min. Samples were then dehydrated with 100% ACN by shaking at RT for 5 min. Sequencing-grade trypsin (Promega, Germany) was added to the gels and incubated at 37°C overnight for in-gel digestion. Peptide products were extracted from the samples by adding 50% (v/v) ACN in 0.1% (v/v) formic acid, incubated at RT for 10 min, and dried at 45°C for 4 h. Tryptic peptides were protonated with 0.1% (v/v) formic acid before operating LC-MS/MS. The analysis was performed with three experimental replications and all experiments were conducted in duplicate.
Protein identification and bioinformatics
Data were quantified by MaxQuant 1.6.1.12 using the Andromeda search engine to correlate MS/MS spectra to the UniProtViridiplantae protein database (Bryant et al. Citation2016). A unique peptide with at least seven amino acids was required for protein identification and further quantification analysis. Biological relationships between proteomic data obtained from different LED treatments were analyzed using a Venn diagram (http://jvenn.toulouse.inra.fr/app/index.html) (Bardou et al. Citation2014). The non-overlapping proteins were further subjected to functional identification and classification based on their biological process and molecular function using UniProt identifiers (https://www.uniprot.org). The raw data were deposited in the jPOST repository (Okuda et al. Citation2016) under reference number PXD027227.
Phytochemical analysis of A. annua leaves by gas chromatography-mass spectrometry (GC-MS)
Fresh leaves of A. annua were ground into a fine powder in liquid nitrogen using a pestle and mortar. The sample powder was transferred to 10 mL glass tubes containing 2 mL of dichloromethane. After mixing, each glass tube containing a sample was sonicated for 15 min and centrifuged at 5000 rpm for 5 min. The supernatant was filtrated with a 0.45 µm syringe filter membrane and stored at −80°C until required for analysis. Phytochemical analysis of the leaf extract was performed by GC-MS under the following condition: the oven was programmed at 100°C for 1 min, then at 5°C/min to 150°C, at 2°C/min to 200°C, at 15°C/min to 300°C, and held for 15 min. Compounds were identified by comparison of retention time and fragments with standards. Mass spectral information was extracted and compared with the Wiley No.7 database to identify unknown compounds. Compounds with at least 80% match quality were relatively quantified against the internal standard (methyl heptadecanoate, C17) and represented as average relative contents (µg mL−1).
Clustering analysis of terpenoid proteins and plant metabolites
To examine the correlation between the abundance of terpenoid compounds and unique terpenoid proteins under a particular light spectrum, hierarchical clustering analysis (HCA) was performed using MultiExperiment Viewer (MeV) version 4.9.0 (Saeed et al. Citation2003). Relative contents of terpenoid compounds were obtained from GC-MS data and normalized to the amount of internal standard, while protein levels were obtained from maximum intensity of LC-MS/MS, normalized to log2 values. Grouping results were used to construct a dendrogram, with a heatmap representing the levels of proteins and terpenoid compounds.
Results
Differential expression of proteins in leaves, stems and roots of A. annua under different LED light spectra
GeLC-MS/MS shotgun proteomics were performed to examine how A. annua plants responded to LED light qualities and the number of proteins in leaves, stems, and roots of 7-day light-treated plants were investigated. The analysis revealed more than 20,000 proteins in each treatment. Highest total number of proteins was detected under red spectrum (25,987 proteins), followed by blue spectrum (24,556 proteins) and white spectrum (22,863 proteins). Comparative analysis using Venn diagrams revealed that numbers of proteins in leaves, stems and roots under white light condition were different. The highest proportion of protein (54.73%) was recorded in leaves of A. annua grown under white light; however, 3282 proteins were also found in stems and roots ((a)). Among the total proteins, 2964, 4414, and 3701 were uniquely detected in leaves, stems, and roots, respectively. Leaves of blue light-treated plants also showed the highest protein response with 66.59% of total proteins detected. Under blue spectrum, 3744 proteins were commonly observed in all organs with 2368, 3522, and 2414 proteins uniquely expressed in leaves, stems, and roots, respectively. Similar to white and blue spectra, 81.53% of total proteins were reported in leaves of red light treatment. A Venn diagram demonstrated that leaves, stems, and roots of A. annua from red light treatment shared 14,462 common proteins with 2644 proteins only detected in leaves. Smaller numbers of organ-specific proteins were found in stems and roots of A. annua.
Figure 1. Proteomic and metabolomic responses of three-month-old A. annua treated with white (W), blue (B) and red (R) LED lamps. (a) Venn diagram showing the number of differentially expressed proteins in leaves, stems and roots. Plants were cultivated in an indoor plant factory under artificial light (PFAL) and illuminated with different spectra of LED lamps as W (445, 554 nm), B (445 nm) and R (660 nm) at light intensity of 200 ± 10 μmol m−2 s−1 photosynthetic photon flux density (PPF) with 16 h photoperiod and incubated under 25.0 ± 0.5°C and 65.0 ± 1.5% relative humidity. Samples were harvested from four biological replicates, each with nine experimental replications, 7 days after treatment and separated into leaves, stems and roots for GeLC-MS/MS analysis. Proteomic data with q-value greater than 0.95 were used for comparative analysis using a Venn diagram. (b) Classification of uniquely expressed proteins in different organs of A. annua under white (W), blue (B) and red (R) light conditions. Proteomic data were functionally classified by UniProt identifiers (https://www.uniprot.org) using biological processes and molecular functions. Asterisks indicate numbers of terpenoid proteins registered in the UniProt database. (c) Venn diagram showing the number of differentially expressed proteins detected in leaves of A. annua grown under white (W), blue (B) and red (R) LED spectra. (d) Hierarchical clustering analysis (HCA) showing the relation between light spectrum and detected volatile terpenoids using MultiExperiment Viewer (MeV) version 4.9.0. High contents of terpenoid compounds are shown in red, with dark blue indicating low content. Each row represents the individual metabolite detected under specific light treatment (shown in the column).
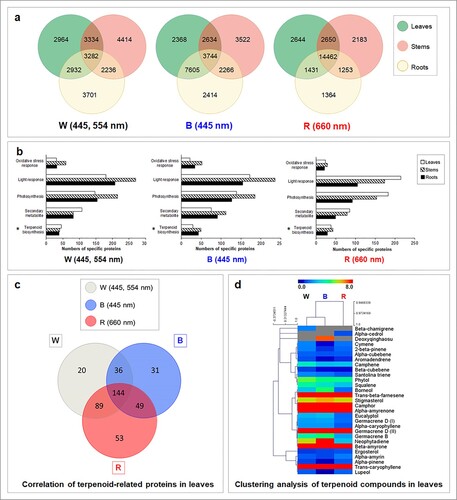
Functional analysis of uniquely expressed proteins in different organs of A. annua treated with different LED light spectra
Uniquely expressed proteins from each treatment were further characterized for their functional identifications and protein classifications using UniProt identifiers (https://www.uniprot.org). Proteins were classified based on their biological process and molecular function. Results revealed abundances of light-responsive proteins, photosynthetic proteins, and secondary metabolite-related proteins ((b)). Light-responsive proteins comprised those with light stimulus-response [GO:0009416, GO:0071482], photosynthesis [GO:0015979], light reaction of photosynthesis [GO:0019684], red or far-red light signaling pathway [GO:0010017], blue light signaling pathway [GO:0009785], UV-B response [GO:0010224], and light intensity response [GO:0071484]. The highest number of proteins in this group was reported in stems of A. annua grown under white and blue spectra. By contrast, leaves showed the highest number of proteins when exposed to red spectrum. Photosynthetic proteins are the largest subdivision of light-responsive proteins, and light has been widely recognized for its central role in the photosynthesis process. Proteins involved in photosynthesis [GO:0015979], dark reaction [GO:0019685], light reaction [GO:0019684], light harvesting in photosystem I [GO:0009768], and the light-independent chlorophyl biosynthetic process [GO:0036068] were assembled in this group. Results revealed the presence of 100–200 photosynthetic proteins in leaves, stems, and roots of A. annua in each treatment. Expression patterns were similar to the spectral response of light-responsive proteins, with the highest numbers found in stems of A. annua under white and blue treatments but not in red treatment. Moreover, proteins related to oxidative stress response [GO:0006979] such as catalase (EC 1.11.1.6), glutathione S-transferase (EC 2.5.1.18), peroxidase (EC 1.11.1.7), superoxide dismutase (EC 1.15.1.1), and other scavenging enzymes were also detected, especially in stems of A. annua.
Proteins involved in plant metabolite production were also explored. This group included sterol [GO:0016126], alkaloid [GO:0009821], flavonoid [GO:0009813], and terpenoid [GO:0016114] biosynthetic proteins. Responses differed among the organs and light spectra. Under white light, leaves showed the highest number of non-overlapping proteins, while stems and roots expressed a similar number. Under blue spectrum, secondary metabolic proteins were highly expressed in stems followed by roots and leaves, respectively. In red light treatment, the highest number was found in leaves, with fewer proteins in stems and roots. Based on biological process and molecular function in UniProt protein identifiers, a total of 502 proteins found in all organs under all light conditions were classified as terpenoid proteins, consisting of 50 proteins in the MVA and MEP pathways, an isoprene synthase, 62 monoterpene synthases, 149 sesquiterpene synthases, 75 diterpene synthases, 74 triterpene synthases, 74 tetraterpene synthases, and 17 other terpene synthases (Table S1). Leaves of A. annua showed 45 unique proteins under white light treatment, while the other organs gave reduced numbers. Expression patterns of terpenoid proteins under blue spectrum were similar to those of secondary metabolic proteins, with the highest number of non-overlaps (49 proteins) found in stems, while red light showed the highest number of terpenoid proteins in leaves.
Accumulation of terpenoid-related proteins in leaves of A. annua under different light spectra
Different light qualities induced different numbers of terpenoid proteins in A. annua, and effects of light qualities were highlighted in leaves where terpenoid compounds were mainly produced. Analysis revealed a total of 422 terpenoid-related proteins. Leaves of A. annua exhibited high sensitivity to red LED light and 335 proteins (79.38% of total terpenoid proteins) were detected. The smallest number (260 proteins) was observed under blue LED light, approximately 61.6% of the total terpenoid proteins in leaves. A Venn diagram revealed 144 common proteins in all light treatments ((c)), with 20, 31, and 53 proteins explicitly expressed under white, blue, and red spectra, respectively.
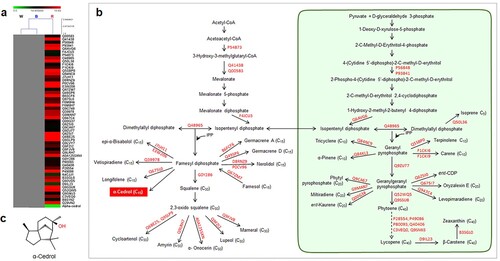
Under white LED condition, 20 unique proteins were reported (), with a high proportion of MVA and MEP pathway-related proteins (25.0%) followed by sesquiterpene synthases (20.0%) and monoterpene synthases (15.0%). Out of five proteins in the MVA and MEP pathways, three related to the cytosolic MVA pathway and one was localized in the plastidic MEP pathway, while the rest were involved in the exchange of the common precursor between both pathways. Among these, diphosphomevalonate decarboxylase (Acc. No. O23722), reported in Arabidopsis thaliana (Cordier et al. Citation1999), presented at the highest level (). This protein catalyzes the first committed step in isopentenyl diphosphate (IPP) biosynthesis, leading to biosynthesis of isoprene-containing compounds such as sterols and terpenoids (Henry et al. Citation2015). The MEP-related protein, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (Acc. No. Q6EPN6) involved in the formation of isoprenoid intermediates, IPP and dimethylallyl diphosphate (DMAPP) via the MEP pathway (Huang et al. Citation2018), was also detected with relative expression value of 14.01. The highest level of terpenoid synthase was vetispiradiene synthase 2 (Acc. No. Q39979) that showed relative expression value of 19.19. This sesquiterpene synthase was previously reported to convert trans, trans-farnesyl diphosphate (FPP) to vetispiradiene in Hyoscyamus muticus (Back and Chappell Citation1995). Moreover, prolycopene isomerase 1 (Acc. No. Q52QW3) involved in lycopene (tetraterpene) biosynthesis was also highly expressed in this treatment.
Table 1. Numbers and proportions of unique terpenoid proteins detected in leaves of A. annua grown under white (W), blue (B) and red (R) light spectra.
Table 2. Identification of uniquely expressed terpenoid proteins and their expression levels in leaves of A. annua grown under different light spectra.
Blue spectrum exhibited several proteins in the MVA and MEP pathways and sesquiterpene synthases. Acetyl-CoA acetyltransferase (Acc. No. Q8S4Y1) showed relative expression level of 15.99 under this spectrum. The functional study of this protein was demonstrated in Arabidopsis thaliana and showed its important role in biosynthesis of mevalonate in the cytosolic MVA pathway (Cordier et al. Citation1999). High level of 5-epi-aristolochene synthase 2 (Acc. No. Q84LF0) was also reported, with relative expression of 15.54. This cyclase protein is involved in the cyclization of FPP to form the bicyclic sesquiterpene, 5-epi-aristolochene in Nicotiana attenuate (Bohlmann et al. Citation2002).
Red LED spectrum demonstrated the highest number of unique terpenoid proteins, suggesting the role of this spectrum in regulating terpenoid proteins. Among several groups of terpenoid proteins, isoprene synthase (Acc. No. Q50L36) was only detected under this spectrum. This protein is responsible for the conversion of DMAPP to isoprene, a building block of the terpenoid biosynthetic pathway. High numbers of functional proteins were involved in the biosynthesis of sesquiterpenes and tetraterpenes. Alpha-bisabolol synthase (Acc. No. E3W206) showed the highest protein level among sesquiterpene synthases. This protein was cloned and characterized from Santalum spicatum and showed its role in the formation of β-, α-bisabolol, α-bisabolene and also farnesene isomers from FPP intermediate (Jones et al. Citation2011). A high proportion of tetraterpene synthases including phytoene synthase, 15-cis-phytoene desaturase, lycopene beta cyclase, beta-carotene 3-hydroxylase, and zeta-carotene desaturase were also detected in this condition.
LED spectra affected plant metabolite production in leaves of A. annua
Light qualities have shown their important role in regulating plant metabolite production in several plant species. Numbers of metabolic proteins were reported to be influenced by light spectra. Here, we determined how plant metabolite production responded to light signals. The presence of mono-, sesqui-, di- and triterpenoids was analyzed by GC-MS. Phytochemical profiles differed depending on light compositions. Analysis results revealed 28 volatile terpenoid compounds with more than 80% match quality to the Wiley 7 No.1 Library, comprising 8 monoterpenes, 11 sesquiterpenes, 2 diterpenes and 7 triterpenes ().
Table 3. Phytochemical profiles obtained from leaf extracts of A. annua grown under white (W), blue (B) and red (R) LED spectra.
Similar numbers of monoterpenoid compounds were observed in all light spectra; however, camphor content was high (23.5 µg mL−1 relative content), especially under blue spectrum. Maximum total content of sesquiterpenes was obtained from blue light treatment. Germacrene-D was the most abundant compound with 24.5 µg mL−1 relative content, approximately 2 times the lowest content under red LED treatment. Variations were observed among the C15 compounds as the artemisinin derivative deoxyqinghaosu only found under blue spectrum irradiation. Two diterpenes, neophytadiene and phytol, also showed highest contents under this spectrum. Similar to other terpenoids, highest content of triterpenoids was found in blue light treatment. Among seven triterpenes reported in this study, beta-amyrone was found with greatest abundance under blue light followed by white and red spectra, respectively. Correlation between the abundance of volatile terpenoids and light spectrum was assessed by hierarchical clustering analysis (HCA) with Pearson correlation ((d)). The heatmap showed that terpenoid profiles of A. annua grown under white and blue spectra were strongly clustered, while the red spectrum profile was separately clustered.
Distribution of functional proteins in artemisinin biosynthetic pathway of A. annua
Light spectrum interfered with the numbers of terpenoid proteins as well as the production of terpenoid compounds. Variations in proteins and terpenoids were observed among different light conditions. Therefore, we presumed that the light spectrum regulated the terpenoid biosynthetic pathway through the activation of particular proteins. To clarify the effect of light spectrum on alteration of terpenoid biosynthesis, localizations of functional proteins were illustrated in the MVA, MEP, and terpenoid biosynthetic pathways of A. annua plants under a particular light spectrum ().
Figure 2. Specific relationships of functional proteins and terpenoid compounds in response to white spectrum (445, 554 nm) in A. annua. (a) Hierarchical clustering analysis showing the correlation of proteins and associated terpenoid compounds detected under white spectrum. (b) Terpenoid biosynthetic pathway showing accession numbers of detected functional proteins and their products. The yellow box indicates the associated terpenoid that uniquely expressed under white spectrum, while (c) displays its chemical structure (β-Chamigrene).
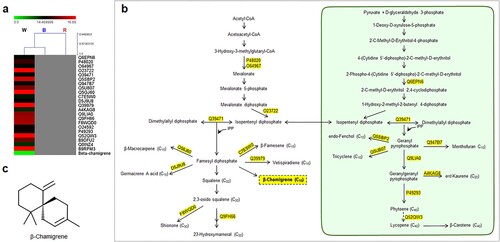
Figure 3. Specific relationships of functional proteins and terpenoid compounds in response to blue spectrum (445 nm) in A. annua. (a) Hierarchical clustering analysis showing the correlation of proteins and associated terpenoid compounds detected under blue spectrum. (b) Terpenoid biosynthetic pathway showing accession numbers of detected functional proteins and their products. The blue box indicates the associated terpenoid that uniquely expressed under blue spectrum, while (c) displays its chemical structure (Deoxyqinghaosu).
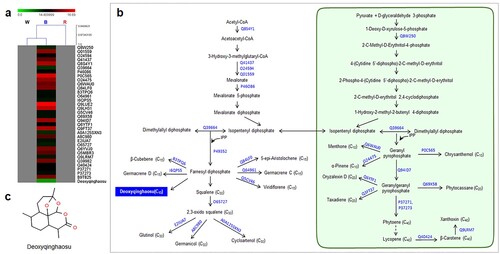
Figure 4. Specific relationships of functional proteins and terpenoid compounds in response to red spectrum (660 nm) in A. annua. (a) Hierarchical clustering analysis showing the correlation of proteins and associated terpenoid compounds detected under red spectrum. (b) Terpenoid biosynthetic pathway showing accession numbers of detected functional proteins and their products. The red box indicates the associated terpenoid that uniquely expressed under red spectrum, while (c) displays its chemical structure (α-Cedrol).
Out of 20 unique proteins in white light treatment (), 18 functional proteins were identified and shown in (b). These proteins were mainly distributed in the MVA, MEP, and sesquiterpene biosynthetic pathways. Three were monoterpene synthases, while the rest comprised two tetraterpene synthases and one each of di- and triterpene synthase. A higher number of unique proteins (31 proteins) were particularly induced under blue spectrum, including 28 functional identified proteins in terpenoid biosynthesis ((b)). Similar to white light treatment, proteins related to the MVA pathway, MEP pathway, and sesquiterpene biosynthesis were the most responsive groups. Equal amounts of biosynthetic enzymes (four proteins each) were di-, tri- and tetraterpene synthases, while the smallest proportion of terpenoid proteins (9.7%) was identified as monoterpene synthases. The most noticeable number was observed in red light treatment, where 53 unique proteins were differentially expressed. Unlike those in white and blue treatments, functional analysis revealed large numbers of proteins related to sesquiterpene and tetraterpene biosynthesis in red light condition ((b)). Approximately 52.4% of unique sesquiterpene synthases were specifically induced by red spectrum. Furthermore, 10 proteins involved in carotenoid biosynthesis were explicitly expressed in this condition. Proteins in the MVA and MEP pathways and di- and triterpene biosynthesis were also induced under this particular wavelength.
Discussion
Light signals play important roles in regulating terpenoid production of A. annua, which could be monitored in terms of transcriptomic expressions correlated with target terpenoid compounds (Zhang et al. Citation2018; Lopes et al. Citation2020), however, there has no obvious report on enzymes (proteins) involved in the metabolic flux of cytosolic MVA and plastidic MEP pathways of terpenoid biosynthesis. Zhang et al. (Citation2018) previously described the effect of LED spectra at 470 and 670 nm on promoting artemisinin and artemisinic acid contents over those illuminated with 770 nm. Transcriptomic analysis showed upregulation of six genes after exposure to 470 and 670 nm; including four genes involved in the cyclization of artemisinin (C15) biosynthesis (ADS, CYP71AV1, DBR2, and ALDH1) and two genes (HMGS and HMGR) in the MVA pathway. On the other hand, Lopes et al. (Citation2020) reported a wide range of secondary metabolites including monoterpenes (C10), sesquiterpenes (C15), and other non-terpenoid compounds after being exposed to 475, 510, 570, or 650 nm of LED light. Although about 40–50 compounds were detected by GC-MS, only two genes involved in the cyclization steps of artemisinin production (ADS and CYP71AV1) were investigated for transcriptomic expression. However, only the expression of ADS gene was correlated with increased artemisinin content in blue light treatment.
Our study firstly demonstrated the evidence of enzymes (proteins) involved in the metabolic flux of cytosolic MVA and plastidic MEP pathways of terpenoid biosynthesis. The different numbers of proteins were detected in leaves, stems, and roots of A. annua at 7 days after LED light exposure ((a)). As expected, variations in proteomic profiles were observed among different organs of A. annua treated with different light spectra. Fully exposed leaves exhibited the highest total number of proteins under LED light spectra irradiation, suggesting leaves as the most responsive organ in response to light exposure. Our results demonstrated that proteins involved in the molecular mechanism of light-sensing and light response such as photoreceptors and photosynthesis were mainly expressed. Although light is an important factor for plant growth and development, the adverse effect of light as an oxidative stress-inducing factor was also reported (Barber and Andersson Citation1992; Li et al. Citation2009; Dinakar et al. Citation2012). The high presence of detoxification enzymes in all light conditions suggested the role of LED spectra as stress-inducing factors. However, oxidative stress that occurred in this study did not interfere with physiological processes since proteins involved in photosynthesis were also noticed.
Since the leaves were considered as the main sources of terpenoid production, the effect of the LED light spectrum on the number of terpenoid proteins in leaves of A. annua was highlighted. A total of 289, 260, and 335 terpenoid proteins were reported in leaves of A. annua treated with white, blue, and red spectra, respectively. Most terpenoid proteins responded to a particular light spectrum in a similar manner but differential proteins were noticeable for 20, 31, and 53 proteins uniquely expressed in white, blue, and red light treatments, respectively ((c)). Interestingly, high numbers of MVA and MEP pathways-related proteins and sesquiterpene synthases were found under white and blue light treatments (). The red spectrum influenced sesquiterpene and tetraterpene (carotenoid) biosynthesis (). Our study demonstrated that the specific wavelength of 445 nm present in white and blue spectra exhibited similar terpenoid profiles, possibly due to the activity of photoreceptors in the explicit region of the light spectrum (Kong and Okajima Citation2016; Mawphlang and Kharshiing Citation2017). Influences of LED light signals on expression patterns of biosynthetic proteins and terpenoid compounds were demonstrated. Among all LED spectra, the red spectrum was the most effective condition for regulating terpenoid proteins. HCA revealed that sesquiterpenes were highly responsible for induction of terpenoid proteins in each light treatment. A bicyclic sesquiterpene, β-chamigrene, showed strong correlation with the expression level of terpenoid proteins under white spectrum ((a,c)). This compound exhibited promising biological properties such as antibacterial activities and cytotoxic activities against HeLa and Hep-2 cancer cell lines (Dias et al. Citation2005; Antonsen et al. Citation2014). Deoxyqinghaosu positively correlated with the expression level of unique terpenoid proteins under blue spectrum ((a,c)). Although it is an inactive form of artemisinin and has no antimalarial activity against Plasmodium berghei (Li Citation2012), our previous study revealed a strong correlation between deoxyqinghaosu and increased antimalarial activity against P. falciparum NF54 (unpublished data). Alpha cedrol was only detected in red light condition ((a,c). This compound has been found to exert antioxidant, anti-inflammatory, antimicrobial, and antifungal activities along with cytotoxic effects against some cancer cells (Loizzo et al. Citation2008; Khoury et al. Citation2014; Wang et al. Citation2019; Mishra et al. Citation2021). Accordingly, specific induction of terpenoid proteins and correlated sesquiterpenes were conceivably caused by LED spectra. These will provide more understanding of light signals on metabolic proteins in regulating the overall pathway of terpenoid biosynthesis under particular light signals. To support and further validate our results, a transcriptomic study of biosynthetic enzymes is required.
Conclusions
This study demonstrated the integration of metabolomic and proteomic analyses to investigate the responses of A. annua plants under different light spectra. An LC-MS/MS platform combined with functional analysis revealed differential inductions of light-responsive proteins, oxidative stress-related proteins, and metabolite biosynthetic enzymes in all tested organs. Among these, 422 terpenoid-related proteins were detected in leaves, with some unique proteins for each treatment. Twenty proteins were explicitly expressed under white spectrum, while 31 proteins were only found in blue spectrum treatment. Unexpectedly, red light had more extensive effects on terpenoid protein inductions, especially the protein involved in sesquiterpene and tetraterpene biosynthesis. Integrated metabolomics and proteomics revealed possible roles of the light spectrum in regulating production of sesquiterpenoid compounds. HCA revealed specific correlations between terpenoid proteins and sesquiterpenes under a particular light spectrum. Our results suggested light quality as an important factor for creating proteomic variations, leading to differences in terpenoid production. This information can be applied for the functional study of light-inducing proteins in terpenoid biosynthesis to provide improvement of important compounds for pharmaceutical use.
Supplemental Material
Download MS Word (613 KB)Acknowledgements
The authors thank Growlab Agritech Company Limited, Thailand for providing the LED lamps. Experimental facilities used in this research were supported by CIF, Faculty of Science, Mahidol University, Thailand and Agricultural Research Development Agency (ARDA) project no. CRP600502035. We also thank the Proteomics Research Laboratory, BIOTEC, NSTDA, Thailand for providing technical support and the facilities for proteomic analysis.
Data availability statement
All data generated in this study were deposited in the jPOST repository at https://repository.jpostdb.org/entry/JPST001250, reference number PXD027227.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Darunmas Sankhuan
Ms. Darunmas Sankhuan is a doctoral candidate under the double degree program between Mahidol University, Thailand (Biotechnology) and Niigata University, Japan (Agriculture and Bioresources). Darunmas got a scholarship from RGJ (Royal Golden Jubilee) of Thailand Research Fund (TRF), as well as co-support from Niigata University under Global Circus program. Her special interest in metabolic bioengineering, transcriptomic analysis, proteomic analysis and plant metabolome.
Sittiruk Roytrakul
Dr. Sittiruk Roytrakul is a head of Functional Ingredients and Food Innovation Research Group and Proteomics Research Laboratory, National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA), Thailand. He specializes in a range of platform to support various proteome-related researches from identification of single proteins to large-scale proteomic studies.
Masaru Nakano
Prof. Dr. Masaru Nakano is working in areas of Agriculture and Bioresource at Department of Life and Food Science, Faculty of Agriculture, Niigata University, Japan. His researches are focusing on horticultural science, especially ornamental and floriculture crops using genetic transformation and molecular breeding technology.
Kanyaratt Supaibulwatana
Assoc. Prof. Dr. Kanyaratt Supaibulwatana has held the position of Director, School of Bioinnovation and Bio-based Product Intelligence and Director of Plant Physiology and Agri-Biotechnology lab., Department of Biotechnology, Faculty of Science, Mahidol University. Her research focuses on agricultural biotechnology, plant physiology under stresses, plant factories, micropropagation, mutation breeding and metabolic bioengineering of medicinal plants. In addition, Dr. Kanyaratt has actively involved in the initiative programs of public-private partnerships to promote innovation and entrepreneurial ecosystems by encouraging various activities including; setup of new graduate programs for R&D and industrialization quality system, joint innovation incubator, and establishment of in-campus Start Up facilities arising from scientific inventions to promote University-Industry Linkages. https://science.mahidol.ac.th/expertise/search.php?q=Kanyaratt%20Supaibulwatana.
References
- Abbas F, Ke Y, Yu R, Yue Y, Amanullah S, Jahangir MM, Fan Y. 2017. Volatile terpenoids: multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta. 246:803–816.
- Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Kim HI, Yoneyama K, Xie X, Ohnishi T, et al. 2014. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. PNAS. 111:18084–18089.
- Aharoni A, Giri AP, Verstappen FW, Bertea CM, Sevenier R, Sun Z, Jongsma MA, Schwab W, Bouwmeester HJ. 2004. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell. 16:3110–3131.
- Antonsen S, Skattebøl L, Stenstrøm Y. 2014. Synthesis of Racemic β-Chamigrene, a Spiro[5.5]undecane Sesquiterpene. Molecules. 19:20664–20670.
- Aracri B, Bartley GE, Scolnik PA, Giuliano G. 1994. Sequence of the phytoene desaturase locus of tomato. Plant Physiol. 106:789.
- Araki T, Saga Y, Marugami M, Otaka J, Araya H, Saito K, Yamazaki M, Suzuki H, Kushiro T. 2016. Onocerin biosynthesis requires two highly dedicated triterpene cyclases in a fern Lycopodium clavatum. Chembiochem. 17:288–290.
- Attia M, Kim SU, Ro DK. 2012. Molecular cloning and characterization of (+)-epi-α-bisabolol synthase, catalyzing the first step in the biosynthesis of the natural sweetener, hernandulcin, in Lippia dulcis. Arch. Biochem Biophys. 527:37–44.
- Attucci S, Aitken SM, Gulick PJ, Ibrahim RK. 1995. Farnesyl pyrophosphate synthase from white lupin: molecular cloning, expression, and purification of the expressed protein. Arch Biochem Biophys. 321:493–500.
- Aubourg S, Lecharny A, Bohlmann J. 2002. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genom. 267:730–745.
- Back K, Chappell J. 1995. Cloning and bacterial expression of a sesquiterpene cyclase from Hyoscyamus muticus and its molecular comparison to related terpene cyclases. J Biol Chem. 270:7375–7381.
- Barber J, Andersson B. 1992. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci. 17:61–66.
- Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. 2014. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 15:1.
- Bartley GE, Scolnik PA. 1993. cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J Biol Chem. 268:25718–25721.
- Basyuni M, Oku H, Tsujimoto E, Kinjo K, Baba S, Takara K. 2007. Triterpene synthases from the Okinawan mangrove tribe, Rhizophoraceae. FEBS. 274:5028–5042.
- Bertea CM, Schalk M, Karp F, Maffei M, Croteau R. 2001. Demonstration that menthofuran synthase of mint (Mentha) is a cytochrome p450 monooxygenase: cloning, functional expression, and characterization of the responsible gene. Arch Biochem Biophys. 390:279–286.
- Bhakuni RS, Jain DC, Sharma RP, Kumar S. 2001. Secondary metabolites of Artemisia annua and their biological activity. Curr Sci. 80:35–48.
- Bleeker PM, Spyropoulou EA, Diergaarde PJ, Volpin H, De Both MT, Zerbe P, Bohlmann J, Falara V, Matsuba Y, Pichersky E, et al. 2011. RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol. Biol. 77:323–336.
- Block AK, Vaughan MM, Schmelz EA, Christensen SA. 2019. Biosynthesis and function of terpenoid defense compounds in maize (Zea mays). Planta. 249:21–30.
- Bohlmann J, Keeling CI. 2008. Terpenoid biomaterials. Plant J. 54:656–669.
- Bohlmann J, Stauber EJ, Krock B, Oldham NJ, Gershenzon J, Baldwin IT. 2002. Gene expression of 5-epi-aristolochene synthase and formation of capsidiol in roots of Nicotiana attenuata and N. sylvestris. Phytochemistry. 60:109–116.
- Bohlmann J, Steele CL, Croteau R. 1997. Monoterpene synthases from grand fir (Abies grandis). cDNA isolation, characterization, and functional expression of myrcene synthase, (-)-(4S)-limonene synthase, and (-)-(1S,5S)-pinene synthase. J. Biol. Chem. 272:21784–21792.
- Bonk M, Hoffmann B, Von Lintig J, Schledz M, Al-Babili S, Hobeika E, Kleinig H, Beyer P. 1997. Chloroplast import of four carotenoid biosynthetic enzymes in vitro reveals differential fates prior to membrane binding and oligomeric assembly. FEBS. 247:942–950.
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248–254.
- Breitenbach J, Kuntz M, Takaichi S, Sandmann G. 1999. Catalytic properties of an expressed and purified higher plant type zeta-carotene desaturase from Capsicum annuum. FEBS. 265:376–383.
- Breitenbach J, Sandmann G. 2005. zeta-carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene. Planta. 220:785–793.
- Bryant L, Patole C, Cramer R. 2016. Proteomic analysis of the medicinal plant Artemisia annua: data from leaf and trichome extracts. Data Brief. 7:325–331.
- Cai Y, Luo Q, Sun M, Corke H. 2004. Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life Sci. 74:2157–2184.
- Chang Z. 2016. The discovery of Qinghaosu (artemisinin) as an effective anti-malaria drug: a unique China story. Sci China Life Sci. 59:81–88.
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD. 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89:789–804.
- Chi YH, Koo SS, Oh HT, Lee ES, Park JH, Phan KAT, Wi SD, Bae SB, Paeng SK, Chae HB, et al. 2019. The physiological functions of universal stress proteins and their molecular mechanism to protect plants from environmental stresses. Front Plant Sci. 10:750.
- Chiou CY, Pan HA, Chuang YN, Yeh KW. 2010. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta. 232:937–948.
- Choi D, Ward BL, Bostock RM. 1992. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell. 4:1333–1344.
- Chye ML, Tan CT, Chua NH. 1992. Three genes encode 3-hydroxy-3-methylglutaryl-coenzyme A reductase in Hevea brasiliensis: hmg1 and hmg3 are differentially expressed. Plant Mol Biol. 19:473–484.
- Colby SM, Crock J, Dowdle-Rizzo B, Lemaux PG, Croteau R. 1998. Germacrene c synthase from Lycopersicon esculentum cv. VFNT cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. PNAS. 95:2216–2221.
- Cordier H, Karst F, Bergès T. 1999. Heterologous expression in Saccharomyces cerevisiae of an Arabidopsis thaliana cDNA encoding mevalonate diphosphate decarboxylase. Plant Mol Biol. 39:953–967.
- Czechowski T, Larson TR, Catania TM, Harvey D, Wei C, Essome M, Brown GD, Graham IA. 2018. Detailed phytochemical analysis of high- and low artemisinin-producing chemotypes of Artemisia annua. Front Plant Sci. 9:641–641.
- Deguerry F, Pastore L, Wu S, Clark A, Chappell J, Schalk M. 2006. The diverse sesquiterpene profile of patchouli, Pogostemon cablin, is correlated with a limited number of sesquiterpene synthases. Arch Biochem Biophys. 454:123–136.
- Dias T, Brito I, Moujir L, Paiz N, Darias J, Cueto M. 2005. Cytotoxic sesquiterpenes from Aplysia dactylomela. J Nat Prod. 68:1677–1679.
- Dinakar C, Djilianov D, Bartels D. 2012. Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci. 182:29–41.
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 361:455–467.
- Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Fäldt J, Miller B, Bohlmann J. 2003. (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell. 15:1227–1241.
- Efferth T, Dunstan H, Sauerbrey A, Miyachi H, Chitambar C. 2001. The anti-malarial artesunate is also active against cancer. Int J Oncol. 18:767–773.
- Fitzpatrick AH, Bhandari J, Crowell DN. 2011. Farnesol kinase is involved in farnesol metabolism, ABA signaling and flower development in Arabidopsis. Plant J. 66:1078–1088.
- Fuglevand G, Jackson JA, Jenkins GI. 1996. UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell. 8:2347–2357.
- Genschik P, Criqui MC, Parmentier Y, Marbach J, Durr A, Fleck J, Jamet E. 1992. Isolation and characterization of a cDNA encoding a 3-hydroxy-3-methylglutaryl coenzyme A reductase from Nicotiana sylvestris. Plant Mol. Biol. 20:337–341.
- Gomez SK, Cox MM, Bede JC, Inoue K, Alborn HT, Tumlinson JH, Korth KL. 2005. Lepidopteran herbivory and oral factors induce transcripts encoding novel terpene synthases in Medicago truncatula. Arch. Insect Biochem. Physiol. 58:114–127.
- Graziose R, Lila MA, Raskin I. 2010. Merging traditional Chinese medicine with modern drug discovery technologies to find novel drugs and functional foods. Curr Drug Discov Technol. 7:2–12.
- Hall DE, Robert JA, Keeling CI, Domanski D, Quesada AL, Jancsik S, Kuzyk MA, Hamberger B, Borchers CH, Bohlmann J. 2011. An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J. 65:936–948.
- Hayashi H, Huang P, Inoue K, Hiraoka N, Ikeshiro Y, Yazaki K, Tanaka S, Kushiro T, Shibuya M, Ebizuka Y. 2001. Molecular cloning and characterization of isomultiflorenol synthase, a new triterpene synthase from Luffa cylindrica, involved in biosynthesis of bryonolic acid. FEBS. 268:6311–6317.
- Heijde M, Ulm R. 2012. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17:230–237.
- Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C. 2004. Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 38:765–778.
- Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, Rohmer M, Bach TJ. 2003. Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J Biol Chem. 278:26666–26676.
- Henry LK, Gutensohn M, Thomas ST, Noel JP, Dudareva N. 2015. Orthologs of the archaeal isopentenyl phosphate kinase regulate terpenoid production in plants. PNAS. 112:10050–10055.
- Hieber AD, Mudalige-Jayawickrama RG, Kuehnle AR. 2006. Color genes in the orchid Oncidium Gower Ramsey: identification, expression, and potential genetic instability in an interspecific cross. Planta. 223:521–531.
- Huang H, Ullah F, Zhou D-X, Yi M, Zhao Y. 2019. Mechanisms of ROS regulation of Plant development and stress responses. Front Plant Sci. 10:800.
- Huang R, Wang Y, Wang P, Li C, Xiao F, Chen N, Li N, Li C, Sun C, Li L, et al. 2018. A single nucleotide mutation of IspF gene involved in the MEP pathway for isoprenoid biosynthesis causes yellow-Green leaf phenotype in rice. Plant Mol Biol. 96:5–16.
- Husselstein-Muller T, Schaller H, Benveniste P. 2001. Molecular cloning and expression in yeast of 2,3-oxidosqualene-triterpenoid cyclases from Arabidopsis thaliana. Plant Mol. Biol. 45:75–92.
- Iijima Y, Davidovich-Rikanati R, Fridman E, Gang DR, Bar E, Lewinsohn E, Pichersky E. 2004. The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physio. 136:3724–3736.
- Irmisch S, Krause ST, Kunert G, Gershenzon J, Degenhardt J, Köllner TG. 2012. The organ-specific expression of terpene synthase genes contributes to the terpene hydrocarbon composition of chamomile essential oils. BMC Plant Biol. 12:84.
- Jenkins GI. 2014. The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell. 26:21–37.
- Jones CG, Moniodis J, Zulak KG, Scaffidi A, Plummer JA, Ghisalberti EL, Barbour EL, Bohlmann J. 2011. Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J Biol Chem. 286:17445–17454.
- Kajiwara S, Kakizono T, Saito T, Kondo K, Ohtani T, Nishio N, Nagai S, Misawa N. 1995. Isolation and functional identification of a novel cDNA for astaxanthin biosynthesis from Haematococcus pluvialis, and astaxanthin synthesis in Escherichia coli. Plant Mol. Biol. 29:343–352.
- Karaket N, Wiyakrutta S, Lacaille-Dubois MA, Supaibulwatana K. 2014. T-DNA insertion alters the terpenoid content composition and bioactivity of transgenic Artemisia annua. Nat Prod Commun. 9:363–366.
- Karvouni Z, John I, Taylor JE, Watson CF, Turner AJ, Grierson D. 1995. Isolation and characterisation of a melon cDNA clone encoding phytoene synthase. Plant Mol Biol. 27:1153–1162.
- Keller Y, Bouvier F, d’Harlingue A, Camara B. 1998. Metabolic compartmentation of plastid prenyl lipid biosynthesis-evidence for the involvement of a multifunctional geranylgeranyl reductase. FEBS. 251:413–417.
- Khoury M, El Beyrouthy M, Ouaini N, Iriti M, Eparvier V, Stien D. 2014. Chemical composition and antimicrobial activity of the essential oil of Juniperus excelsa M. Bieb. Growing Wild in Lebanon. Chem Biodivers. 11:825–830.
- Klayman DL. 1985. Qinghaosu (artemisinin): an antimalarial drug from China. Science. 228:1049–1055.
- Köllner TG, Gershenzon J, Degenhardt J. 2009. Molecular and biochemical evolution of maize terpene synthase 10, an enzyme of indirect defense. Phytochemistry. 70:1139–1145.
- Köllner TG, Schnee C, Li S, Svatos A, Schneider B, Gershenzon J, Degenhardt J. 2008. Protonation of a neutral (S)-beta-bisabolene intermediate is involved in (S)-beta-macrocarpene formation by the maize sesquiterpene synthases TPS6 and TPS11. J Biol Chem. 283:20779–20788.
- Kong SG, Okajima K. 2016. Diverse photoreceptors and light responses in plants. J Plant Res. 129:111–114.
- Korth KL, Stermer BA, Bhattacharyya MK, Dixon RA. 1997. HMG-CoA reductase gene families that differentially accumulate transcripts in potato tubers are developmentally expressed in floral tissues. Plant Mol. Biol. 33:545–551.
- Koschmieder J, Fehling-Kaschek M, Schaub P, Ghisla S, Brausemann A, Timmer J, Beyer P. 2017. Plant-type phytoene desaturase: functional evaluation of structural implications. PloS one. 12:e0187628.
- Kranz-Finger S, Mahmoud O, Ricklefs E, Ditz N, Bakkes PJ, Urlacher VB. 2018. Insights into the functional properties of the marneral oxidase CYP71A16 from Arabidopsis thaliana. Biochim Biophys Acta. 1866:2–10.
- Lange BM, Croteau R. 1999. Isopentenyl diphosphate biosynthesis via a mevalonate-independent pathway: Isopentenyl monophosphate kinase catalyzes the terminal enzymatic step. PNAS. 96:13714–13719.
- Laule O, Fürholz A, Chang H-S, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M. 2003. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. PNAS. 100:6866–6871.
- Lee S, Chappell J. 2008. Biochemical and genomic characterization of terpene synthases in Magnolia grandiflora. Plant Physiol. 147:1017–1033.
- Legris M, Ince YÇ, Fankhauser C. 2019. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun. 10:5219.
- Li G, Köllner TG, Yin Y, Jiang Y, Chen H, Xu Y, Gershenzon J, Pichersky E, Chen F. 2012. Nonseed plant Selaginella moellendorffi has both seed plant and microbial types of terpene synthases. PNAS. 109:14711–14715.
- Li Y. 2012. Qinghaosu (artemisinin): chemistry and pharmacology. Acta Pharmacol Sin. 33:1141–1146.
- Li Z, Wakao S, Fischer BB, Niyogi KK. 2009. Sensing and responding to excess light. Annu Rev Plant Biol. 60:239–260.
- Li ZH, Matthews PD, Burr B, Wurtzel ET. 1996. Cloning and characterization of a maize cDNA encoding phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Plant Mol Biol. 30:269–279.
- Lin C. 2002. Blue light receptors and signal transduction. Plant Cell. 14(Suppl):207–225.
- Lluch MA, Masferrer A, Arró M, Boronat A, Ferrer A. 2000. Molecular cloning and expression analysis of the mevalonate kinase gene from Arabidopsis thaliana. Plant Mol Biol. 42:365–376.
- Loizzo MR, Tundis R, Menichini F, Saab AM, Statti GA, Menichini F. 2008. Antiproliferative effects of essential oils and their major constituents in human renal adenocarcinoma and amelanotic melanoma cells. Cell Prolif. 41:1002–1012.
- Lopes EM, Guimarães-Dias F, Gama TdSS, Macedo AL, Valverde AL, de Moraes MC, de Aguiar-Dias ACA, Bizzo HR, Alves-Ferreira M, Tavares ES, et al. 2020. Artemisia annua L. and photoresponse: from artemisinin accumulation, volatile profile and anatomical modifications to gene expression. Plant Cell Rep. 39:101–117.
- Martin DM, Fäldt J, Bohlmann J. 2004. Functional characterization of nine Norway Spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 135:1908–1927.
- Mawphlang OIL, Kharshiing EV. 2017. Photoreceptor mediated plant growth responses: implications for photoreceptor engineering toward improved performance in crops. Front Plant Sci. 8:1181.
- Mishra SK, Bae YS, Lee Y-M, Kim J-S, Oh SH, Kim HM. 2021. Sesquiterpene alcohol cedrol chemosensitizes human cancer cells and suppresses cell proliferation by destabilizing plasma membrane lipid rafts. Front Cell Dev. 8.
- Montamat F, Guilloton M, Karst F, Delrot S. 1995. Isolation and characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl-coenzyme A synthase. Gene. 167:197–201.
- Morita M, Shibuya M, Kushiro T, Masuda K, Ebizuka Y. 2000. Molecular cloning and functional expression of triterpene synthases from pea (Pisum sativum) new alpha-amyrin-producing enzyme is a multifunctional triterpene synthase. FEBS. 267:3453–3460.
- Muhlemann JK, Klempien A, Dudareva N. 2014. Floral volatiles: from biosynthesis to function. Plant Cell Environ. 37:1936–1949.
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 15:473–497.
- Nguyen DT, Göpfert JC, Ikezawa N, Macnevin G, Kathiresan M, Conrad J, Spring O, Ro DK. 2010. Biochemical conservation and evolution of germacrene A oxidase in Asteraceae. J Biol Chem. 285:16588–16598.
- Niehaus TD, Okada S, Devarenne TP, Watt DS, Sviripa V, Chappell J. 2011. Identification of unique mechanisms for triterpene biosynthesis in Botryococcus braunii. PNAS. 108:12260–12265.
- Okada A, Shimizu T, Okada K, Kuzuyama T, Koga J, Shibuya N, Nojiri H, Yamane H. 2007. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Mol Biol. 65:177–187.
- Okuda S, Watanabe Y, Moriya Y, Kawano S, Yamamoto T, Matsumoto M, Takami T, Kobayashi D, Araki N, Yoshizawa AC, et al. 2016. jPOSTrepo: an international standard data repository for proteomes. Nucl Acids Res. 45:1107–1111.
- Opitz S, Nes WD, Gershenzon J. 2014. Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochemistry. 98:110–119.
- Otomo K, Kenmoku H, Oikawa H, König WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T. 2004. Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J. 39:886–893.
- Peters RJ. 2006. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry. 67:2307–2317.
- Phillips MA, Wildung MR, Williams DC, Hyatt DC, Croteau R. 2003. cDNA isolation, functional expression, and characterization of (+)-alpha-pinene synthase and (-)-alpha-pinene synthase from loblolly pine (Pinus taeda): stereocontrol in pinene biosynthesis. Arch Biochem Biophys. 411:267–276.
- Rai R, Meena RP, Smita SS, Shukla A, Rai SK, Pandey-Rai S. 2011. UV-B and UV-C pre-treatments induce physiological changes and artemisinin biosynthesis in Artemisia annua L. – An antimalarial plant. J Photochem Photobiol. B. 105:216–225.
- Ringer KL, McConkey ME, Davis EM, Rushing GW, Croteau R. 2003. Monoterpene double-bond reductases of the (-)-menthol biosynthetic pathway: isolation and characterization of cDNAs encoding (-)-isopiperitenone reductase and (+)-pulegone reductase of peppermint. Arch Biochem Biophys. 418:80–92.
- Riou C, Tourte Y, Lacroute F, Karst F. 1994. Isolation and characterization of a cDNA encoding Arabidopsis thaliana mevalonate kinase by genetic complementation in yeast. Gene. 148:293–297.
- Rivera SB, Swedlund BD, King GJ, Bell RN, Hussey CE, Shattuck-Eidens DM, Wrobel WM, Peiser GD, Poulter CD. 2001. Chrysanthemyl diphosphate synthase: isolation of the gene and characterization of the recombinant non-head-to-tail monoterpene synthase from Chrysanthemum cinerariaefolium. PNAS. 98:4373–4378.
- Rohdich F, Wungsintaweekul J, Luttgen H, Fischer M, Eisenreich W, Schuhr CA, Fellermeier M, Schramek N, Zenk MH, Bacher A. 2000. Biosynthesis of terpenoids: 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase from tomato. PNAS. 97:8251–8256.
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 34:374–378.
- Sasaki K, Ohara K, Yazaki K. 2005. Gene expression and characterization of isoprene synthase from Populus alba. FEBS Lett. 579:2514–2518.
- International Rice Genome Sequencing Project, Sasaki T. 2005. The map-based sequence of the rice genome. Nature. 436:793. (1476–4687).
- Sawai S, Uchiyama H, Mizuno S, Aoki T, Akashi T, Ayabe S, Takahashi T. 2011. Molecular characterization of an oxidosqualene cyclase that yields shionone, a unique tetracyclic triterpene ketone of Aster tataricus. FEBS Lett. 585:1031–1036.
- Schäfer UA, Reed DW, Hunter DG, Yao K, Weninger AM, Tsang EW, Reaney MJ, MacKenzie SL, Covello PS. 1999. An example of intron junctional sliding in the gene families encoding squalene monooxygenase homologues in Arabidopsis thaliana and Brassica napus. Plant Mol Biol. 39:721–728.
- Schepmann HG, Pang J, Matsuda SP. 2001. Cloning and characterization of Ginkgo biloba levopimaradiene synthase which catalyzes the first committed step in ginkgolide biosynthesis. Arch Biochem Biophys. 392:263–269.
- Sharopov FS, Salimov A, Numonov S, Safomuddin A, Bakri M, Salimov T, Setzer WN, Habasi M. 2020. Chemical composition, antioxidant, and antimicrobial activities of the essential oils from Аrtemisia annua L. growing wild in Tajikistan. Nat Prod Commun. 15:1934578X20927814.
- Sharrock RA. 2008. The phytochrome red/far-red photoreceptor superfamily. Genome Biol. 9:230.
- Supaibulwatana K, Banyai W, Cheewasakulyong P, Kirdmanee C, Kamchonwongpaisan S, Yuthavong Y. 2004. Effect of culture conditions, elicitation and induced mutagenesis on plant growth and productions of antimalarial agents in Artemisia annua L. In: Jonas R, Pandy A, Tharun G, editors. Proceedings of the biotechnological advances and application in bioconversion of renewable raw materials; Braunschweig, Germany. p. 240–249.
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. 2009. CYP76M7 is an ent-cassadiene C11alpha-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell. 21:3315–3325.
- Takaya A, Zhang YW, Asawatreratanakul K, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Koyama T. 2003. Cloning, expression and characterization of a functional cDNA clone encoding geranylgeranyl diphosphate synthase of Hevea brasiliensis. Biochim Biophys Acta. 1625:214–220.
- Wang G, Tian L, Aziz N, Broun P, Dai X, He J, King A, Zhao PX, Dixon RA. 2008. Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 148:1254–1266.
- Wang J-w, Chen S-s, Zhang Y-m, Guan J, Su G-Y, Ding M, Li W, Zhao Y-Q. 2019. Anti-inflammatory and analgesic activity based on polymorphism of cedrol in mice. Environ Toxicol Pharmacol. 68:13–18.
- Wang Q, Jia M, Huh JH, Muchlinski A, Peters RJ, Tholl D. 2016. Identification of a Dolabellane type Diterpene synthase and other root-expressed Diterpene synthases in Arabidopsis. Front Plant Sci. 7:1761.
- Wang Z, Yeats T, Han H, Jetter R. 2010. Cloning and characterization of oxidosqualene cyclases from Kalanchoe daigremontiana: enzymes catalyzing up to 10 rearrangement steps yielding friedelin and other triterpenoids. J Biol Chem. 285:29703–29712.
- Wu Y, Wang Q, Hillwig ML, Peters RJ. 2013. Picking sides: distinct roles for CYP76M6 and CYP76M8 in rice oryzalexin biosynthesis. Biochem J. 454:209–216.
- Xie H-T, Wan Z-Y, Li S, Zhang Y. 2014. Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in arabidopsis. Plant Cell. 26:2007–2023.
- Xie X, Kirby J, Keasling JD. 2012. Functional characterization of four sesquiterpene synthases from Ricinus communis (castor bean). Phytochemistry. 78:20–28.
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya NM, Coates RM, Peters RJ. 2007. Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry. 68:312–326.
- Yasumoto S, Fukushima EO, Seki H, Muranaka T. 2016. Novel triterpene oxidizing activity of Arabidopsis thaliana CYP716A subfamily enzymes. FEBS Lett. 590:533–540.
- Yazaki K, Arimura G-i, Ohnishi T. 2017. ‘Hidden’ terpenoids in plants: their biosynthesis, localization and ecological roles. Plant Cell Physiol. 58:1615–1621.
- Yu B, Liu Y, Pan Y, Liu J, Wang H, Tang Z. 2018. Light enhanced the biosynthesis of terpenoid indole alkaloids to meet the opening of cotyledons in process of photomorphogenesis of Catharanthus roseus. Plant Growth Regul. 84:617–626.
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al. 2005. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38.
- Zhang D, Sun W, Shi Y, Wu L, Zhang T, Xiang L. 2018. Red and blue light promote the accumulation of artemisinin in Artemisia annua L. Molecules. 23:1329.
- Zhou X, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, Fish T, Liu J, Thannhauser TW, Li L. 2015. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. PNAS. 112:3558–3563.
- Zhu C, Yamamura S, Nishihara M, Koiwa H, Sandmann G. 2003. cDNAs for the synthesis of cyclic carotenoids in petals of Gentiana lutea and their regulation during flower development. Biochim Biophys Acta. 1625:305–308.
