ABSTRACT
Tomato is one of the major economically domesticated crops, and it is extensively used in different ways and purposes worldwide. Cell metabolism is the central core of all the biological processes to sustain life including cell growth, differentiation, maintenance, and response to environmental stress. To evaluate how genetic engineering can improve tomato fruit metabolome, the transcriptomic and metabolomic datasets of two transgenic tomatoes (SlMX1 overexpression and RNAi lines) have been compared with wild-type. The combined results demonstrated that the constitutive expression of SlMX1 not only increased trichome formation, carotenoids, and terpenoids as has been stated in several studies, but has also up- and down-regulated the expression of multiple genes related to cell growth (cell wall turnover), primary (carbohydrates, vitamins, and phytohormones), and secondary (phenylpropanoids, carotenoids, and terpenoids) metabolism, cell signaling, and stress responses. These changes in gene expression due to the constitutive expression of SlMX1 promote the most important agroeconomic traits such as fruit yield and quality, biosynthesis of health-promoting phytochemicals (including phenolic acids, flavonoids, and anthocyanins), and finally, activate resistance to Botrytis cinerea and repress the expression of over-ripening-related genes, thus extending the fruit shelf-life. In conclusion, the traits improvement achieved by SlMX1 overexpression can be harnessed in molecular breeding programs to engineer fruit size and yield, induce health-promoting secondary metabolites, promote fungal resistance, and finally extend the fruit shelf-life.
Introduction
Tomato fruit is one of the major economically domesticated crops and an excellent source of health-promoting compounds such as mineral nutrients, vitamins, and other active secondary metabolites that serve as antioxidant compounds associated with reduced risk of inflammatory process and chronic diseases such as cardiovascular and neoplasic diseases (Ewas et al. Citation2016; Lozano-Milo et al. Citation2020). The relationship between health and diet has encouraged the people to consume the fresh fruit and vegetables (García-Pérez and Gallego Citation2022). However, to ensure adequate availability of high-quality plant products for the ever-growing world population, traits including fruit yield, quality, health-beneficial compounds, and pathogens’ defense resistance need to be manipulated in crops individually and in combination (Ewas et al. Citation2016; Ewas, Gao, et al. Citation2017).
The fruit size and weight are positively correlated with tomato yield, whereas the fruit number is mostly negatively correlated (Guo and Simmons Citation2011). Fruit size is the result of the coordination of cell division and expansion, controlled mainly by plant hormones which control both pericarp cell size and number, respectively (Gan et al. Citation2022). At molecular level, cell cycle regulators such as cyclin and cyclin-dependent kinase (CDK) are key factors that control cell division (Carneiro et al. Citation2021). The expression of cell size regulator (CSR) is associated with fruit development in tomato. Previous studies characterized CSR gene to regulate cell size, leading to the enlargement of fruit size and increases the final yield (Van der Knaap et al. Citation2014).
Fruit quality includes several organoleptic characteristics (sugars, organic acids, volatile compounds, and so on), firmness, and health-related compounds. Among them, fruit firmness is a complex trait which involves numerous physical properties, including cell wall structure, cellular turgor, and cuticle characteristics (Li et al. Citation2020). At the onset of ripening, the disintegration and degradation of the cell wall soften the whole fruit and limit its quality and shelf-life (Romero and Rose Citation2019). Fruit softening starts with the activation of cell wall-degrading enzymes such as β-galactosidase and polygalacturonase among others (Zhang et al. Citation2015). Suppression of the expression of these enzymes in climacteric fruit delayed the fruit softening (Meli et al. Citation2010). Various ripening-deficient mutants have been used to extend fruit shelf-life, including nonripening (nor), never-ripe (nr), alcobaca, and ripening inhibitor (rin). Unfortunately, an accompanying loss of flavor results in most of these mutants (Osorio et al. Citation2020). Several molecular genetic strategies have been explored to reduce fruit softening (loss of firmness) in tomato by regulating different transcription factors (TFs) (Liu et al. Citation2016). Some recent studies have also demonstrated that plant TFs can positively regulate fruit ripening or delay this process by altering the ethylene biosynthesis pathway genes, namely ACS2, ACS4, ACO1, and ACO3 (Zhu et al. Citation2014; Jian et al. Citation2021).
TFs are proteins that recognize a certain sequence of deoxyribonucleic acid on the promoter locus of genes and control the gene expression either positively (activator) or negatively (repressor). Previous studies clearly indicate that TFs serve as key regulators of diverse plant functions and characterized to be major regulators in several metabolic pathways (Iwase et al. Citation2009). Manipulation of plant TFs to induce secondary metabolism has also provided efficient strategies for crop enrichment with health-promoting compounds (Butelli et al. Citation2008).
Large class of health-related secondary metabolites is the phenylpropanoids that are synthesized from the carbon skeleton of phenylalanine through seven steps in the shikimate pathway (Zhang et al. Citation2015). Phosphoenolpyruvate, produced from glycolysis, and erythose-4-phosphate, from the oxidative pentose phosphate (OPPP), are main precursors for phenylalanine. Phenylpropanoids mainly include phenolic acids, lignans, coumarins, stilbenes, and flavonoids (chalcones, flavones, flavanones, isoflavonoids, flavonols, flavanols, and anthocyanins) (García-Pérez et al. Citation2020). It plays vital roles in plant defense, floral pigments, survival, and structural support (Deng and Lu Citation2017). Susceptibility/resistance to opportunistic pathogens such as Botrytis cinerea and fruit-softening rate are great processes in shelf-life determination of tomato fruit (Cantu et al. Citation2009). In addition, several health-beneficial features of flavonoids have been reported including antioxidant, antiosteoporosis, and anticancer activities (Pandey et al. Citation2015; García-Pérez et al. Citation2018, Citation2019).
RNA sequencing could be a useful technique to shed light on tomato fruit metabolome, yield, and quality characteristics. In previous studies (Ewas et al. Citation2016; Ewas, Gao, et al. Citation2017), it has been reported that SlMX1 gene encoding a MIXTA-like MYB TF, which can positively regulate trichome formation, and show enhanced resistance to different biotic and abiotic stress factors. However, the mechanism of SlMX1 to regulate these processes remains unknown. Here, a large dataset spanning the transcriptome and metabolome has been generated for tomato SlMX1 overexpression and RNAi lines. The current study reported that SlMX1 is more than a trichome regulator. New roles of SlMX1 gene in tomato fruit metabolism, development, and ripening have been explored. In this work, the relations between SlMX1 expression and the improvement in biosynthesis of primary and secondary metabolites have been demystified. Our study paves the way toward further tomato improvement and for decoding the mechanisms underlying the myriad of other agronomic traits that can be improved by SlMX1 TF.
Materials and methods
Plant materials and growth conditions
Six-week-old healthy seedling of wild-type (WT) tomato (Solanum lycopersicum L. ‘Ailsa Craig’) and transgenic tomato (three SlMX1 overexpression and two RNAi lines) were obtained from the seed bank of Huazhong Agriculture University, China. Plants were transplanted in germination trays and then transferred for cultivation in tomato pots containing peat moss, compost, vermiculite, perlite, and sand in a climate-controlled greenhouse at 24°C during the day and 18°C during night, with day-neutral photoperiod (12 h light/dark), during the second week of April for two successive seasons.
Tomato genetic transformation
SlMX1 has been amplified from the cDNA and genomic DNA of tomato, using specific primers based on the sequence, Solyc01g010910. The amplified PCR product has been cloned into the pMD18-T vector, and the sequence has been confirmed by sequencing. The pMD18-T vector containing the SlMX1 cDNA has been double digested using XbaI and SacI and ligated into the plant binary vector pBI121 under the control of a strong constitutive promoter (CaMV35S), cleaved with XbaI and SacI.
Suppression of SlMX1 expression has been performed using the RNAi vector pK7GWIWG2 (II), into which has been cloned a fragment of SlMX1 from pDONR207 using the Clonase BP reaction (Invitrogen, NY, USA). Both constructs of SlMX1 have been transformed into tomato, mediated by Agrobacterium tumefaciens strain C58. Empty vectors have been transformed as controls.
Fourteen SlMX1 OE lines and seven RNAi lines have been obtained as revealed by real-time PCR analyses. Tomato wild-type (WT), SlMX1 overexpressed lines (OE2, OE5, and OE11) with a high transcript level, and knock-out lines (RNAi2 and RNAi7) with a low transcript level, have been used for further experiments.
RNA Seq library preparation and sequencing from tomato leaves
To isolate total RNA from 2-month-old tomato leaves of WT, SlMX1 OE, and RNAi lines (two biological replicates each) samples (approximately 20 mg) have been frozen in liquid nitrogen. Using mortar and piston, samples have been homogenized and transferred to a 1.5-mL centrifuge tubes. Further processing has been performed using the RNeasy R Plant Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. The RNA has been eventually eluted with RNase-free water.
cDNA has been synthesized using SuperScript TMII Reverse Transcriptase (RT) (Invitrogen) following the supplier’s instructions. In short, 1 μL Oligo (dT) primers, 500–1000 ng RNA, and 1 μL dNTP mix have been incubated for 5 min at 65°C. 1× Reaction buffer and 10 μM DTT have been added. Samples have been incubated at 42°C before addition of the RT. The synthesis has been performed over 60 min with heat inactivation of the enzyme for 15 min at 70°C.
Transcript analysis has been performed by using cDNA corresponding to 500 ng RNA from three biological replicates. SYBR green assays have been developed using iQTM SYBRR® Green Supermix (Bio-Rad, Hercules, USA) with gene-specific primers. The reaction setup has been adjusted to a total volume of 25 μL with 12.5 μL iQ SYBR Green Supermix, 1 μL of each primer at 10 μM, and 10 μL cDNA template. PCR has been performed on ‘iQ5 multicolor real-time PCR detection system’ (Bio-Rad). Expressions have been calculated using the CT method (Livak and Schmittgen Citation2001). The Actin gene was used as an internal control. All primers used in this analysis are listed in Supplementary Table 1.
Total RNA has been extracted and treated with Dnase I to degrade any possible DNA contaminants. RNA libraries with inserts approximately 250–500 bp in size have been prepared (BGI, China) for 50 bp single-end sequencing on the Illumina HISeq 2000, at a depth of ≈50 million reads per library.
Nucleotide sequences of the contigs have been blasted against the tomato database (ftp.solgenomics.net/unigene_builds/combined_species_assemblies/tomato_species). The annotation was performed using a local Eblast tool (E-value 1e–9) (Conesa et al. Citation2005). Further analyses with this tool included functional annotation by Gene Ontology (GO) terms and Enzyme Commission numbers (EC code), InterPro terms (InterProScan), and metabolic pathways (Kyoto Encyclopedia of Genes and Genomes, KEGG) (Ogata et al. Citation1998).
Tomato fruit yield parameters
Ten tomato plants from WT and transgenic lines have been selected randomly at red ripening stage to determine the next fruit growth parameters: number of fruits per plant, average fruit size (cm), weight (g), and fruit yield per plant (kg).
Cell number and cell size of tomato fruits have been also evaluated. Three cross-sections of the mesoderm per fruit have been used for cytological evaluation, according to the method described previously by Cheniclet et al. (Citation2005).
Main phytohormones (auxin, cytokinins, gibberellins, and abscisic acid) related with fruit growth and development have been analyzed following the procedure described elsewhere (Durgbanshi et al. Citation2005), with slight modifications. Briefly, frozen fresh tissue (0, 2 g) has been spiked with 100 ng of d6-ABA, 100 ng of dihydrojasmonic acid, and 100 ng of d6-SA, and homogenized with 5 mL of distilled water. After centrifugation at 4000×g at 4°C, supernatants have been recovered and pH adjusted to 3 with 30% acetic acid. The acidified water extract has been partitioned twice against 3 mL of diethyl ether. The organic upper layer has been recovered and vacuum evaporated in a centrifuge concentrator (Speed Vac, Jouan, Saint Herblain Cedex, France). The dry residue has been then resuspended in a 10% MeOH solution by gentle sonication. The resulting solution has been passed through 0.22 μm regenerated cellulose membrane syringe filters (Albet S.A., Barcelona, Spain) and directly injected into a UPLC system (Acquity SDS, Waters Corp., Milford, MA, USA). Analyses have been separated by reversed-phase (Nucleodur C18, 1.8 μm 50 × 2.0 mm, Macherey-Nagel, Barcelona, Spain) using a linear gradient of ultrapure H2O (A) and MeOH (B) (both supplemented with 0.01% acetic acid) at a flow rate of 300 μl min−1. The gradient used was as follows: (0–2 min) 90:10 (A:B), (2–6 min) 10:90 (A:B), and (2–6–7 min) 90:10 (A:B). Hormones have been quantified with a Quattro LC triple quadruple mass spectrometer (Micromass, Manchester, UK) connected online to the output of the column through an orthogonal Z-spray electrospray ion source. Quantitation of auxins (Indole Acetic Acid, IAA), cytokinins (trans-Zeatin, tZ), gibberellins (Gibberellic acid, GA3), the ethylene precursor ACC, and the abscisic acid (ABA) have been achieved by external calibration with standards of known amount.
Tomato fruit quality and ripening parameters
Fruits of WT and transgenic tomato have been harvested at red ripening stage, then surface-washed in large volumes of water containing 2.5 mL commercial bleach per liter, rinsed with tap water and finally, surface-dried and sorted. Fruits with any abnormalities or damage have been discarded.
The next organoleptic parameters: fruit dry matter (%), titratable acidity, and total soluble solids (TSS, %) have been determined according to AOAC procedures (Citation2003). Fruit firmness has been determined using fruit firmness tester (GY-2, China). The maximum force recorded at 10 mm of compression has been used as an estimation of the fruit firmness from the averaged value of at least three tested fruits with a minimum of three compressions per fruit. The significance has been calculated by unpaired t-test.
Fruits stored at room temperature have been used for shelf-live determination. Fruits have been assessed for deterioration on a five-point scale twice per week. At the first day, all fruits had a score of 0. A fruit deterioration index of 1.0 has been taken as the limit of marketable quality.
Fruit dehydration analysis has been performed using 10 fruits that were harvested at the breaker-ripening stage. The fruits have been kept at room temperature for 30, 40, and 50 days. The fresh weights have been recorded every 10 days. The fresh weight of the fruit at the breaker ripening stage has been recorded as the starting point. The water loss has been calculated as the percentage difference in weight between the starting weight and each subsequent measurement.
Ethylene, a well-known phytohormone related with fruit ripening, content has been also determined from 15 tomato fruits from each WT, three SlMX1 OE, and two RNAi lines at breaker stage. Fruits have been dipped in 0.1 mL aqueous solution of ethephon for 10 min, and then immediately fruits have been sealed in a 2.5 l plastic container for 2 h. An Agilent 7890 series gas chromatograph (Agilent Technologies) with a flame ionization detector has been used to gauge ethylene liberate. 1 mL of gas sample has been injected into a headspace coupled to a Trace GC Ultra gas chromatography (Thermo Electron) fitted with a flame ionization detector (GC-FID) using an RT-alumina Plot column (Restek Corporation). Nitrogen has been used as the carrier gas at a flow rate of 3 mL min−1, and commercial standard mixtures of ethylene have been used for the calibration curves. Column, injector, and detector temperatures have been 34°C, 250°C, and 250°C, respectively, and this has been replicated three times for each biological sample. Ethylene production has been expressed as µl kg–1 h–1.
Tomato fruit health-promoting characteristics
Tomato fruits are well-known as important source of several natural phytochemicals or bioactive compounds such as minerals, antioxidants (phenolic compounds, vitamins, and carotenoids), and biological activities (against cancer, inflammation, obesity, hypertension, and so on). Tomato fruits have been collected at ripening stage, then tomato fruits have been cut using a sharp knife and frozen immediately (−18°C). All tomato samples have been freeze-dried, then crushed to a fine powder, and stored at −20°C.
Mineral content (Ca, Fe, Zn, P, Mg, Na, Cu, Mn, and K) has been determined by atomic absorption spectrophotometry (Mod AAnalyst 800, Perkin Elmer, CT, USA) using the corresponding standards.
Vitamin content, including retinol (A), thiamine (B1), riboflavin (B2), niacin (B3), choline (B4), pyridoxine (B6), pantothenic acid (B5), folate (B9), ascorbic acid (C), and alpha-tocopherol (E) content, have been determined as described elsewhere (Datta et al. Citation2019). All powder samples of tomato fruit have been added separately at 10% of a standard diet, with consideration of their nutritional values.
Total phenolic content and antioxidant activity have been determined as follow. Tomato fruits and leaves samples (1 g fw) finely ground in liquid nitrogen has been suspended in 20 mL of methanol and stirred for 3 h at 23°C in the dark, and then centrifuged for 15 min at 12,000×g. Total phenolic content has been determined using Folin–Ciocalteau reagent, and then 2 mL saturated sodium carbonate solution (about 75 g L−1) has been added into the reaction mixture, as described earlier by Shahzad, Harlina, et al. (Citation2021). Optical density has been measured at 765 nm after a 2 h incubation at 25°C. Gallic acid has been used as a reference standard, and the results have been expressed in gallic acid equivalents (mg 100 g−1 fw). Individual phenolic compounds have been quantified using the HPLC system as described above, and the UV detector has been set at 280 nm. The elution gradient was as follows: solvent A (water and acetic acid; 99:1) and solvent B (methanol), starting at 100% A, going to 60% A in 25 min, maintaining this proportion for two more minutes, then to 95% A in 37 min, and maintaining this proportion for 5 min and a re-equilibration time of 3 min. Flow rate has been set as 0.9 mL min−1. Analysis and identification of phenolic compounds have been performed by comparison of their retention time and UV spectra with those from standards p-coumaric, caffeic, chlorogenic, quercetin, kaempferol, naringerin, cis-resveratrol, and trans-resveratrol (Sigma–Aldrich), as described in a previous study (Shahzad, Ewas, et al. Citation2021). The results have been represented as mg 100 g−1fw. Concentration range, regression equation, linear range, correlation coefficient (R2), limit of detection (LOD), and limit of quantification (LOQ) of the HPLC analysis of the phenolic compounds quantified are presented in Supplementary Table 2.
The fruit antioxidant capacity has been determined following the trolox equivalent antioxidant capacity (TEAC) assay. Five tomato fruits from WT and transgenic lines have been harvested at 2 weeks after breaker-ripening stage. Fruits have been blended in a metal blender. After centrifugation, supernatants have been taken for TEAC measurement. The method is based on the ability of antioxidant molecules to quench the long-lived ABTS+, a blue–green chromophore with characteristic absorption at 734 nm, compared with that of Trolox, a water-soluble vitamin E analog. The addition of antioxidants to the preformed radical cation reduces it to ABTS, determining a decolorization. A stable stock solution of ABTS+ has been produced by reacting a 7 mmol L−1 aqueous solution of ABTS with 2.45 mmol L−1 potassium persulfate (final concentration) and allowing the mixture to stand in the dark at room temperature for 12–16 h before use. At the beginning of the analysis day, an ABTS+ working solution has been obtained by the dilution in ethanol of the stock solution to an absorbance of 0.70 ± 0.02 AU at 734 nm, verified by a Hewlett-Packard 8453 Diode Array spectrophotometer (HP, Waldbronn, Germany), and used as mobile phase in a flow-injection system. Results have been expressed as TEAC in mmol kg−1 DW of sample. All extracts, obtained by water, acetone, and chloroform, have been analyzed by this assay.
Tomato fruit fungal resistance
The necrotrophic pathogen B. cinerea (B05.10) was grown and collected as described by Stefanato et al. (Citation2009). WT (Ailsa Craig) and SlMX1 OE and RNAi tomatoes were harvested 14 days after breaker and surface sterilized. Intact WT and transgenic fruits were sprayed thoroughly with spores (2.5 9 105 spores mL−1) three times in the flow cabinet and kept at 20°C, in high humidity. Infection symptoms were observed at 3- and 7-day post-inoculation (dpi). To quantify B. cinerea growth using quantitative polymerase chain reaction (qPCR), 1 cm samples of infected fruit tissues were harvested 3 days after inoculation. Seeds were removed and samples were freeze-dried. Total DNA was isolated and qPCR was performed as described previously (Zhang et al. Citation2013).
Cuticle plays important roles protecting fruits of surrounding biotic (biological) and abiotic (mechanical, chemical, or physical) injuries during growth and ripening. The cuticle composition from breaking ripening fruits has been determined as follow. For wax and cutin analysis, the cuticle has been extracted from manually dissected fruit peels as described in earlier report (Hovav et al. Citation2007). Cuticular waxes have been extracted by immersing the peels (30–40 mg per sample) twice for 1–2 h into 5 mL of CHCl3 at room temperature. Both CHCl3 solutions have been combined, and n-tetracosane have been added as an internal standard. The solvent has been removed under a gentle stream of nitrogen, and the remaining wax mixture has been re-dissolved in 200 mL of CHCl3 and stored at 4°C until use. After the wax extraction, the remaining peels have been depolymerized by the addition of 2 mL of BF3/methanol for 3 h at 70°C, and n-tetracosane has been added as an internal standard. Then, after the addition of 2 mL of water, the cutin mixture has been extracted three times with diethylether. The solvent of the combined extracts has been removed under nitrogen, and the remaining cutin mixture has been re-dissolved in 1,000 mL of CHCl3 and stored at 4°C until use. Fifty microliters of the wax or cutin samples have been used for independent GC analyses. CHCl3 has been evaporated from the samples under a nitrogen stream while heating to 50°C. Then, the wax or cutin mixtures have been treated with bis-N,N-(trimethylsilyl) trifluoroacetamide (Sigma–Aldrich) in pyridine (30 min at 70°C) to transform all hydroxyl-containing compounds into the corresponding trimethylsilyl derivatives. The qualitative composition has been studied with capillary GC (5890 II; Hewlett-Packard; 30-m DB-1, 0.32-mm i.d., film thickness = 1 µm; J&W Scientific) with He carrier gas inlet pressure for a constant flow of 1.4 mL min−1 and a mass spectrometric detector (5971; Hewlett-Packard). GC has been carried out with temperature-programed injection at 50°C, 2 min at 50°C, raised by 40°C min−1 at 200°C, held for 2 min at 200°C, raised by 3C min−1 to 320°C, and held for 30 min at 320°C. The quantitative composition of the mixtures has been studied using capillary GC-FID under the same GC conditions as described above, but the H2 carrier gas inlet pressure has been programed for a constant flow of 2 mL min−1. Single compounds have been quantified against the internal standard by manual integration of peak areas.
Accession numbers
Sequence data from this article can be found in the Sol Genomics Network (SGN) under accession number Solyc02g088190 (SlMX1 cDNA) and (SlMX1 gDNA) from S. lycopersicum.
Statistical analysis
Mean and standard error have been calculated for all parameters. Data have been analyzed using student t-test. Statistical differences at α = 0.05, α = 0.01, and α = 0.005 have been denoted as single (*), double (**), and triple (***) asterisks.
Results
Tomato transcriptome reveals that SlMX1 is more than a trichome regulator
Frozen leaves from 2-month-old plant have been used to prepare the transcriptome sequencing libraries. SlMX1 OE, RNAi, and WT libraries have been separately sequenced using the Illumina high-throughput second-generation sequencing platform. Subsequently, all possible contaminants and low-quality reads have been excluded. Around, 40 million total and 39 million mapped reads have been acquired from SlMX1 OE, RNAi, and WT, respectively (). The GC percentages have been between 38 and 41% in the three libraries. The ratio of nucleotides with a quality value >30 (Q 30%) ranged from 92 to 93%. The raw read numbers also ranged 39–40 million reads, whereas the clean reads ranged between 38 and 39 million reads with percentages of 96–97% for all libraries. Finally, there has been no proportion of anonymous nucleotides (N, %) in any of the thoughtful libraries ().
Table 1. Transcriptome sequencing statistical summary of sequenced and assembled results.
To recognize the functions of novel tomato genes, differentially expressed genes (DEGs) have been elucidated using a BLASTX search versus the NR NCBI protein database content with a cut-off E-value ≥10-5 relying on sequence similarity. RNA Seq data revealed 23,442 unigenes, including 14,003 (63.71%) with uncharacterized proteins or unknown function. Moreover, out of the total obtained genes, the sequences of 3767 genes have been subjected to GO analysis to determine the main functions of the novel genes (Supplementary Figure 1). Approximately, 8643 (33.12%) obtained sequences have been submitted to the Clusters of Orthologous Groups/eukaryotic Orthologous Groups (COG/KOG) database, which indicated that the highest number of genes have been classified as ‘posttranslational modification, protein turnover and chaperones.’ Furthermore, 7635 (31.21%) sequences had matches in the KEGG database, 23,362 (97.99%) had matches in the Protein families (Pfam) database, and 22,131 (94.43%) matched to NCBI nonredundant (Nr) protein sequences (Supplementary Figure 1).
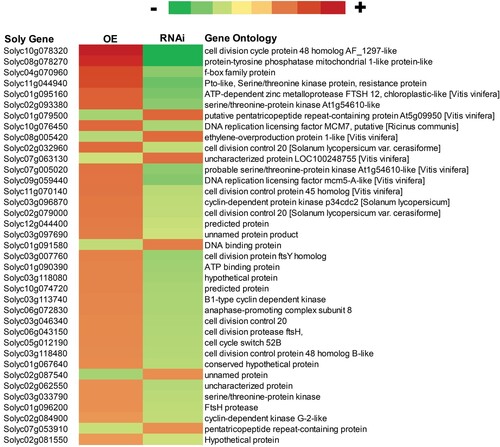
To study genome-wide modulation by SlMX1 in tomato, RNA sequencing of leaf samples from SlMX1 OE, WT, and RNAi plants has been carried out to divulge the underlying genetics. Nearly, 31 genes that involved in cell division pathway have been expressed more highly in SlMX1 OE lines than WT plants and down-regulated in RNAi lines, whereas six other genes that were involved in cell division pathway have been down-regulated in SlMX1 OE lines and up-regulated in RNAi lines compared to WT plants ().
Figure 1. Differential expression of genes involved in cell division pathway in WT, SlMX1, and RNAi tomato. Heat maps display the up-regulated (red bars) or down-regulated (green bars) tomato genes. Genes with expression ratios ≥2 and Padj <0.05 (n = 3) are depicted.
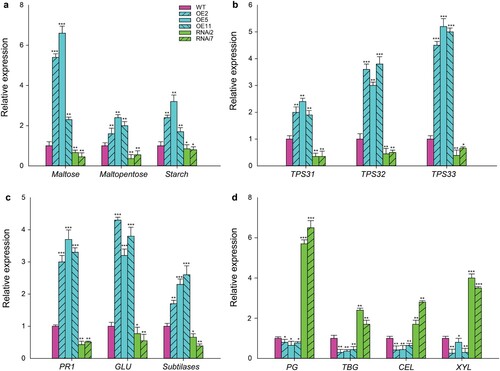
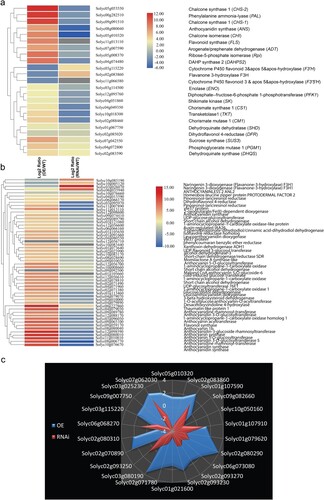
Transcriptomic analysis revealed a large number of genes which have been involved in defense, terpenoid, carbohydrate, and cell wall turnover, were up- and down-regulated in SlMX1 OE and RNAi fruit, respectively, compared to WT fruit (). The deep analysis including maltose, maltopentose, and starch known as carbohydrate biosynthetic genes ((a)). Also, TPS31, TPS32, and TPS33 known as terpenoid biosynthetic genes ((b)) and pathogenesis-related protein 1 (PR1), beta-1,3-glucanase (GLU), and a subtilisin-like protease (Subtilases) that known as defense-related genes against B. cinerea and Alternaria solani ((c)) were found. Finally, transcription level of cell wall turnover-related genes PG (polygalacturonase), TBG (tomato β-galactosidase), CEL (cellulase), and XYL (β-d-xylosidase) have been reduced and induced in SlMX1 OE and RNAi fruit, respectively compared to WT fruit ((d)).
Figure 2. Differential expression of genes involved in defense, terpenoid, carbohydrate, and cell wall turnover in tomato fruit of WT, SlMX1 OE, and RNAi lines. (a) Quantitative expression analysis of the carbohydrate biosynthetic genes. (b) Quantitative expression analysis of the terpenoid biosynthetic genes. (c) Quantitative expression analysis of the defense-related genes against Botrytis cinerea and Alternaria solani. PR1, pathogenesis-related protein 1; GLU, beta-1,3-glucanase; Subtilases, subtilisin-like protease. (d) Quantitative expression analysis of cell wall turnover-related genes. PG, polygalacturonase; TBG, tomato β-galactosidase; CEL, cellulase; XYL, β-D-xylosidase. Mean ± SEM; two biological and three technical replicates. Single (*P < 0.05), double (**P < 0.01) and triple (***P < 0.001) asterisks denote statistically significant differences between the transgenic and wild-type lines.
These findings confirmed the essential role of SlMX1 on several key metabolic pathways related with fruit growth and ripening, including fruit yield, quality, health-promoting, fungal resistance, and delay ripening processes.
Overexpression of SlMX1 improves fruit yield
The role of SlMX1 in regulating and improving the fruit growth compared transgenic and WT tomato, as can easily be observed in (a,b). To explore the cellular mechanism of fruit enlargement regulated by SlMX1, the cell number and cell size have been measured in tomato pericarp cross-sections. No significant difference has been found in the number of cell layers between all transgenic and WT lines (data not shown). Conversely, the average pericarp cell size of SlMX1 OE has been significantly increased compared to those of WT fruit while it decreased remarkably in RNAi lines ((c)).
Figure 3. Overexpression of SlMX1 regulates fruit yield in tomato. (a) Fruit-specific phenotypes of wild-type tomatoes (cv. Ailsa Craig), overexpression (SlMX1 OE) under the control of the 35S promoter, and RNAi lines. (b) Representative pictures of fruit size of WT, SlMX1 OE, and RNAi tomatoes. (c) A transverse section of the pericarp at ripening stage. Size bar is 50 μm. (d) Fruit size (cm2). (e) Fruit number per plant. (f) Fruit weight (g). (g) Fruit yield per plant (kg). Data are mean values ± SD; n = 20 (d, e, f and g). Student’s t-test has been performed. Single (*P < 0.05) and double (**P < 0.01) asterisks denote statistically significant differences between the transgenic and wild-type lines.
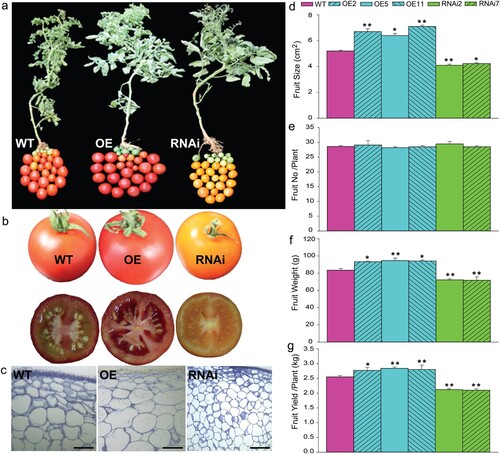
The average fruit size and fruit weight of SlMX1 OE lines have been significantly (p < 0.05) increased compared to those of WT and RNAi lines; however, no significant differences in the number of fruit per plant between transgenic and WT lines were detected ((d–f)). The fruit weight for SlMX1 OE fruits has been increased significantly in 10 units (94.61 g) compared to those of WT (83.42 g), whereas a considerable decrease has been recorded for RNAi fruits (72.24 g) ((f)). This increase in the average fruit size and fruit weight resulted in a significant gain in fruit yield per plant of SlMX1 OE (up to 2.84 kg per plant) as compared to WT (up to 2.51 kg per plant) and RNAi lines (up to 2.12 kg per plant) ((g)).
Some phytohormones play a key role promoting cell and organ division and elongation during growth stages, particularly auxins, cytokinins, and gibberellins, whereas other promote fruit ripening, senescence, and abscission, such as ethylene and abscisic. Therefore, targeted phytohormones profiling analysis has been carried out in SlMX1 OE, RNAi, and WT growing fruits to explore the metabolic effect of SlMX1 gene on the fruit sink activity. The results showed that concentrations of the active cytokinin tZ, the auxin IAA, and the gibberellin GA3 have been significantly increased (up to 25.2, 783, and 96 ng g−1 FW, respectively), in the fruits of SlMX1 OE lines with respect to WT (up to 17.1, 592, and 64 ng g−1 FW, respectively), whereas they decreased sharply (up to 14.2, 209, and 43 ng g−1 FW, respectively), in the fruits of RNAi lines compared to WT plant ((a–c)).
Figure 4. Phytohormone contents in tomato fruits of WT, SlMX1 and RNAi lines at breaking-ripening stage. (a) The active cytokinin tZ; (b) The auxin IAA; (c) The gibberellin GA3; (d) The ethylene precursor ACC; (e) The abscisic acid ABA. The data shown are the mean ± SE (n = 3). Single (*P < 0.05) and double (**P < 0.01) asterisks denote statistically significant differences between the transgenic and wild-type lines.
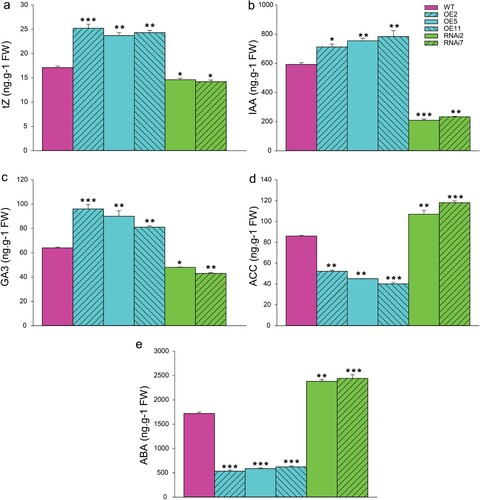
In contrast, ripening-associated phytohormones, such as the ethylene (the precursor ACC has been measured) and the abscisic acid ABA have been decreased significantly (up to 40.1 and 533.6 ng g−1 FW, respectively), in the fruits of SlMX1 OE lines compared to those of WT (up to 86 and 1,720 ng g−1 FW), whereas a significant increase (118 and 2,442 ng g−1 FW, respectively) has been noted in the fruits of RNAi lines ((d,e)).
Overexpression of SlMX1 enhances quality and delay ripening of fruits
The effect of SlMX1 expression on the organoleptic parameters of tomato fruits can be seen in . Fruit dry matter content of SlMX1 OE lines has been significantly (p < 0.05) higher (around 6%) as compared to WT fruits (4.5%), whereas it significantly decreased in RNAi fruits (4%). On the other hand, titratable acidity of tomato fruit was not significantly influenced between all transgenic and WT lines. Finally, overexpression of SlMX1 resulted in significant increase (p < 0.05) of total soluble solids of SlMX1 OE fruits (5.4%) as compared to WT fruits (4%), whereas it significantly decreased in RNAi fruits (3.5%) ().
Table 2. Fruit quality parameters of SlMX1 OE, RNAi, and WT tomato plants.
To explore the role of SlMX1 to delay fruit ripening in tomato, other post-harvest features including fruit firmness, ethylene production, and respiration rate have been investigated. The evolution of fruit firmness determined at ripening stage showed that SlMX1 RNAi fruits underwent significantly faster softening (4.5 N/mm) than WT fruits (6 N/mm). In contrast, firmness value of SlMX1 OE fruits has been approximately 9 N/mm ((a)).
Figure 5. Overexpression of SlMX1 extends shelf-life of tomato fruit. (a) Fruit firmness of SlMX1 OE, WT, and RNAi fruits has been evaluated at the ripening stage. (b) Ethylene emission (µL h−1 kg−1) for SlMX1 OE, WT, and RNAi fruits at breaker stage. (c) Fruit shelf-life of WT, SlMX1 OE, and RNAi fruits at 25° ± 1°C in three time points after ripening stage (1, 20, and 50 d). (d) Changes in fruit respiration rate of WT, SlMX1 OE, and RNAi lines during storage. (e) Transpiration water loss (100%) of WT, SlMX1 OE, and RNAi lines. Data are the mean values ± SD of at least 10 individual fruits. The asterisks indicate statistically significant differences between transgenic and WT fruits (*P < 0.05, **P < 0.01).
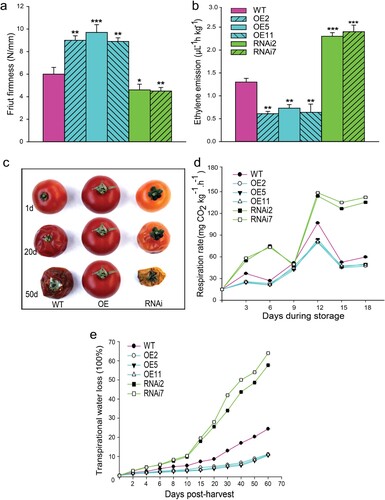
Given that fruit ripening is highly regulated by ethylene production, ethylene emission associated with fruit ripening has been assessed in SlMX1 OE, RNAi, and WT lines. The results reveal that the softening acceleration observed in RNAi fruits has been dramatically associated with the increase in climacteric ethylene emission, which has been approximately two times higher than that in WT fruit. Conversely, ethylene production of SlMX1 OE fruit markedly decreased about three times than that of WT fruit ((b)).
To demonstrate the effects of SlMX1 on shelf life, fruits from WT and transgenic lines have been harvested at ripening stage and stored at room temperature (23–25°C and 55–60% relative humidity). Ten days later, RNAi lines showed early signs of fruit deterioration, whereas WT fruits showed no signs of deterioration upto 15 days. In contrast, OE lines showed prolonged shelf life with no signs of fruit deterioration and retained their texture upto 50 days ((c)).
To gain further insight into SlMX1 effects on the two basic aspects (transpirational water loss and respiration rates) that control fruit shelf-life have been determined in storage fruits of transgenic and WT plants. Dramatic increases have been revealed in both respiration and transpiration rates in RNAi fruits compared with those of WT. However, all the SlMX1 OE fruits showed low values of both respiration and transpiration rates compared with WT fruits ((d,e)).
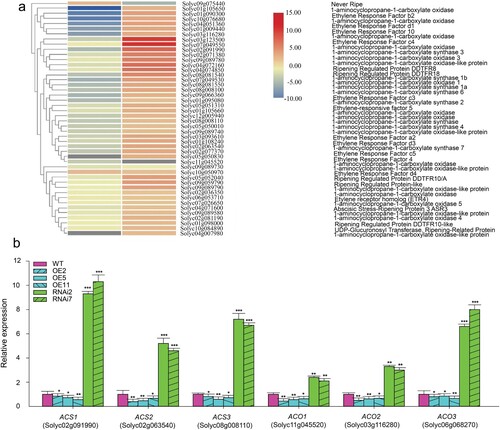
At the molecular level, two important enzymes in ethylene biosynthesis that affects the fruit’s shelf life are ACS (1-aminocyclopropane-1-carboxylate synthase) and ACO (1-aminocyclopropane-1-carboxylate oxidase). To explore the SlMX1 repression mechanism in ethylene synthesis, RNA Seq data have been analyzed (). The expression levels of DEGs involved in ethylene synthesis and signal transduction showed that eight ACS genes and 18 ACO genes have been differently down-regulated in leaves of SlMX1 OE and up-regulated in RNAi compared to WT. On the other hand, five ACO genes have been significantly up-regulated in SlMX1 OE, although ethylene synthesis has been repressed in SlMX1 OE fruits ((a)). To verify this hypothesis, the expression levels of three ACS genes (Solyc02g091990, Solyc02g063540, and Solyc08g008110) and three ACO genes (Solyc11g045520, Solyc03g116280, and Solyc06g068270) have been estimated in WT and transgenic fruits at both breaker and ripening stages. No significant differences (p < 0.05) have been found between WT and transgenic fruits at breaker stage ((b)). Conversely, at the ripening stage, the expression levels of ACS and ACO genes have been differently down-regulated in SlMX1 OE compared with WT fruits, whereas these genes have been significantly up-regulated in RNAi fruits compared to WT fruits ((b)).
Figure 6. Overexpression of SlMX1 modulates the ethylene biosynthesis pathway in tomato. (a) Differential expression of genes involved in ethylene synthesis and signaling pathway in tomato (Solanum lycopersicum). Heat maps display the up-regulated (red bars) or down-regulated (blue bars) tomato genes. Genes with expression ratios ≥2 and Padj <0.05 (n = 3) are depicted. (b) Validation of differential expression of genes involved in ethylene pathway (ACS, 1-aminocyclopropane-1-carboxylate synthase; ACO, 1-aminocyclopropane-1-carboxylate oxidase) in WT, SlMX1 OE and RNAi lines. The data shown are the mean ± SE (n = 3). Single (*P < 0.05) and double (**P < 0.01) asterisks denote statistically significant differences between the transgenic and wild-type lines.
Overexpression of SlMX1 stimulates health-promoting compounds
Tomato fruit is well-known by its high mineral, vitamins, and antioxidant content. In order to evaluate the effect of the overexpression of transgenic and WT on those health-promoting compounds, the corresponding phytochemical analyses have been carried out in tomato fruits at the ripening stage.
SlMX1 OE fruits showed a remarkable increase in minerals content of Ca, Fe, K, and Mg, whereas RNAi fruits exhibited low amounts as compared to WT fruits (). In contrast, Zn has been markedly decreased in SlMX1 OE and increased RNAi fruits as compared to WT fruit. Meanwhile, there have been no significant differences (p > 0.05) in P, Na, Mn, and Cu concentrations between transgenic and WT fruits ().
Table 3. Mineral content in SlMX1 OE, RNAi, and WT tomato fruits.
Also, a significative increase (P < 0.05) has been observed in the antioxidant vitamins retinol (A), Riboflavin (B2), ascorbic acid (C), and α-tocopherol (E) contents of SlMX1 OE fruits up to (2.3, 2, 1.3, and 2.9-folds, respectively) higher than WT. While a significant decrease has been recorded in RNAi fruits (1.8, 1.75, 1.9, and 2-folds, respectively) lower than WT fruits ().
Table 4. Vitamin content in SlMX1 OE, RNAi, and WT tomato fruits.
To explore the effects of SlMX1 on antioxidant phytochemicals, quantification of total polyphenolic and individual phenolic acids contents in methanolic extracts of SlMX1 OE, WT, and RNAi leaves and fruits have been analyzed using gallic acid equivalent. Total polyphenolic content of SlMX1 OE leaves and fruits have been higher (1.84 and 0.74 mg g−1 FW, respectively) as compared to WT leaves and fruits (0.24 and 0.18 mg g−1 FW, respectively), whereas itsignificantly decreased in RNAi fruits (0.10 and 0.05 mg g−1 FW, respectively) ((a)).
Figure 7. Overexpression of SlMX1 enhanced accumulation of polyphenolic compounds (a) Quantification of total polyphenolic contents in methanolic extracts of SlMX1 OE, WT, and RNAi leaves and fruits using gallic acid equivalent. (b) Quantification of total flavonoid content in methanolic extracts of SlMX1 OE, WT, and RNAi fruit using quercetin equivalent. The data shown are the mean ± SE (n = 3). Single (*P < 0.05), double (**P < 0.01), and triple (***P < 0.001) asterisks denote statistically significant differences between the transgenic and wild-type lines.
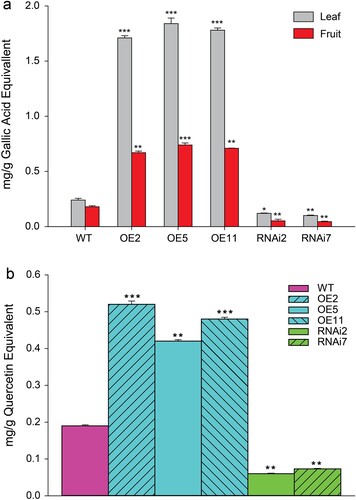
SlMX1 has been identified also as a TF that specifically activates carotenoids accumulation in tomato in previous studies. To determine whether SlMX1 could also activate flavonoids synthesis, quantification of total flavonoid content in methanolic extracts of SlMX1 OE, WT, and RNAi fruit have been carried out using quercetin equivalent. Total flavonoid of SlMX1 OE fruit has been higher (0.52 mg g−1 FW) as compared to WT fruit (0.2 mg g−1 FW). Conversely, a marked decrease in total flavonoids has been observed in the fruit of SlMX1 RNAi lines (0.06 mg g−1 FW) compared to WT fruit ((b)).
Consistently, the content of individual phenolic acids (caffeic acid, p-coumaric acid, ferulic acid, and sum of all identified phenolic acids) has been significantly increased in SlMX1 OE leaves (53.2, 16.4, 5.7, and 72.6 mg 100 g−1 FW) respectively, as compared to WT leaves (29.2, 9.3, 2.8, and 30.2 mg 100 g−1 FW, respectively), wheras RNAi leaves showed very low content of all individual phenolic acids (12.2, 3.4, 0.9, 11.7 mg 100 g−1 FW) as compared to WT leaves ((a)). Similar to leaf tissue, a significant enhancement in individual phenolic acid content has also been observed in SlMX1 OE fruit tissue while it is significantly reduced in RNAi lines as compared to WT fruit ((b)).
Figure 8. Overexpression of SlMX1 modulates the phenylpropanoid compounds in tomato. (a) Quantification of leaf phenolic acids (mg/100 g) in WT, SlMX1 and RNAi lines. (b) Quantification of fruit phenolic acids (mg/100 g) in WT, SlMX1, and RNAi lines. (c) Quantification of flavonoid species content (mg/g Fw) in the methanolic extracts of tomato fruits of WT, SlMX1 OE, and RNAi lines. (d) Quantification of anthocyanin (A535/100 mg Fw) in the methanolic extracts of tomato fruits of WT, SlMX1 OE, and RNAi lines. (e) Measurement of antioxidant activity TEAC (mmol/kg–1 DW) in the methanolic extracts of tomato fruits of WT, SlMX1 OE, and RNAi lines. (f) Quantitative expression analysis of the structural genes of phenylpropanoid pathway/flavonoid pathway in fruit of the WT, SlMX1 OE, and RNAi lines at breaker ripening stage. GT, Glucosyltransferase; PAL, Phenylalanine amonia lyase; 4CL, 4-coumaroyl CoA ligase; CHS, Chalcone synthase; CHI, Chalcone isomerase; F3H, Flavonone-3-hydroxylase; DFR, Dihydroflavonol 4-reductase; ANS, Anthocyanidine synthase. The data shown are the mean ± SE (n = 3). Single (*P < 0.05), double (**P < 0.01) and (**P < 0.001) asterisks denote statistically significant differences between the transgenic and wild-type lines.
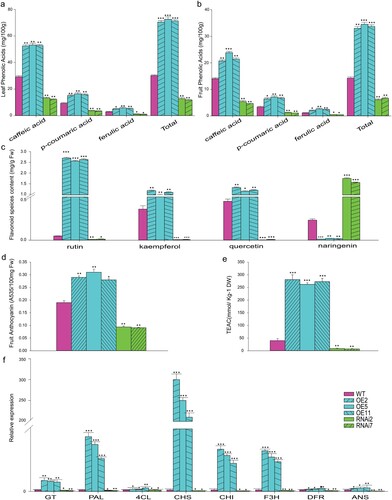
Analysis of the different flavonoid species revealed elevated levels of major flavonoids including rutin, kaempferol, and quercetin in SlMX1 OE fruits (2.7, 1.2, and 1.3 mg g−1 FW) respectively, which have been about 73, 42, and 37-fold higher as compared to WT fruit and the marked reduction of all of them in RNAi fruits. Surprisingly, the naringenin content in RNAi fruit has been significantly increased (1.75 mg g−1 FW) compared to WT (0.18 mg g−1 FW), whereas it is strongly decreased in SlMX1 OE fruit (0.01 ± mg g−1 FW) as compared to WT fruit ((c)). This result could explain the yellowish color of RNAi fruit ((a,b)).
Spectrophotometric analysis of total anthocyanin confirmed a significant increase in anthocyanin accumulation in SlMX1 OE fruit approximately 1.5 fold compared to WT fruit, whereas opposite result has been noted with RNAi fruit that recorded reduction in anthocyanin content >50% as compared to WT fruit ((d)). These results also explain the cherry color of SlMX1 OE fruit ((a,b)).
The total antioxidant capacity was determined as trolox (TEAC) equivalents. The results demonstrated that the total antioxidant capacity of SlMX1 OE fruit has been enhanced significantly (p < 0.005) by 7-fold ((e)). Conversely, a marked decrease in total antioxidant capacity has been observed in RNAi fruit up to 5-fold compared to WT fruit ((e)).
To study how SlMX1 modulates the phenylpropanoid pathway, expression profile of phenylpropanoid pathway genes encoding GT, PAL, 4CL, CHS, CHI, F3H, DFR, and ANS has been examined at ripening stage by a quantitative real-time PCR. In SlMX1 OE fruit, more than 150-fold induction of the genes encoding PAL and CHS, between 50- and 100-fold induction of genes encoding GT, CHI, and ANS, between 10- and 50-fold induction of genes encoding F3H and 4CL have been observed relative to WT fruit ((f)). However, there has been no significant difference in the relative expression level of the gene encoding for DFR in transgenic and WT fruit ((f)).
All these results have been proved by transcriptomic data from RNA Seq analysis, which showed significant induction in most flavonoid and anthocyanin biosynthetic genes in SlMX1 OE tomato and marked reduction in RNAi plants compared with WT tomato ((a,b)). These results indicate that fruit-specific expression of SlMX1 in tomato leads to induction of all the biosynthetic genes required for the production of flavonols and their derivatives, and, in addition, those required for the synthesis of CGA and its derivatives. In a similar manner, RNA Seq data showed 22 different genes that were involved in phenylpropanoid pathway. Eighteen phenylpropanoid synthetic genes have been highly expressed in SlMX1 OE lines compared with WT plants, whereas only four phenylpropanoid synthetic genes have been up-regulated in RNAi lines compared with WT plants ((c)).
Figure 9. Differential expression of genes in leaves of tomato plants. (a) Differential expression of genes involved in flavonoid biosynthesis. (b) Differential expression of genes involved in anthocyanin biosynthesis. Heat maps display the up-regulated (red bars) or down-regulated (blue bars) tomato genes. Genes with expression ratios ≥2 and Padj <0.05 (n = 3) are depicted. (c) Validation of differential expression of genes involved in phenylpropanoid pathway in tomato leaves of WT, SlMX1 OE, and RNAi lines. The data shown are the mean ± SE (n = 3).
Overexpression of SlMX1 induces fungal resistance
The susceptibility/resistance of transgenic and WT tomato fruit to the necrotrophic gray mold fungus (B. cinerea Pers.) has been examined with the aim to test whether there was any correlation with the increase in anthocyanins in SlMX1 OE fruits. At 3- and 7-day post-inoculation (dpi), the proportion of SlMX1 OE fruits showed symptoms of significant infection smaller than that in WT and RNAi fruits ((a)). In contrast with SlMX1 OE fruits, which showed more resistance to the necrotrophic pathogen B. cinerea, RNAi fruits showed a significant increase in the lesion area caused by this fungus compared to those in WT fruits ((a)). The results of quantitative PCR with DNA extracted from infected tomatoes fruits confirmed significant increase in B. cinerea growth rate on RNAi fruits than on WT fruits at 3 dpi, whereas a significant reduction in fungus growth rate on SlMX1 OE fruit compared to WT fruit at 3 dpi ((b)).
Figure 10. Overexpression of SlMX1 extends shelf-life of tomato fruit. (a) Accumulation of anthocyanins in SlMX1 OE tomatoes (Solanum lycopersicum) reduces susceptibility to Botrytis cinerea. Symptoms of sprayed tomatoes fruits of WT, SlMX1 OE, and RNAi lines after inoculation with B. cinerea B05.10. (b) Quantitative PCR revealed more B. cinerea growing on the regions of RNAi fruits than regions on WT fruits, whereas few B. cinerea growing on the regions of SlMX1 OE fruits compared with those on WT fruits at 3- and 7-day post-inoculation (dpi). B. cinerea growth has been calculated by comparing the ratio of B. cinerea DNA to tomato DNA. Error bars + standard error of the mean (SEM) (n ≥ 3). *P < 0.05; **P < 0.01, compared with control WT regions under natural light at mature ripening stage. (c) Overexpression of SlMX1 delays fruit ripening in tomato. Average initiation time of ripening in WT, SlMX1 OE, and RNAi lines. (d) Ripening speed (dpbr, days post breaker to the ripening) in WT, SlMX1 OE, and RNAi lines. Values are mean values AE SD of three biological repetitions. Single (*P < 0.05) and double (**P < 0.01) asterisks denote statistically significant differences between the transgenic and wild-type lines. (e) Fruit-ripening phenotypes, the time course of tomato fruit ripening, (br + d) in WT, SlMX1 OE, and RNAi lines. Data are the mean values ± SD of at least 10 individual fruits. The asterisks indicate statistically significant differences between transgenic and WT fruits (*P < 0.05, **P < 0.01).
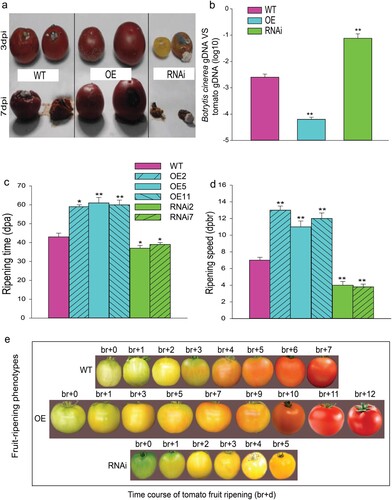
Dramatic changes have been recorded considering initiation of ripening and speed of ripening in fruits of SlMX1 OE and RNAi lines compared with WT fruits. The onset of ripening occurred with more than 2 weeks delay in SlMX1 OE lines (60 dpa) compared to WT (43 dpa), whereas RNAi lines exhibited acceleration in fruit ripening (38 dpa) compared to WT ((c)). Furthermore, once ripening process began at breaker stage, the color of SlMX1 OE fruits changed to cherry color very slowly (br + 12) compared to WT which reached to red-ripe stage 7 days post-br (br + 7). Conversely, RNAi fruits reached to its yellowish-ripe stage rapidly (br + 5) compared to WT fruits ((d,e)).
At fruit-ripening stage, the Sudan Red staining and SEM results showed greater cuticle deposition in peel of SlMX1 OE fruit compared to WT fruit, whereas the opposite has been observed for the fruit of SlMX1 RNAi plants (Ewas et al. Citation2016). To test the hypothesis that the cuticle thickness variations have been caused by differences in cuticular components, which could affect tomato yield, the chemical analysis of cuticular components has been carried out for transgenic and WT fruits in breaking-ripening stage. A significative increase (p < 0.01) in the total wax amount has been recorded in SlMX1 OE fruit (upto 1500 µg dm−2) compared with those of WT fruit (upto 1030 µg dm−2), whereas obvious reduction has been seen in the total wax amount of RNAi fruit (upto 750 µg dm−2).
In the similar manner, a significant increase in individual composition of wax including (fatty acids, aldehydes, alkan-1-ols, alken-1-ols, alkenes, alkanes, iso-alkanes, anteiso-alkanes and triterpenoids/sterols) of SlMX1 OE fruit at breaking-ripening stage (upto 66, 31, 103, 309, 83, 694, 38, 7.2, and 532, respectively) compared with those of WT fruit (37, 25, 52, 47, 102, 521, 28, 5.7, and 351, respectively) ((a)). While significant decrease has been recorded in most of individual composition of wax (upto 19, 28, 25, 51, 118, 272, 16, 4.5, and 138, respectively) of RNAi fruit compared with WT fruits ((a)). In the same context, at breaking ripening stage, overexpression of SlMX1 results in a significant increase in cutin monomer acid composition including (hexadecanoic acid, 16-OH hexadecanoic, 9(10),16-diOH hexadecanoica, hexadecane-1,16-dioic, octadecanoic acid, 18-OH octadecanoic, 9,18-diOH octadecanoic, 9,10,18-triOH octadecanoic, 9,10,18-triOH octadecenoicb, p-Coumaric acid, m-Coumaric acid, and total identified) of SlMX1 OE fruit compared with those of WT fruit, whereas a marked reduction has been reported in the cutin monomer acid composition of RNAi fruit ().
Figure 11. Overexpression of SlMX1 modified cuticle and wax in tomato fruit. (a) Fruit cuticle wax class composition of normal ripening tomato cultivar AC as wild-type, SlMX1 OE, and RNAi lines. (b) Relative expression of SlWIN1 and SlSHIN2 the positive regulators of wax and cutin formation in tomato fruit. Wax and cutin have been sampled at ripening stage. Mean values (μg dm−2) of individual wax class amounts are presented with SE (n = 3). (c) Relative expression of cell size regulators CSRs and related genes in tomato fruit. Single (*P < 0.05) and double (**P < 0.01) asterisks denote statistically significant differences between the transgenic and wild-type lines. Vertical bars represent SD, the mean values of three biologically independent experiments.
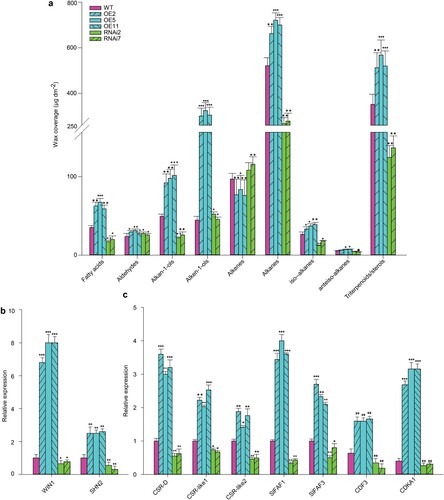
Table 5. Cutin monomer acid composition in SlMX1 OE, RNAi, and WT tomato fruits.
The transcriptomic analysis revealed that WIN1 and SHN2 (key genes in wax and cutin deposition) have been up-regulated in SlMX1 OE fruits upto 8 and 2.5 folds, respectively, higher than WT, whereas they have been down-regulated in RNAi fruits upto 1.4 and 1.6 folds, respectively lower than WT fruits ((b)). Expression analysis of cell size regulator CSR and related genes have been carried out in transgenic and WT fruit at turning stage. The transcript accumulations of CSR-D, CSR-like1, CSR-like2, SlFAF1, SlFAF3, CDKA1, and CDF3 have been significantly up-regulated in SlMX1 OE fruit compared with WT, and down-regulated in RNAi fruit ((c)).
Discussion
In spite of the numerous reports that focused on the genetic roles for improving many agro-traits, it still lacks explanation that could demonstrate improvement mechanisms of fruit quantity and quality simultaneously. In the current study, an multilevel analysis, from molecular to agronomic traits, of the relationship between gene expression, using omics technologies, with increasing fruit quality and yield, promoting healthy and organoleptic alternatives, as well as extending the fruit shelf life through manipulation of SlMX1TF in tomato is presented.
SlMX1 is a member of the MIXTA group within the MYB gene family. MIXTA is distinguished by its unique roles in regulating cell growth and development, as has been described elsewhere (Zhang et al. Citation2019). This raises several important questions about the relevant role of transcriptional regulation of cell growth, structure, and its metabolism (discussed below).
Role of SlMX1 in tomato cell growth and fruit yield
The results of current study confirmed that the overexpression of SlMX1 TF results in a significant increase in tomato yield. This increase was followed by stimulating elongation in SlMX1 OE fruit cells as shown in (c), thus significantly increasing fruit size and fruit weight ((a,b,d,f)). Although no significant increase was reported in the number of tomato fruits due to SlMX1 overexpression, the final yield was markedly increased simultaneously with the increase in the size and weight of SlMX1 OE fruits ((g)). Previous studies reported a positive correlation between fruit yield and average of both of fruit size and fruit weight of tomato (Isa et al. Citation2017).
Phytohormones are paramount factors controlling cell size that ultimately lead to different organ sizes (Nitsch et al. Citation2012). This hypothesis has been proven correct through our results related to phytohormones estimation in transgenic and WT tomato. This phenomenon confirmed a significant increase in active cytokinin tZ, auxin IAA, and gibberellin GA3 concentrations in SlMX1 OE lines, and a marked decrease in RNAi lines compared to the WT fruits ().
The era of science is switching toward the promise of next-generation RNA sequencing (NGS) technology for agricultural applications (Hu et al. Citation2018). The current study serves as a foundation and landmark in both directions by combining the advantages of NGS technology and the efficiency, accuracy, and speed of genetic engineering. The remarkable enrichment of transcriptomic data as well as the induction of expression levels related to cell division genes in SlMX1 OE lines unveils the pivotal role of SlMX1 in extending cell size and structure ultimately boosting the yield of tomato. Several genes including CSR-D, CSR-like1, CSR-like2, SlFAF1, and SlFAF3 play an important role in improving fruit size (Van der Knaap et al. Citation2014; Czerednik et al. Citation2015; Renau-Morata et al. Citation2017). In this work, the up-regulation of these genes due to SlMX1 overexpression in tomato have been observed.
Role of SlMX1 in fruit quality and ripening
A highly significant correlation was observed for the organoleptic parameters of tomato fruits and SlMX1 expression. Fruit dry matter and total soluble solids contents were increased significantly in SlMX1 OE fruits compared to WT fruits, whereas it significantly decreased in RNAi fruits ().
In addition to the aforementioned results, many previous studies indicated a close correlation between plant yield and water content of plant cells (Chavez et al. Citation2020). This correlation is attributed to the effect water as driving force for cell wall turnover, cell expansion, and enlargement (Gallego and Zarra Citation1998), and thus ends with a significant increase in fruit size and yield (Raja et al. Citation2021). Data presented in this study regarding fruit dry matter and total soluble solids exhibited an excellent model for close correlation between water content and fruit quality parameters.
Moreover, the low ethylene rate emitted from SlMX1 OE tomato fruits coincided with a delay in both the onset and the speed of fruit ripening (Ambawat et al. Citation2013) ((b); (c,d,e)). Previous studies reported the negative effects of water loss on fruit shelf-life (Gebeye Citation2018; Lara et al. Citation2019). The results in present study indicated that the values of water loss have been higher in RNAi fruits than those of SlMX1 OE lines compared to WT fruits ((e)).
Role of SlMX1 in health-promoting characteristics
Phenylpropanoids represent one of the three major classes of secondary metabolites in plant kingdom, where they function as fundamental components of several structural polymers (Sharma et al. Citation2019). Although several studies in recent decades have focused on the formation of phenylpropanoids, still the underlying molecular mechanism behind both the synthesis and regulation in cell growth and development has not been fully understood (Vogt Citation2010). Moreover, the cross-talk through the intersection between related pathways of the final yield quantity and quality along with the phenylpropanoids synthesis still not elaborated extensively and thoroughly (Vogt Citation2010; Sharma et al. Citation2019). Several studies referred to close relation between main phenylpropanoid processor and synthesis of carotenoids and terpenoids (Sharma et al. Citation2019; Chalvin et al. Citation2020). However, the molecular mechanism needs to be clarified in more detail. SlMX1 TF has been indicated as trichome regulator that directly manipulates secondary metabolism in tomato fruit including carotenoids and terpenoids (Ewas et al. Citation2016; Ewas, Gao, et al. Citation2017). Our current study revealed the intersection of phenylpropanoid synthesis mechanism with both carotenoid and terpenoid synthesis in tomato. The increase or decrease in total polyphenolic and individual phenolic acids contents due to inducing or silencing SlMX1 TF, respectively, in tomato leaves and fruits interprets the metabolic synthesis association of phenylpropanoids, and both carotenoids and terpenoids.
In parallel, several MYB TFs, including SlMYB12, VvMYB5b, MdMYB1, MdMYB10, AtMYB75, AtMYB114, AmROS1, AmROS2, SlANT1, and SlANT2 have been reported as positive regulators for secondary metabolism and cell development in tomato (Cheminant et al. Citation2011; Appelhagen et al. Citation2018; Colanero et al. Citation2019). The MYB TFs thus play a significant role in rewiring the plant metabolome by inducing transcription of the flavonoids late biosynthetic genes (LBGs) (Appelhagen et al. Citation2018). Also, it is certain that phenylpropanoid compounds are the main classes of flavonoids compounds synthesized via the flavonoid biosynthetic pathway (Cheminant et al. Citation2011). This is also confirmed through results obtained in our study which show that by increasing the synthesis of phenylpropanoid, the total and individual flavonoid contents increase in methanolic extracts of SlMX1 OE fruit compared to those of WT fruit, whereas decreased in RNAi lines ((b); (c)). In the same context, an increase in the accumulated flavonoids, which are the precursor compounds for anthocyanidins synthesis (Liu et al. Citation2018; Jian et al. Citation2019), lead to a significant increase in the anthocyanins synthesis in fruits and leaves of SlMX1 OE lines ((d)). Several phenolic compounds are classified as non-enzymatic antioxidants (Shoeva et al. Citation2016). In line with this fact, our results confirmed that improved metabolism of phenolic compounds has been reflected in a significantly enhanced antioxidant capacity of SlMX1 OE fruit while decreased in RNAi fruits ((e)).
Since snapdragon R2R3MYB TF has been identified as the first MIXTA gene, many homologs MIXTA have been characterized in several species, including arabidopsis, maize, cotton, rice, potato, and tomato (Du et al. Citation2012; Ambawat et al. Citation2013; Oshima et al. Citation2013; Yang et al. Citation2015; Ewas et al. Citation2016). Many members of the MIXTA group play diverse roles in plant species related to development, growth, and resistance as well as stimulating the biosynthesis mechanism of primary and secondary metabolites (Ewas et al. Citation2016; Ewas, Gao, et al. Citation2017). From our transcriptomic results, it is clear that SlMX1 TF plays various roles at the molecular and metabolic levels to regulate cell growth and development as well as biosynthesis. The RNA Seq data indicated that SlMX1 modulates phenylpropanoid pathway by activating most of the related biosynthetic genes. The RNA Seq data indicated that SlMX1 modulates phenylpropanoid pathway by activating most of the related biosynthetic genes ((a,b)). This modulation starts from the stimulation of primary genes responsible for general phenylpropanoids synthesis, including PAL, C4H, and 4CL, then this alteration extends to activate the expression levels of subsequent genes in flavonoids pathway, such as CHS, CHI, F3H, F3′5′H, FLS, and DFR until this modulation ends with inducing anthocyanin biosynthetic genes ‘ANS’ (). Modulation of phenylpropanoid pathway in RNA Seq data has been validated through the results of transcript analysis, which indicates that SlMX1 regulates the expression of GT, PAL, 4CL, CHS, CHI, F3H, DFR, and ANS genes in tomato ((f)). The alteration in the expression level of phenylpropanoid biosynthetic genes is correlated with alteration in the synthesis of phenolic acids, flavonoids, and anthocyanin compounds ( and ).
Role of SlMX1 in fungal resistance
The results of present study strongly confirm the positive role of SlMX1 expression in fungal resistance. SlMX1 OE fruits showed more resistance to the necrotrophic pathogen B. cinerea, whereas RNAi fruits showed a significant increase in the lesion area caused by this fungus compared to those in WT fruits ((a)). Additionally, the results of quantitative PCR with DNA extracted from infected tomato fruits confirmed a significant increase in B. cinerea growth rate on RNAi fruits than on WT fruits, whereas a significant reduction in fungus growth rate on SlMX1 OE fruit compared to WT fruit ((b)). These results can be explained based on the positive correlation between SlMX1 overexpression and the increased anthocyanins accumulation and cuticular wax biosynthesis in tomato fruits.
In general, a good balance between both primary and secondary metabolite synthesis provides a better plant growth and development as well as for adaptation to adverse environmental conditions (Ewas et al. Citation2016; Ewas, Gao, et al. Citation2017; Ewas, Khames, et al. Citation2017; Shahzad, Ewas, et al. Citation2021). The effect of cuticular wax biosynthesis on plant organs size has been reported previously (Shi et al. Citation2011; Nitsch et al. Citation2012). It can be summarized that the induction of SlMX1 results in a significant increase in wax and cutin accumulation, hence this increase in wax and cutin deposition requires an increase in both the cell expansion and cell size, which promoted a significant enhancement in fruit size, fruit weight, and consequently a marked increase in tomato yield.
In the Arabidopsis thaliana genome sequence, AtMYB16 and AtMYB106 genes are highly similar to SlMX1 (Ewas et al. Citation2016). AtMYB16 and AtMYB106 have been reported to regulate cuticle development through the induction of SHN1 and WIN1, which promote cutin and wax formation, respectively (Oshima et al. Citation2013; Ewas et al. Citation2016). Our results indicated that WIN1 and SHIN2 transcripts have been induced in SlMX1 overexpression and repressed in SlMX1-silenced lines. This regulation led to increased wax and cutin in SlMX1 OE fruit. Previous reports showed that wax and cutin affects plant development especially epidermal cell shape, cell elongation, and cell size (Shi et al. Citation2011; Cai et al. Citation2017). The significant role of SHN clade members in regulating the biosynthesis of cuticular lipids for surface formation has also been reported (Shi et al. Citation2011). This alteration in the cell structure and composition has been accompanied by a change in the cell shape and elongation of the cells (Shi et al. Citation2011).
Decrease or increase in ABA concentration through induction or reduction of SlMX1, respectively, can alter cuticular wax biosynthesis and hence affect fruit tolerance to biotic and abiotic stresses.
It has been clearly proven that the increased accumulation of cutin, wax (Bemer et al. Citation2012), flavonoids (Zhang et al. Citation2015), and anthocyanin (Zhang et al. Citation2013) compounds in the peel of tomato fruits led to a relative delay in ripening and reduced post-harvest softening and thus prolonging the shelf-life. The accumulation of cutin, wax, flavonoids, and anthocyanin in the thick skin of SlMX1 OE tomato extends the fruit shelf-life compared to WT tomato has been showed. In contrast, SlMX1 RNAi tomato with thin skin and less accumulation of cutin, wax, flavonoids, and anthocyanin had a short life compared to WT fruit (; ; ; ). Furthermore, the current study showed that the anthocyanin-enriched fruits of SlMX1 OE tomato are less susceptible to B. cinerea infection compared to WT fruit ((a,b)) and this is correlated to the higher antioxidant capacity of SlMX1 OE fruit ((e)). Contrary, a recent report revealed that inhibiting the antioxidant activities increase the B. cinerea infection (Shu et al. Citation2021).
The increased wax and cutin deposition in the thick peel of SlMX1 OE tomato fruits positively affects the reduction of water loss and thus prolonging the shelf-life of the fruits after harvesting. At molecular level, overexpression of SlMX1 TF suppresses the two most important enzymes in ethylene biosynthesis pathway, ACS (1-aminocyclopropane-1-carboxylate synthase) and ACO (1-aminocyclopropane-1-carboxylate oxidase) (Yokotani et al. Citation2009), which affect the fruit’s shelf-life of SlMX1 OE tomato plants ((c); (a,b)). Based on multi-omics analysis, this study revealed the intersection between thick tomato skin and modification of pathways related to quality and yield of tomato through regulation of TF SlMX1.
Post-harvest infection with various pathogens and fruit softening are the most important challenges so far facing by tomato producers. Extending fruit shelf-life is often at the expense of aroma, flavor, and texture as well as fruit size through the adopted biotechnological strategies (Baldwin et al. Citation2011). To our knowledge, this study is the first research shedding light on the intersection between cell size and structure with fruit ripening and shelf-life.
Conclusion
The importance of this study is summarized in understanding the new insights into tomato essential genes based on integrated multi-omics analysis. Transcriptomic analysis illustrated that SlMX1 modulates fruit metabolism of cell walls, carbohydrates, vitamins, phytohormones, phenylpropanoids, carotenoids, and terpenes, as well as boosting tomato yield, quality, health-promoting components, fungal resistance, and extended fruit shelf-life through regulation of key structural and regulatory genes of the corresponding pathways. This study paves the way toward further tomato improvement and for deciphering the mechanisms underlying myriad of other agronomic traits that can be improved with SlMX1 TF. This work can be a distinctive strategy to improve other economically important crops.
Supplemental Material
Download Zip (268.1 KB)Acknowledgements
The authors would like to thank Prof. Dr Jie Luo, Dr Lianghuan Qu, and Dr Yanqian Gao for their suggestions and technical support to the research and for advice on statistics.
Data availability statement
All the data supporting the findings of this study are available within manuscript or supplementary file.
Additional information
Funding
Notes on contributors
Mohamed Ewas
Mohamed Ewas works at Department of Genetic Resources, Desert Research Center, Cairo, Egypt. His work focuses on molecular and genetic mechanisms of stress tolerance in plants.
Putri Widyanti Harlina
Putri Widyanti Harlina is an assistant professor at Department of Food Technology, Universitas Padjadjaran, Indonesia. She focuses on lipidomics and antioxidation functions in foods.
Raheel Shahzad
Raheel Shahzad is an assistant professor at Department of Biotechnology, Universitas Muhammadiyah Bandung. His work focuses on molecular mechanisms of plant tolerance / adaptations to various environmental stresses.
Eman Khames
Eman Khames works at Department of Biochemistry, Faculty of Pharmacy, Tanta University, Tanta, Egypt.
Farhan Ali
Farhan Ali works as a researcher in Cereal Crops Research Institute (CCRI), Nowshera, Pakistan.
Elsayed Nishawy
Elsayed Nishawy works at Department of Genetic Resources, Desert Research Center, Cairo, Egypt. His work focuses on molecular and genetic mechanisms of stress tolerance in plants.
Nagwa Elsafty
Nagwa Elsafty works at Plant Genetics Resources Department, Desert Research Center, Cairo, Egypt.
Hamdy M. Ibrahim
Hamdy M. Ibrahim works at Tissue Culture Unit, Plant Genetics Resources Department, Desert Research Center, Cairo, Egypt.
Pedro P. Gallego
Pedro P. Gallego is a full professor at Department of Plant Biology and Soil Science, Faculty of Biology, University of Vigo, Vigo, Spain. He focuses on plant biotechnology and physiology of woody plants, particularly with economic importance.
References
- Ambawat S, Sharma P, Yadav NR, Yadav RC. 2013. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 19:307–321.
- Appelhagen I, Wulff-Vester AK, Wendell M, Hvoslef-Eide AK, Russell J, Oertel A, Martens S, Mock HP, Martin C, Matros A. 2018. Colour bio-factories: towards scale-up production of anthocyanins in plant cell cultures. Metab Eng. 48:218–232.
- [AOAC] Association of Official Analytical Chemists. 2003. Official methods of analysis of the Association of Official’s Analytical Chemists. Helrich K, editor. 17th ed. Arlington (VA): Association of Official Analytical Chemists.
- Baldwin EA, Plotto A, Narciso J, Bai J. 2011. Effect of 1-methylcycloprene on tomato flavor components, shelf life and decay as influenced by harvest maturity and storage temperature. J Sci Food Agric. 91:969–980.
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto Pde B, Angenent GC, de Maagd RA. 2012. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell. 24:4437–4451.
- Butelli E, Titta L, Giorgio M, Mark HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, et al. 2008. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol. 26:1301–1308.
- Cai S, Chen G, Wang Y, Huang Y, Marchant B, Wang Y, Yang Q, Dai F, Hills A, Franks PJ, et al. 2017. Evolutionary conservation of ABA signaling for stomatal closure in ferns. Plant Physiol. 174(2):732–747.
- Carneiro AK, Montessoro PF, Fusaro AF, Araujo BG, Hemerly AS. 2021. Plant CDKs-driving the cell cycle through climate change. Plants. 10(9):1804.
- Cantu D, Blanco-Ulate B, Yang L, Labavitch JM, Bennett AB, Powell ALT. 2009. Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 150:1434–1449.
- Chalvin C, Drevensek S, Dron M, Bendahmane A, Boualem A. 2020. Genetic control of glandular trichome development. Trends Plant Sci. 25(5):477–487.
- Chavez FJ, Berry PM, Foulkes MJ, Sukumaran S, Reynols MP. 2020. Identifying quantitative trait loci for lodging-associated traits in the wheat doubled-haploid population Avalon × Cadenza. Crop Sci. 61:2371–2386.
- Cheminant S, Wild M, Bouvier F, Pelletier S, Renou JP, Erhardt M, Hayes S, Terry MJ, Genschik P, Achard P. 2011. DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell. 23:1849–1860.
- Cheniclet C, Rong WY, Causse M, Frangne N, Bolling L, Carde JP, Renaudin JP. 2005. Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiol. 139:1984–1994.
- Colanero S, Tagliani A, Pierdomenico P, Gonzali S. 2019. Alternative splicing in the anthocyanin fruit gene encoding an R2R3 MYB transcription factor affects anthocyanin biosynthesis in tomato fruits. Plant Commun. 1:100006.
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21:3674–3676.
- Czerednik A, Busscher M, Angenent GC, de Maagd RA. 2015. The cell size distribution of tomato fruit can be changed by overexpression of CDKA1. Plant Biotechnol J. 13(2):259–268.
- Datta S, Sinha BK, Bhattacharjee S, Seal T. 2019. Nutritional composition, mineral content, antioxidant activity and quantitative estimation of water soluble vitamins and phenolics by RP-HPLC in some lesser used wild edible plants. Heliyon. 5(3):e01431.
- Deng Y, Lu S. 2017. Biosynthesis and regulation of phenylpropanoids in plants. Crit Rev Plant Sci. 36:257–290.
- Du H, Feng BR, Yang SS, Huang YB, Tang YX. 2012. The R2R3-MYB transcription factor gene family in maize. PLoS One. 7:e37463.
- Durgbanshi A, Arbona V, Pozo O, Miersch O, Sancho JV, Gómez-Cadenas A. 2005. Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography–electrospray tandem mass spectrometry. J Agric Food Chem. 53:8437–8442.
- Ewas M, Gao Y, Ali F, Nishawy EM, Shahzad R, Subthain H, Amar M, Martin C, Jie L. 2017. RNA-seq reveals mechanisms of SlMX1 for enhanced carotenoids and terpenoids accumulation along with stress resistance in tomato. Sci Bull. 62:476–485.
- Ewas M, Gao Y, Wang S, Liu X, Zhang H, Nishawy EME, Ali F, Shahzad R, Ziaf K, Subthain H, et al. 2016. Manipulation of tomato SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Sci Bull. 61:1413–1418.
- Ewas M, Khames E, Ziaf K, Shahzad R, Nishawy E, Ali F, Subthain H, Amar MH, Ayaad M, Ghaly O, et al. 2017. The tomato DOF daily fluctuations 1, TDDF1 acts as flowering accelerator and protector against various stresses. Sci Rep. 7:10299.
- Gallego PP, Zarra I. 1998. Cell wall autolysis during kiwifruit development. Ann Bot. 81:91–96.
- Gan L, Song M, Wang X, Yang N, Li H, Liu X, Li Y. 2022. Cytokinins are involved in regulation of tomato pericarp thickness and fruit size. Hortic Res. 9:uhab041.
- García-Pérez P, Barreal ME, Rojo-De Dios L, Cameselle-Teijeiro JF, Gallego PP. 2019. Bioactive natural products from the genus Kalanchoe as cancer chemopreventive agents: a review. In: Rahman A, editor. Studies in natural products chemistry, vol. 61. Amsterdam: Elsevier; p. 49–84. DOI:10.1016/B978-0-444-64183-0.00002-6.
- García-Pérez P, Gallego PP. 2022. Plant phenolics as dietary antioxidants: insights on their biosynthesis, sources, health-promoting effects, sustainable production, and effects on lipid oxidation. In: Bravo-Diaz C, editor. Lipid oxidation in food and biological systems. Cham: Springer; p. 405–426. DOI:10.1007/978-3-030-87222-9_18.
- García-Pérez P, Losada-Barreiro S, Bravo-Díaz C, Gallego PP. 2020. Exploring the use of Bryophyllum as natural source of bioactive compounds with antioxidant activity to prevent lipid oxidation of fish oil-in-water emulsions. Plants. 9:1012.
- García-Pérez P, Lozano-Milo E, Gallego PP, Tojo C, Losada-Barreiro S, Bravo-Díaz C. 2018. Plant antioxidants in food emulsions. In: Milani J, editor. Some new aspects of colloidal systems in foods. Rijeka: IntechOpen; p. 11–29. DOI:10.5772/intechopen.79592.
- Gebeye AH. 2018. Shelf life and quality of tomato (Lycopersicon esculentum Mill.) fruits as affected by different packaging materials. African J Food Sci. 12(2):21–27.
- Guo M, Simmons CR. 2011. Cell number counts – the fw2.2 and CNR genes and implications for controlling plant fruit and organ size. Plant Sci. 181:1–7.
- Hovav R, Chehanovsky N, Moy M, Jetter R, Schaffer AA. 2007. The identification of a gene (Cwp1), silenced during solanum evolution, which causes cuticle microfissuring and dehydration when expressed in tomato fruit. Plant J. 52:627–639.
- Hu H, Scheben A, Edwards D. 2018. Advances in integrating genomics and bioinformatics in the plant breeding pipeline. Agriculture. 8:75.
- Isa HM, Manga AA, Hussaini MA. 2017. Correlation and contribution of some growth and yield components to fruit yield of tomato (Solanum lycopersicum L.). J Dryland Agri. 3(1):94–98.
- Iwase A, Matsui K, Ohme-Takagi M. 2009. Manipulation of plant metabolic pathways by transcription factors. Plant Biotech. 26(1):29–38.
- Jian W, Cao H, Yuan S, Liu Y, Lu J, Lu W, Li N, Wang J, Zou J, Tang N, Xu C. 2019. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic Res. 6:22.
- Jian W, Zheng YX, Yu TT, Cao HH, Chen Y, Cui QY, Xu C, Li ZG. 2021. SlNAC6, A NAC transcription factor, is involved in drought stress response and reproductive process in tomato. J Plant Physiol. 264:153483.
- Lara I, Heredia A, Dominguez E. 2019. Shelf life potential and the fruit cuticle: the unexpected player. Front Plant Sci. 10:770.
- Li R, Sun S, Wang H, Wang K, Yu H, Zhou Z, Xin P, Chu J, Zhao T, Wang H, et al. 2020. FIS1 encodes a GA2-oxidase that regulates fruit firmness in tomato. Nat Commun. 11:5844.
- Liu DD, Zhou LJ, Fang MJ, Dong QL, An XH, You CX, Hao YJ. 2016. Polycomb-group protein SlMSI1 represses the expression of fruit-ripening genes to prolong shelf life in tomato. Sci Rep. 6:31806.
- Liu Y, Tikunov Y, Schouten RE, Marcelis LFM, Visser RGF, Bovy A. 2018. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: a review. Front Chem. 6:52.
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 25:402–408.
- Lozano-Milo E, García-Pérez P, Gallego PP. 2020. Narrative review of production of antioxidants and anticancer compounds from Bryophyllum spp. (Kalanchoe) using plant cell tissue culture. Longhua Chin Med. 3:18.
- Meli VS, Ghosh S, Prabha TN, Chakraborty N, Datta A. 2010. Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc Natl Acad Sci USA. 107:2413–2418.
- Nitsch L, Kohlen W, Oplaat C, Charnikhova T, Cristescu S, Michieli P, Wolters-Arts M, Bouwmeester H, Mariani C, Vriezen WH, Rieu I. 2012. ABA-deficiency results in reduced plant and fruit size in tomato. J Plant Physiol. 169:878–883.
- Ogata H, Goto S, Fujibuchi W, Kanehisa M. 1998. Computation with the KEGG pathway database. Biosystems. 47:119–128.
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M. 2013. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell. 25:1609–1624.
- Osorio S, Carneiro RT, Lytovchenko A, McQuinn R, Sorensen I, Vallarino JG, Giovannoni JJ, Fernie AR, Rose JKC. 2020. Genetic and metabolic effects of ripening mutations and vine detachment on tomato fruit quality. Plant Biotechnol J. 18(1):106–118.
- Pandey A, Misra P, Choudhary D, Yadav R, Goel R, Bhambhani S, Sanyal R, Trivedi R, Trivedi PK. 2015. AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues. Sci Rep. 5:12412.
- Raja V, Wani UM, Wani ZA, Jan N, Kottakota C, Reddy MK, Kaul T, John R. 2021. Pyramiding ascorbate–glutathione pathway in Lycopersicum esculentum confers tolerance to drought and salinity stress. Plant Cell Rep. 41:619–637.
- Romero P, Rose JKC. 2019. A relationship between tomato fruit softening, cuticle properties and water availability. Food Chem. 295:300–310.
- Renau-Morata B, Molina RV, Carrillo L, Cebolla-Cornejo J, Sanchez-Perales M, Pollmann S, Dominguez-Figueroa J, Corales AR, Flexas J, Vincente-Carbajosa J, et al. 2017. Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front Plant Sci. 8:660.
- Shahzad R, Ewas M, Harlina PW, Khan SU, Zhenyuan P, Nie X, Nishawy E. 2021. β-Sitosterol differentially regulates key metabolites for growth improvement and stress tolerance in rice plants during prolonged UV-B stress. J Genet Eng Biotechnol. 19:79.
- Shahzad R, Harlina PW, Ewas M, Zhenyuan P, Nie X, Gallego PP, Khan SU, Nishawy E, Khan AM, Jia H. 2021. Foliar applied 24-epibrassinolide alleviates salt stress in rice (Oryza sativa L.) by suppression of ABA levels and upregulation of secondary metabolites. J Plant Interact. 16:533–549.
- Sharma A, Shahzad B, Rehman A, Bhardwaj R, Landi M, Zheng B. 2019. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules. 24:2452.
- Shi JX, Malitsky S, Oliveira SD, Branigan C, Franke R, Schreiber L, Aharoni A. 2011. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 7:e1001388.
- Shoeva OY, Mock HP, Kukoeva TV, Börner A, Khlestkina EK. 2016. Regulation of the flavonoid biosynthesis pathway genes in purple and black grains of Hordeum vulgare. PLoS ONE. 11:e0163782.
- Shu P, Zhang S, Li Y, Wang X, Yao L, Sheng J, Shen L. 2021. Over-expression of SlWRKY46 in tomato plants increases susceptibility to Botrytis cinerea by modulating ROS homeostasis and SA and JA signaling pathways. Plant Physiol Biochem. 166:1–9.
- Stefanato FL, Abou-Mansour E, Buchala A, Kretschmer M, Mosbach A, Hahn M, Bochet CG, Metraux JP, Schoonbeek HJ. 2009. The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J. 58:499–510.
- Van der Knaap E, Chakrabarti M, Chu YH, Clevenger JP, Illa-Berenguer E, Huang Z, Keyhaninejad N, Mu Q, Sun L, Wang Y, Wu S. 2014. What lies beyond the eye: the molecular mechanisms regulation tomato fruit weight and shape. Fron Plant Sci. 5:227.
- Vogt T. 2010. Phenylpropanoid biosynthesis. Mol Plant. 3:2–20.
- Yang C, Gao Y, Gao S, Yu G, Xiong C, Chang J, Li H, Ye Z. 2015. Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genomics. 16:868.
- Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y. 2009. Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J Exp Bot. 60:3433–3342.
- Zhang L, Pan J, Wang G, Du H, He H, Pan J, Cai R. 2019. Cucumber CsTRY negatively regulates anthocyanin biosynthesis and trichome formation when expressed in tobacco. Front Plant Sci. 10:1232.
- Zhang N, De Stefano R, Robine M, Butelli E, Bulling K, Hill L, Rejzek M, Martin C, Schoonbeek HJ. 2015. Different reactive oxygen species scavenging properties of flavonoids determine their abilities to extend the shelf life of tomato. Plant Physol. 169:1568–1583.
- Zhang Y, Butelli E, De Stefano R, Schoonbeek HJ, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A, et al. 2013. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol. 23:1094–1100.
- Zhu MK, Hu ZL, Zhou S, Wang LL, Dong TT, Pan Y, Chen GP. 2014. Molecular characterization of six tissue-specific or stress-inducible genes of NAC transcription factor family in tomato (Solanum lycopersicum). J Plant Growth Regul. 33:730–744.
