Abstract
In a scenario of climate change and risk of losing biodiversity, the safeguard of locally adapted breeds and their genetic reservoirs assumes a key role. Furthermore, these breeds are linked to their rearing territory maintenance also through the nutritional, economic or social value of their products. This study focussed on the analysis of milk protein genetic variability of the Piedmontese, the most abundant beef Italian breed, and the Barà-Pustertaler breed, through the testing of individual milk samples by isoelectrofocusing. Allelic and haplotypes frequencies were analysed and the presence of rare variants (CSN1S1*G, 0.7%, and CSN2*C, 0.7%, in Barà and Piedmontese breeds, respectively) was detected. The analysed breeds showed allelic frequencies close to the one found in breeds with good cheesemaking aptitude, like the Italian Brown and Reggiana breeds (e.g. CSN2*B, 20.7% and 16.4%, and CSN3*B, 41,1% and 35%, in Piedmontese and Barà breeds, respectively), and are a good reservoir of alleles that are decreasing (e.g. CSN2*B) in Italian Holstein. Moreover, they have higher frequencies of interesting haplotypes (e.g. BA1B, 8% and 11.2%, and BA2B, 20.8 and 13.1% in Piedmontese and Barà breeds, respectively) that could positively influence composition, nutritional, and technological properties of milk. These breeds could be used to further investigate the effects of variants and haplotypes that could help support the evolution and the changing demands of the dairy sector.
In order to address the problem of climate change effects on animal biodiversity, local breeds should be safeguarded and valorised.
The analysed breeds have discrete frequencies of milk protein variants and casein haplotypes associated with good technological properties.
Knowing the genetic background of non-dairy commercial breeds is the first step to preserve potentially useful genetic variability.
Highlights
Introduction
Over the last years, climate change and its impact are hot topics of discussion for scientists and the media. To address these problems, it is necessary to maintain plants and animals biodiversity and ensure their safeguard. After several years of selection of breeds specialised in milk, meat, or both productions, the market is now oriented towards highly specialised breeds, reserving low attention to local breeds. Of the 1,224 cattle breeds contained in the Domestic Animal Diversity Information System (DAD-IS) developed by the Food and Agriculture Organisation of the United Nations (FAO), 181 are extinct (14.8%), while 245 are endangered (20%). Nowadays, it is more important than ever to know and to enhance the value of local breeds, that are sources of genetic variability (Anderson Citation2003; Ovaska and Soini Citation2017, Senczuk et al. Citation2020). These breeds have not been thoroughly studied and, consequently, are poorly selected. It is for this reason that they represent an almost untouched reservoir of genetic variability. Local breeds are adapted to different landscapes and situations in which they were historically bred, and are characterised by rusticity, thus bypassing limits met by commercial breeds. Furthermore, safeguarding local breeds allows them to maintain the link with their original rearing area. Also, the European Union (EU) supports and encourages the safeguard of local breeds through economic interventions intended for breeders, in order to maintain and enhance these breeds and populations, their traditional rearing area, and their typical productions (Gandini et al. 2010). Thus, local breeds could have better economic performances than commercial breeds thanks to the higher value and selling price given to their products (Martin-Collado et al. Citation2010).
Among the Italian local cattle breeds in need of a safeguard program there are the Barà or Pustertaler Sprinzen (BP), and the Piedmontese (PI). BP is a dual-purpose cattle breed bred in Val Pusteria, Val Badia and Valle Isarco valleys (in the Alps in Northern Italy), well known for both winter and summer tourism. PI is an autochthonous cattle breed of Piedmont region (Northern Italy), well known because of the double muscling trait that confers higher meat yield, a higher proportion of expensive cuts of lean and very tender meat (Dunner et al. Citation2003). Even though PI is the most important and abundant Italian beef breed, and thus not strictly a local breed, it is worth mentioning that it keeps a strong link with the region: indeed, the overwhelming majority of Piedmontese cattle (96%) are reared in the Piemonte region (Technical Report of the Piedmontese National Breeders Association, ANABORAPI Citation2019). Furthermore, acknowledged that PI is a beef breed, the use of its milk in derived dairy products has never been abandoned. The increasing attention to the potential link between local dairy products and uniqueness of a breed is also showing its effects on PI. The official data about milk and controlled cows during the last 10 years show that numbers of controlled Piedmontese cows raised from few heads in 2010, to 210 in 2013, up to about 700 in 2016, with the last available data of almost 500 cows (AIA Citation2019). These numbers, although limited, confirm a renewed attention of farmers about PI milk and support the importance of studying milk traits.
Milk from BP and PI is used to produce typical dairy products, but few is known about the genetic background of these two breeds in relation with milk production. Genetic variability at breed or individual level is one of the factors responsible for the changes of milk properties, technological characteristics, and final quality of milk and dairy products. Local breeds could be characterised by a higher quality milk production (e.g. locally adapted Modenese cows in comparison with Italian Holstein), as demonstrated by Mariani (Citation1985) and Summer et al. (Citation2002).
Due to the well-known effect of milk protein genetic polymorphism on milk nutritional and technological properties, aim of the present work was to characterise milk proteins polymorphisms of Barà-Pustertaler and Piedmontese breed as a first step towards a better comprehension of the real value of these local breeds as genetic resource and in terms of valorisation of the breeding areas, as well as the importance of maintenance of the link between the products and the territory.
Materials and methods
Breeds context
Pustertaler breed was derived by crossbreeding between the Austrian Pinzgau breed, and black and red pied cattle. During the 60’s, the Barà breed, already bred in Piedmont, was recognised as closely related to the Pustertaler breed. Therefore, the two breeds were indicated together as Barà-Pustertaler, a single population officially recognised and included in the registry of local cattle. BP live weight ranges between 400 and 800 kg in females and between 700 and 1,000 kg in males. BP is usually fed a diet based on local hay, pulses, cereals and by-products, to which the breed adapts due to its rusticity. The type of rearing includes grazing during the warm season, therefore breeding occurs in mountain environment. This linkage between environment, breed and milk production gives higher quality and nutraceutical effect to its products, enriched by conjugates of linoleic acid (CLA), thanks to the typical pasture rearing (Chilliard et al. Citation2007). Meat is characterised by an excellent marbling fat well distributed at muscle level, and milk is used to produce typical dairy products such as Toma cheese, butter, ricotta cheese and ‘Cevrin di Coazze’ cheese, made from a mix of Camosciata goat breed and Barà cattle breed milk. In 2018, the milk production was 3,059 kg/lactation, with a percentage fat and protein content of 3.60 and 3.35, respectively (AIA Citation2018). Piedmontese breed, autochthonous of Piedmont region (Northern Italy), was reared in the past for its triple aptitude: milk, meat and work. With the introduction of machines in agriculture, the need for work animals ended and, therefore PI was defined as a dual-purpose breed in 1958, with dairy purpose prevailing on the meat one. Starting from 1976, PI is officially recognised as a beef breed, taking advantage of a genetic modification (missense mutation C313Y, coding region of myostatin gene on chromosome 2, Dunner et al. Citation2003) that leads to muscle hypertrophy and hyperplasia improving global tenderness and final meat yield. PI live weight ranges from 520 to 550 kg for females and from 700 to 850 kg for males (Renna and Cornale Citation2011). Feeding it’s mainly characterised by fresh and conserved forages with cereals- and legumes-based concentrate. Rearing system ranges from permanent housing, typical of traditional farms, to semi-extensive or extensive systems. Although PI is a specialised breed for meat production, in the last years, some farmers keep milking PI cows and using the milk to produce typical dairy products such as ‘Bra’, ‘Castelmagno’ and ‘Raschera’ cheeses. For all these products, PI maintains a strong link with territory. In 2018, the milk production was 1,732 kg/lactation, with a percentage fat and protein content of 3.70 a 3.57, respectively (bulletin of the Italian Breeders Association - AIA).
Sampling
A total of 142 individual milk samples belonging to BP and PI breeds (70 and 72, respectively), were analysed in this study. The sampling was carried out between July and September 2019 in a total of 18 different farms (7 for BP and 11 for PI) distributed in the traditional rearing area, with the aim of obtaining a picture of the genetic variability of the two breeds in Piedmont region. Milk samples (1.5 mL each) were conferred by the Piedmontese Regional Breeders Association (ARAP), based in Cuneo, Piedmont. Samples were collected during functional controls carried out routinely by ARAP, added with Bronopol 0.02% (w/v) and stored at −18 °C until analysed. In particular, 10 and a mean of 6.5 individual samples were collected in each farm for BP and PI, respectively. Daily milk production of Piedmontese breed samples and percentages of protein and fat content for both PI and BP were also available and the average values were in accordance with the data reported by the AIA bulletins, with small deviation explained by the sampling period.
Genotyping
The isoelectrofocusing (IEF) technique was used to analyse the genetic variability of milk proteins, following the protocol described by Erhardt et al. (Citation1998). This biochemical method allows to separate macromolecules on a polyacrylamide gel with a discrimination based on their isoelectric point. IEF is applied to simultaneously display the main milk protein genetic variants of each individual milk sample, through a single analysis. For the rapid and low-cost screening of milk protein variability of breeds and populations, especially where breed characterisation is still limited, IEF is still the most effective method (Caroli et al. Citation2009), since it does not require the veterinary intervention for blood sampling, nor DNA extraction from blood or other tissues. A first analysis of the gels was made during the fixation phase with trichloroacetic acid (TCA) and confirmed after staining with Coomassie Blue G-250. As demonstrated by Caroli et al. (Citation2016), β-casein it’s more sensible to TCA fixation than other milk proteins and therefore it is possible to clearly identify β-casein variants in this early phase.
Software used for data analyses
Results of genotyping were analysed using two different software: Genepop (Rousset Citation2019) and Phase 2.1 (Stephens Citation2001). Genepop was used to calculate variant frequency and to test Hardy-Weinberg equilibrium at all loci in the two breeds. Phase 2.1 was used to estimate casein haplotypes frequencies.
Results
Allelic frequencies
Milk samples were analysed through IEF method, which allowed identifying polymorphisms at CSN1S1, CSN2, CSN3 and LGB loci, encoding respectively for αS1-, β-, and κ-casein, and β-lactoglobulin. In particular, beside the most common alleles, two rare alleles were also detected: G variant at CSN1S1 in BP and the C variant at CSN2 in PI. Allelic frequencies calculated using Genepop software are shown in . In PI, CSN1S1 is mostly represented by the B variant (85.5%). A similar scenario was found in BP, where the B variant frequency was 82.2%. The G variant was found just in one sample at the heterozygous state as shown in . Regarding CSN2, in PI the variant with higher frequency was A2 (50.0%), followed by A1, B, and C variants (28.6%, 20.7%, and 0.7%, respectively). The same situation occurred in BP, though the difference between A2 and A1 variant frequencies were not as evident as in PI (43.6% versus 40.0%); C variant in BP was not detected. CSN3 locus showed a higher frequency of A than B variant, both in PI and in BP; the E variant of CSN3 was not detected. LGB locus was similar in both PI and BP, with the B variant showing a higher frequency than A (57.0% versus 43.0% in PI, and 60.0% versus 40.0% in BP). All the loci in PI were in Hardy Weinberg equilibrium, whereas in BP CSN2 and LGB loci did not respect the equilibrium condition, maybe because of some differences in the original frequencies of the two populations that are now considered as a unique breed.
Figure 1. Results of the analysis of 8 individual milk samples by the isoelectrofocusing (IEF) technique. The dots represent the IEF patterns corresponding to the B (black), C (grey) and G (white) variants.
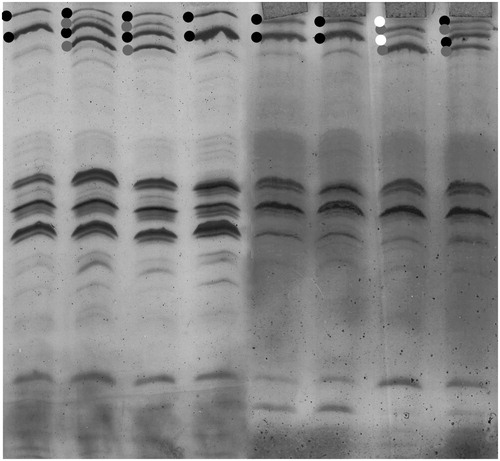
Table 1. Comparison between the allelic frequencies of the main milk protein in Piedmontese (PI), and Barà-Pustertaler (BP) breeds analysed within the present work and Italian Holstein (IH), Italian Brown (IB), and Reggiana (RE) breeds (Caroli et al. Citation2004; Chessa et al. Citation2020).
Haplotypes
Haplotypes frequencies are reported in . The total number of haplotypes detected in PI was 16 and the haplotypes combinations showing a higher frequency were BA2B (20.8%), BA2A (19.1%), BBA (16.2%), and BA1A (16.1%). These results are generally consistent with the allelic frequencies obtained with Genepop software. In fact, the greatest presence of CSN1S1*B variant on total haplotypes (present in at least 13 of 16 total haplotypes combinations), is justified by its high allelic frequency (85.5%). In the same way, the presence of A2 variant in haplotypes combinations is due to its frequency in β-casein gene (50.0%). Differently, despite the high frequency of κ-casein A variant (58.8%), the most represented haplotype was not BA2A, but BA2B. PI breed is characterised by 4 haplotypes combinations absent in BP: BCA, BCB, CCA, and CCB.
Table 2. Comparison between the casein haplotype frequencies in Piedmontese (PI), and Barà-Pustertaler (BP) breeds analysed within the present work and Italian Holstein (IH), Italian Brown (IB), and Reggiana (RE) breeds (Caroli et al. Citation2004; Chessa et al. Citation2020).
In BP, 14 haplotypes were detected and those characterised by a higher frequency were BA1A (24.2%), BA2A (18.6%), BA2B (13.1%), BA1B (11.2%), and BBA (10.5%). Although the frequencies of the alleles at the different loci were not so different between PI and BP, the haplotype frequencies were quite different. Considering only the four most frequent haplotypes in the two breeds, only three were in common and with a different order in term of frequency, except for the second haplotype (BA2A) which was the same for both breeds and showed also similar frequencies (19.1 and 18.6% in PI and BP, respectively). Obviously, being the two breeds characterised by the presence of two different rare alleles at two different casein genes, they had the relative rare haplotypes not in common (BCA, BCB, CCA and CCB in PI and GA1A and GA2A in BP, respectively).
Discussion
Allelic frequencies
The dairy sector is continuously growing, stimulated by the demand of milk, dairy products and by the need to offer products that meet the consumer demand, in terms of organoleptic and nutritional properties. Casein variants are known to be related with nutritional, nutraceutical, and technological quality of milk. In particular, β-casein presents two variants (namely A1 e A2), which are objects of debate because of their effect on human digestion and health. Specifically, A1 variant is responsible for the release of β-casomorphin-7 (BCM7), an opioid peptide (Hartwig et al. Citation1997; Jinsmaa and Yoshikawa Citation1999) considered responsible for neurological disorders and other human diseases, and that seems to be related with the slowness of the gastrointestinal tract after ingestion of milk containing this variant (Summer et al. Citation2020). Recently, Chessa et al (Citation2020), analysed the changes in casein allelic frequencies in the Holstein breed and noticed a reduction of B variant of β-casein, and the important decrease in the frequency of some haplotypes associated with good technological properties. Thus, locally adapted breeds, especially those used to produce typical dairy products, could be an important reservoir of genetic variability. In order to better understand the quality of milk production of the two breeds under analysis, we compared their milk protein allelic frequencies with those of other Italian breeds: namely Italian Holstein (IH), Italian Brown (IB), and Reggiana (RE) breeds (Caroli et al. Citation2004; Chessa et al. Citation2020). shows that, in both PI and BP, the main variant of αS1-casein was B, as expected by the fact that it is the most diffused allele in all breeds (Caroli et al. Citation2009). Anyway, in both PI and BP, the C variant was found with a frequency closer to that found in breeds with a better cheesemaking aptitude, such as IB and RE breeds (Caroli et al. Citation2004). In the BP breed the G variant was also found. The presence of this variant is associated with a reduced content of αS1-casein in milk and consequently to a greater concentration of other caseins, with possible repercussions on the cheesemaking process. In IEF, G variant migrates in the same position as B variant (Mariani et al. Citation1995; Rando et al. Citation1992), and is therefore difficult to be detected since the only difference is a lower intensity of the corresponding IEF pattern, and could be easily detected only at the heterozygous state associated with a variant migrating elsewhere, such as the C variant, as in the case reported in . Thus, to assess the correct frequency of this variant other molecular approaches should eventually be used. Moreover, due to the limitations of the IEF method used, it could be useful to verify at the molecular level also the presence of CSN2*I and to confirm the absence of CSN3*E.
As for β-casein, in PI individuals the A2 variant was present with the highest frequency, almost equal to the sum of frequencies of A1 and B variants, both carriers of BCM7. Similarly, in BP individuals the A2 variant showed a higher frequency than other variants but the difference with the frequency of A1 variant was smaller than in PI. In both breeds the B variant, associated with better milk technological properties (Comin et al. Citation2008; Chessa et al. Citation2014), was present with a discrete frequency, definitively higher than in the IH, and again, more similar to the IB and RE breeds. Analysing the κ-casein fraction, the A variant was the most represented in both breeds, but the B variant, positively associated with cheese making properties, was more frequent than in IH. Observing these results, it is possible to predict quite good technological traits from the genetic profile of the sampled animals. In particular, the milk produced by PI should have an intermediate value between IH, IB, and RE breeds in terms of consistence and formation of curd, considering the role of CSN3*B variant in dairy characteristics (Saccà et al. Citation2003), since it reached the frequency of 41.1%. The same occurs for β-LG, where the presence of B variant, the most represented in both PI and BP breeds, was described as an advantage with 2% more of cheese yield due to better coagulation and technological quality (Mariani et al. Citation1979; Chouteau and Denieul Citation1986), compared to milk characterised by A variant.
Haplotypes frequencies
A considerable number of haplotypes were found in the investigated breeds, demonstrating that they could be an interesting reservoir of genetic variability. Comparing haplotypic frequencies obtained in PI and BP breeds to those observed by previous works in IH, IB, and RE breeds (Caroli et al. Citation2004; Chessa et al. Citation2020), 8 haplotypes were found with a frequency greater than 3%, while in IH and IB only 6 haplotypes showed a frequency higher than 3% in 2004, and only 5 in IH in 2020. Considering the BA1B haplotypes and BB β/κ-casein combination, already known for their positive association with milk coagulation properties (Comin et al. Citation2008; Chessa et al. Citation2014), they occur with a greater frequency in BP (11.2% and 4.9%, respectively) and PI (8.0% and 5.3%, respectively), than in IH, where the frequency of about 7% of both combination in 2004 is now of 8.3% for BA1B and of 1.2% of the BB β/κ-casein combination, with a strong reduction of the latter (Chessa et al. Citation2020). Thus, BP and PI are placed in an intermediate position between IH/IB and RE (11.6% and 8.6%) for the frequency of haplotypes associated with good cheese making properties, and for this reason they could be considered as objects of a possible selection. Moreover, BA2B is the most represented haplotype in PI. In this breed, although A and B variants of k-casein were found in 8 different haplotype combinations, the B variant was mostly found in the BA2B haplotype, whereas the A variant was found in 3 haplotypes with similar frequencies (BA1A, BA2A e BBA). This is quite interesting, since a selection favouring k-casein B in this breed would also in parallel increase the β-casein A2 variant, which would be advantageous if confirmed that β-casein A2 variant is associated with a higher digestibility of milk than β-casein A1.
Conclusions
Livestock genetic diversity and its conservation has received wide recognition in recent years. One way to address this issue is to know and to enhance the value of local breeds, that could be a valuable reservoir of specific alleles. In this study we analysed and compared milk protein allelic and haplotypic frequencies of Piedmontese and Barà-Pustertaler Italian breeds with those of other Italian dairy commercial breeds and assessed the cheesemaking potential merit of the local ones. These breeds are characterised by allelic frequencies similar to commercial breeds and, as a bonus value, they carry interesting variants (i.e. CSN1S1*G and CSN2*C) which are rare in other breeds and that could lead to positive repercussions on cheesemaking. Furthermore, the frequencies found in this study of the haplotypes with a positive influence on the technologic characteristics of milk leave large space for genetic improvement. This work is a practical example of how the strategy of enhancing research on locally adapted breeds could preserve biodiversity and better exploit the natural resources and conserving and managing habitat diversity.
Ethical approval
The experimental method was performed in accordance with Italian Animal Protection Law (Legislative Decree No. 26/2014, https://www.gazzettaufficiale.it).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- [AIA] Associazione Italiana Allevatori. 2018. Bollettino Controlli produttività sul latte.
- [AIA] Associazione Italiana Allevatori. 2019. Bollettino Controlli produttività sul latte.
- [ANABORAPI] Associazione Nazionale Allevatori dei Bovini di Razza Piemontese. 2019. Technical report
- Anderson S. 2003. Animal genetic resources and sustainable livelihoods. Ecol Econ. 45:331–339.
- Caroli AM, Chessa S, Bolla P, Budelli E, Gandini GC. 2004. Genetic structure of milk protein polymorphisms and effects on milk production traits in a local dairy cattle. J Anim Breed Genet. 121:122–125.
- Caroli AM, Chessa S, Erhardt GJ. 2009. Invited review: milk protein polymorphisms in cattle: effect on animal breeding and human nutrition. J Dairy Sci. 92:5335–5349.
- Caroli AM, Savino S, Bulgari O, Monti E. 2016. Detecting β-casein variation in bovine milk. Molecules. 21:141–146.
- Chessa S, Bulgari O, Rizzi R, Calamari L, Bani P, Biffani S, Caroli AM. 2014. Selection for milk coagulation properties predicted by Fourier transform infrared spectroscopy in the Italian Holstein-Friesian breed. J Dairy Sci. 97:4512–4521.
- Chessa S, Gattolin S, Cremonesi P, Soglia D, Finocchiaro R, Van Kaam J-T, Marusi M, Civati G. 2020. The effect of selection on casein genetic polymorphisms and haplotypes in Italian Holstein cattle. Ital. J. Anim. Sci. 19 :833–839.
- Chilliard Y, Glasser F, Ferlay A, Bernard L, Rouel J, Doreau M. 2007. Diet, rumen biohydrogenation and nutritional quality of cow and goat milk fat. Eur J Lipid Sci Technol. 109:828–855.
- Chouteau Y, Denieul F. 1986. Influence du polymorphisme genetique de deux proteines du lait de vache (b-Lactoglobuline, k-caseine) sur sa composition et son l’aptitude fromagere. Angers: ESA; p. 1–94.
- Comin A, Cassandro M, Chessa S, Ojala M, Dal Zotto R, De Marchi M, Carnier P, Gallo L, Pagnacco G, Bittante G. 2008. Effects of composite beta- and kappa-casein genotypes on milk coagulation, quality, and yield traits in Italian Holstein cows. J Dairy Sci. 91:4022–4027.
- Domestic Animal Diversity Information System (DAD-IS) Food and Agriculture Organization of the United Nations (http://www.fao.org/dad-is)
- Dunner S, Miranda ME, Amigues Y, Cañón J, Georges M, Hanset R, Williams J, Ménissier F. 2003. Haplotype diversity of the myostatin gene among beef cattle breeds. Genet Sel Evol. 35:103–118.
- Erhardt G, Juszczak J, Panicke L, Krick-Saleck H. 1998. Genetic polymorphism of milk proteins in Polish Red Cattle: a new genetic variant of β-lactoglobulin. J Anim Breed Genet. 115:63–71.
- Gandini G, Avon L, Bohte-Wilhelmus D, Bay E, Colinet FG, Choroszy Z, Díaz C, Duclos D, Fernández J, Gengler N, et al.; the EURECA Consortium. 2010. Motives and values in farming local cattle breeds in Europe: a survey on 15 breeds. Anim Genet Resour. 47:45–58.
- Hartwig A, Teschemacher H, Lehmann W, Gauly M, Erhardt G. 1997. Influence of genetic polymorphisms in bovine milk on the occurence of bioactive peptides. Proceedings of the IDF “Milk Protein Polymorphism Seminar II”. Brussels (Belgium): International Dairy Federation; p. 459–460.
- Jinsmaa Y, Yoshikawa M. 1999. Enzymatic release of neocasomorphin and beta-casomorphin from bovine beta-casein. Peptides. 20:957–962.
- Mariani P. 1985. Observations on content and distribution of micellar constituents of milk in four cattle breeds. Ann Fac Vet Med Univ Parma. 5:173–183.
- Mariani P, Morini D, Losi G, Castagnetti GB, Fossa E, Russo V. 1979. Ripartizione delle frazioni azotate del latte in vacche caratterizzate da genotipo diverso nel locus della lattoglobulina. Scie Tecn Latt Cas. 30:153.
- Mariani P, Summer A, Anghinetti A, Senese C, Di Gregorio P, Rando A, Serventi P. 1995. Effetti dell’allele as1Cn G sulla ripartizione percentuale delle caseine as1, as2, b e k in vacche di razza. Bruna Ind Latte. 31:3–13.
- Martin-Collado D, Gandini G, de Haas Y, Dìaz C. 2010. Decision-making tools for the development of breed strategies. Local cattle breeds in Europe. EU GENRES 870/04 project EURECA. Wageningen (the Netherlands): Academic Publishers; p. 120–140.
- Ovaska U, Soini K. 2017. Local breeds – rural heritage or new market opportunities? Colliding views on the conservation and sustainable use of landraces. Sociol Rural. 57:709–729.
- Rando A, Ramunno L, Di Gregorio P, Davoli R, Masina P. 1992. A rare insertion in the bovine as1-casein gene. Anim Genet. 23:55.
- Renna M, Cornale P. 2011. A zootechnical overview of Piemontese cattle breed. 10th International Meeting on Mountain Cheese; Dronero, Italy.
- Rousset F. 2019. Package genepop- Population genetic data analysis using genepop. 2, 9–11.
- Saccà E, Bovolenta S, Venutra V, Giaiarin G. 2003. Coagulation properties and Nostrano di Primiero cheese yield of milk from Brown grazing cows of different k-casein genotype. Ital J Anim Sci. 2:284–286.
- Senczuk G, Mastrangelo S, Ciani E, Battaglini L, Cendron F, Ciampolini R, Crepaldi P, Mantovani R, Bongioni G, Pagnacco G, et al. 2020. The genetic heritage of Alpine local cattle breeds using genomic SNP data. Genet Sel Evol. 52:40.
- Stephens M, Smith N, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68:978–989.
- Summer AD, Frangia F, Ajmone Marsan P, De Noni I, Malacarne M. 2020. Occurrence, biological properties and potential effects on human health of β-casomorphin 7: current knowledge and concerns. Crit Rev Food Sci. 60:3705–3723.
- Summer A, Malacarne M, Martuzzi F, Mariani P. 2002. Structural and functional characteristics of Modenese cow milk in Parmigiano-Reggiano cheese production. Proceedings of the 53rd Annual Meeting of the European Association for Animal Production; Cairo, Egypt.
