Abstract
Lactobacillus is a genus of gram-positive bacteria of high probiotic value. The bacteria of this genus are responsible for maintaining and promoting the microbial balance of the human gut, where they along with promoting better digestion and absorption, also help in suppressing many infectious diseases. Iranian traditional yogurt (Mast) is a rich source of Lactobacillus microorganisms and their identification can be explored for their use in industrial probiotics. This study is aimed at isolation and identification of different Lactobacillus strains present in the Mast. Forty samples of Mast from eight different regions of the Iran were cultured in De Man, Rogosa and Sharpe agar (MRS) media for isolation. The isolated strains were identified by morphological and biochemical methods using gram staining, catalase, oxidase, motility and indole tests. For biochemical identification, the isolates were subjected to carbohydrate fermentation test using phenol red broth base. Overall eight strains were majorly identified based on biochemical and morphological screening which were further confirmed using 16S rDNA-based identification. Two strain of Lactobacillus acidophilus, two strains of Lactobacillus bulgaricus, two strains of Lactobacillus delbrueckii and two strains of Lactobacillus casei were identified. Lactobacillus delbrueckii subsp. Bulgaricus isolated from Bushkan showed higher bile salt resistant activities (18.92, 13.56 and 10.22%) than that of the available Lactobacillus bulgaricus strain (3.08, 1.87 and 1.44%) in 0.3, 0.5 and 1.0% bile salt. This study thus provides insights into probiotic potential of Mast which could be explored in industrial yogurt and cheese production.
Insight into Lactic Acid Bacteria profile of traditional yogurt of Iran.
The Lactic Acid Bacteria profile varies from region to region.
Proposes possible probiotics for commercial yogurt production from traditional yogurt of Iran.
Highlights
Introduction
Dietary products have nutritional value and any supplementary positive health effect to it in modern society is considered to be an added advantage. The use of probiotics in food for the development of functional foods with added advantages is a universally accepted trend (Granato et al. Citation2010). Probiotics are live microorganisms, which are introduced to the food in a specified manner and number, for their ability to induce nutritional and health benefits to the consumer. Probiotics as dietary supplements impart positive health benefits in digestive and immune system in general. In digestive system, the probiotics improve digestion of food, absorption of nutrients, reducing cholesterol levels and preventing growth of undesirable microorganisms in the digestive tract. Immune system is modulated by probiotics; it is known to control allergic inflammation (Tomaro-Duchesneau et al. Citation2014). The most common probiotics in use are lactic acid bacteria (LAB) of Lactobacillus species (Fijan Citation2014).
LAB have been used for fermentation from ages and are still prominently used in food industry for their probiotic nature. Probiotic LAB used in functional foods play an essential role in its fermentation, extending its shelf life, imparting beneficial influence on its nutritional value, and on its healthy and sensory characteristics (Abd El Gawad et al. Citation2010). Probiotic LAB have no pathogenic or toxic effect, can survive in the gastrointestinal tracts and are tolerant to biliary salts (Hasani et al. Citation2011), these properties make them the desired probiotics in food industry. Dairy products made from locally produced raw milk constitute an important part of the daily diet, with their different inherent characteristics; fermentation by LAB is a common feature among them (Abd El Gawad et al. Citation2010). This makes dairy products a rich source for LAB screening. Traditional yogurts constitute a major source for the isolation of these probiotic microorganisms (Simova et al. Citation2009; Abd El Gawad et al. Citation2010; Nishimura et al. Citation2016; Sharifi Yazdi et al. Citation2017).
Mast, is a traditional fermented milk product (yogurt), popular in rural Iran, it constitutes a main source of dairy product to a large population. Mast is made from raw cow or sheep milk, where the raw milk is subjected to spontaneous or starter fermentation to yield this traditional yogurt. Mast production starts with boiling milk and concentrating it to nearly two third of the original volume, and then on attaining the ambient temperature of 40–45 °C the inoculation is done with previous batch of mast. The result after 12 h is natural yogurt with firm consistency and cooked flavour. This method used to ferment the milk is common throughout Iran. Mast is a rich source for identifying new probiotic bacteria for industrial dairy use (Sharifi Yazdi et al. Citation2017). Considering the fixed place of Mast in Iranian diet and it being rich probiotic bacterial source, we collected samples from different regions of Iran for isolating and identifying potential probiotic Lactobacillus strain from them.
Material and methods
Sampling
A total of 40 Mast samples, five each from Ahmadi, Halileh, Bushkan, Fariab, Tang Zard, Dehrood, Khosh Ab and Khosh Makan regions of Bushehr province of Iran were collected.
Identification of lactic acid bacteria
Ringer solution was used for gradual dilution of samples before being enriched in MRS broth (Scharlau, Spain) for 24 h at 37 °C under anaerobic condition. The screening process from each sample is time-consuming process and to overcome this, we acidified (pH 2.0) the medium in order to eliminate non-probiotic and non-resistant to acid and biliary salts. Lactobacillus strains were isolated by morphologic characteristics and catalase test. The gram-positive and catalase negative colonies were subjected to acid and bile tolerance tests.
Tolerances to acid and bile
The colony-forming unit (CFU) was calculated for pure isolates dividing the number of colonies by the dilution factor. The isolates were classified in four groups: (1) resistant to acid and biliary slats (oxagall); (2) resistant to oxagall and sensitive to acid; (3) resistant to acid and sensitive to oxagall and (4) sensitive to both oxagall and acid. Only isolates falling in group four were subjected to further screening. For temperature tolerance, isolates were divided as groups of bacteria that survive temperature of 15, 45, both 15 and 45 °C and neither 15 nor 45 °C. Acid tolerance was tested in MRS medium adjusted pH to 3.0 during incubation for 2 h at 37 °C. For bile salt tolerance test, 2% of the inoculation amount of LAB isolates was inoculated into MRS-THIO medium. The MRS-THIO medium contains MRS medium and 0.2% sodium acetate. The media was divided into four concentrations, containing 0.0, 0.3, 0.5 and 1.0% oxagall. LAB isolates in MRS-THIO medium were incubated at 37 °C for 6 h, the blank medium (0.0% cattle bile salt) was used as control, for all LAB isolates bile salt tolerance activity was determined at 600 nm using formula: bile salt-containing culture medium OD600 nm/blank culture medium OD600 nm ×100%.
Identification and characterisation of LAB
For identification of LAB, the bacteria were isolated were isolated from yogurt samples by appropriate dilutions with 0.9% salt solution. The pH of the media was adjusted to 6.5 to allow growth of Lactobacilli. The plates were aerobically incubated at 37 °C for 48 h. Finally, the single colony of Lactobacillus was isolated by observing their colony morphology and identified using biochemical tests. Well isolated colonies (with cfu 2.5 × 106 cfu/mL) were picked up and transferred to MRS broth for enrichment of Lactobacillus at 37 °C which were finally subjected to molecular characterisation using 16 rDNA analysis.
Molecular identification
The molecular identification was conducted using genomic procedure of 16S rDNA (Boom et al. Citation1990). First, single colonies of isolates were prepared and then their DNA was extracted using DNA extraction kit (GeNet Bio, Daejeon, Korea). The concentration and purity of the extracted DNA were measured using Nanodrop apparatus (Thermo Fisher, Waltham, MA). Specific primers (Cinnagen, Tehran, Iran) were used for replication of extracted 16S rRNA segments. Of 0.2 μmol of each primer F: 5′ACTCCTACGGGAGGCAGCAG-3′ and R: 5′TGACGGGCGGTGTGTACAAG-3′ were used in a PCR reaction mixture containing 2.5 μL of isolated DNA, 200 μmol of dNTP Mix, 1.5 μmol of MgCl2, 2.5 μL of PCR buffer and 2.5 unit Taq DNA polymerase enzyme. Thermal cycling programme was set to include an initial denaturation at 94 °C for 3 min followed by 32 cycles; each cycle consisted of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and elongation at 72 °C for 1 min, final elongation was carried out at 72 °C for 10 min. The PCR products were analysed in 1.5% agarose gel and images were taken using Gel Documentation device under UV rays. Purification of PCR product was performed using Agarose Gel DNA Extraction Kit (Roche, Fishers, IN). The purified DNA from the gels was outsourced to South Korea for sequencing.
Evaluation of probiotic characteristics by biochemical identification
As biochemical identification, the isolates were subjected to carbohydrate fermentation test using phenol red broth base, where the LAB isolates were tested for fermentation of the following 18 sugars (Merck, Kenilworth, NJ): Amygdalin, cellobiose, esculin, fructose, galactose, glucose, lactose, maltose, mannose, salicin, sucrose, sorbitol, arabinose, gluconate, mannitol, rhamnose and ribose. L-arabinose, ribose in this test, phenol-red broth base was used as basal medium. A 1% filter-sterilised sugar solution, using 0.2 mm filter (Minisartplus, Sartorius, Göttingen, Germany), was added aseptically into autoclaved phenol-red broth base before inoculation with overnight culture of each LAB strain. The results were assessed with reference to the available LAB strain (Lactobacillus bulgaricus) control after anaerobic incubation for 5 d.
Data analysis
Data were analysed using SPSS version 20 (SPSS Inc., Chicago, IL) software package and grouping was performed. The BLAST bioinformatics platform in NCBI Bank was used for comparison and arrangement of the 16S rDNA sequences.
Result and discussion
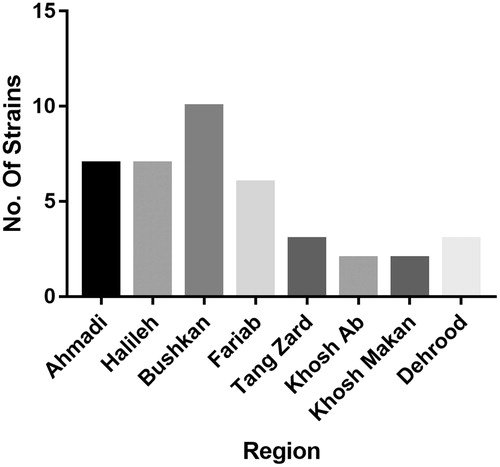
Overall 38 isolates of LAB were classified based upon morphological and biochemical screening from the 40 selected samples of Mast from the eight regions of Bushehr province in Iran (). Bushkan Mast showed maximum number of LAB isolates (n = 10). From Ahmadi and Halileh region, seven LAB isolates were found, Fariab showed six LAB isolates whereas three isolates of LAB were from each Tang Zard and Dehrood regions. Khosh Ab and Khosh Makan region least number of LABs (n = 2). These LAB isolates were subjected to acid and biliary salt (oxagall) screening, the two most common attributes used for easier elimination of non-probiotic growth from samples (Dunne et al. Citation2001). The results gave us eight strains of LAB based upon the criteria set in Bergey’s Manual of Determinative Bacteriology which were further characterised by 16S rDNA analysis.
All the detailed findings of the biochemical and morphological studies of the isolates are presented in . The results identified a total of eight gram-positive probiotic microorganisms. We targeted 16S rDNA for clonal identification as described in materials and methods. Briefly, the target 16S DNA from each LAB isolate were amplified using gene-specific primers. The amplified PCR product of these isolates was sent for sequencing. The resultant sequences were subjected to nBLAST algorithm at the NCBI website http://blast.ncbi.nlm.nih.gov/Blast.cgi for strain identification. The outputs from the BLAST searches were sorted on the basis of the maximum identity and were recorded according to their coverage. Sequence similarity with a cut-off of 90% or greater was considered significant, and the best hit was defined as the sequence with the highest maximum identity to the query sequence. A total of eight probiotic LAB isolate were observed include two strain of Lactobacillus acidophilus, two strains of Lactobacillus bulgaricus, two strains of Lactobacillus delbrueckii and two strain of Lactobacillus casei (). A plethora of studies have identified these probiotic LAB isolates from dairy products in different countries and conditions (Beukes et al. Citation2001; Mathara et al. Citation2004; Ayhan et al. Citation2005; Dorri et al. Citation2013). From Ahmadi region, Lactobacillus casei, Lactobacillus reuteri and Lactobacillus delbrueckii subsp. Bulgaricus were isolated, from Bushkan, Lactobacillus delbrueckii subsp. Bulgaricus, Lactobacillus casei and Lactobacillus acidophilus were isolated and from Fariab Lactobacillus reuteri and Lactobacillus acidophilus were isolated. Their carbohydrate fermentation test showed that they can ferment carbohydrates like Amygdalin, cellobiose, esculin, fructose, galactose, glucose, lactose, maltose, mannose, salicin, sucrose, sorbitol; however, they could not ferment arabinose, gluconate, mannitol, rhamnose and ribose.
Figure 2. Phylogenetic tree of the four native lactic acid bacteria strains found in Mast based on rRNA sequence.

Table 1. The biochemical and morphological characteristics of the isolates.
The isolated Lactobacillus acidophilus has shown the best growth under micro aerophilic condition at 37 °C for 72 h. These bacteria can growth under 45 °C but not less than 15 °C. The Lactobacillus casei shows a variant growth behaviour, when compared to Lactobacillus acidophilus and failed to survive at 45 °C. Lactobacillus delbrueckii subspecies bulgaricus grows at 45 °C but not 15 °C; and Lactobacillus reuteri could not survive 45 °C, but showed growth at 15 °C. The Lactobacillus acidophilus survived 45 °C while Lactobacillus casei shown a different behaviour and could not survive at 45 °C. These findings of variability of growth at different temperature are supported by previous studies (Vanderhoof and Young 2008).
The acid tolerance of the eight isolates at pH 3.0 decreased showed decrease in growth. shows the bile tolerance activity, of the eight isolates and of the available LAB strain (Lactobacillus bulgaricus). Strain L6 showed greater bile salt resistant activity than other strains. Strain L6 is Lactobacillus delbrueckii subsp. Bulgaricus isolated from Bushkan. For a probiotic organism, the tolerance to acid and bile are important features. Lactobacillus delbrueckii subsp. Bulgaricus isolated from Bushkan has the ability to survive in conditions mimicking gastric environment (pH 3.0 and presence of bile salts) and as a probiotic to be used in the food industry it is considered as an important feature.
Table 2. Bile salt resistant activity of Isolated LAB and reference Lactobacillus bulgaricus.
Conclusions
Our findings showed that traditional yogurt called Mast is an important source of probiotics bacteria Lactobacilli. Lactobacillus native to Iran, have probiotic characteristics, tolerate acid and resist biliary salts; therefore, are feasible strains for probiotics in industrial dairy products. Bushkan Mast showed maximum number of LAB strains (n = 8), followed by Ahmadi and Halileh region (n = 4), Fariab and Tang Zard and Dehrood regions (n = 3). Khosh Ab and Khosh Makan region least number of LABs (n = 2). Isolation and identification of these bacteria for industrial yogurt and cheese production can improve the quality of these products in the market and will have an overall effect in improving gastrointestinal health and boosting immune system. The phenotypic method and biochemical tests used in this study can be used for identification process in other test groups.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abd El Gawad I, Abd El Fatah A, Al Rubayyi K. 2010. Identification and characterization of dominant lactic acid bacteria isolated from traditional rayeb milk in Egypt. J Am Sci. 6:728–735.
- Ayhan K, Durlu ‐Özkaya F, Tunail N. 2005. Commercially important characteristics of Turkish origin domestic strains of Streptococcus thermophilus and Lactobacillus delbrueckii ssp. Bulgaricus. Int J Dairy Technol. 58:150–157.
- Beukes EM, Bester BH, Mostert JF. 2001. The microbiology of South African traditional fermented milks. Int J Food Microbiol. 63:189–197.
- Boom RC, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, Van der Noordaa JP. 1990. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 28:495–503.
- Dorri K, Namdar N, HemayatkhahJahromi V. 2013. Isolation of lactobacilli from dairy products and their effects on the main pathogenic bacteria in stomach and intestine. Med Lab J. 7:22–28.
- Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, et al. 2001. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 73:386s–392s.
- Fijan S. 2014. Microorganisms with claimed probiotic properties: an overview of recent literature. Int J Environ Res Public Health. 11:4745–4767.
- Granato D, Branco GF, Nazzaro F, Cruz AG, Faria JA. 2010. Functional foods and nondairy probiotic food development: trends, concepts, and products. Compr Rev Food Sci Food Saf. 9:292–302.
- Hasani M, Hesari J, Farajnia S, Moghadamvahed M. 2011. Technological properties of Lactobacillus species predominate in traditional cheese Lighvan. J Food Ind Res. 21:535–545.
- Mathara JM, Schillinger U, Kutima PM, Mbugua SK, Holzapfel WH. 2004. Isolation, identification and characterisation of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. Int J Food Microbiol. 94:269–278.
- Nishimura M, Ohkawara T, Tetsuka K, Kawasaki Y, Nakagawa R, Satoh H, Sato Y, Nishihira J. 2016. Effects of yogurt containing Lactobacillus plantarum HOKKAIDO on immune function and stress markers. J Tradit Complement Med. 6:275–280.
- Sharifi Yazdi MK, Davoodabadi A, Khesht Zarin HR, Tajabadi Ebrahimi M, Soltan Dallal MM. 2017. Characterisation and probiotic potential of lactic acid bacteria isolated from Iranian traditional yogurts. Ital J Anim Sci. 16:7185–188.
- Simova E, Beshkova D, Dimitrov ZP. 2009. Characterization and antimicrobial spectrum of bacteriocins produced by lactic acid bacteria isolated from traditional Bulgarian dairy products. J Appl Microbiol. 106:692–701.
- Tomaro-Duchesneau C, Jones ML, Shah D, Jain P, Saha S, Prakash S. 2014. Cholesterol assimilation by Lactobacillus probiotic bacteria: an in vitro investigation. BioMed Res Int. 2014:1.
- Vanderhoof JA, Young R. 2008. Probiotics in the United States. Clinical infectious diseases. 46: S67–S72.