Abstract
Intracellular antibodies (intrabodies) are recombinant antibody fragments that bind to target proteins expressed inside of the same living cell producing the antibodies. The molecules are commonly used to study the function of the target proteins (i.e., their antigens). The intrabody technology is an attractive alternative to the generation of gene-targeted knockout animals, and complements knockdown techniques such as RNAi, miRNA and small molecule inhibitors, by-passing various limitations and disadvantages of these methods. The advantages of intrabodies include very high specificity for the target, the possibility to knock down several protein isoforms by one intrabody and targeting of specific splice variants or even post-translational modifications. Different types of intrabodies must be designed to target proteins at different locations, typically either in the cytoplasm, in the nucleus or in the endoplasmic reticulum (ER). Most straightforward is the use of intrabodies retained in the ER (ER intrabodies) to knock down the function of proteins passing the ER, which disturbs the function of members of the membrane or plasma proteomes. More effort is needed to functionally knock down cytoplasmic or nuclear proteins because in this case antibodies need to provide an inhibitory effect and must be able to fold in the reducing milieu of the cytoplasm. In this review, we present a broad overview of intrabody technology, as well as applications both of ER and cytoplasmic intrabodies, which have yielded valuable insights in the biology of many targets relevant for drug development, including α-synuclein, TAU, BCR-ABL, ErbB-2, EGFR, HIV gp120, CCR5, IL-2, IL-6, β-amyloid protein and p75NTR. Strategies for the generation of intrabodies and various designs of their applications are also reviewed.
Abbreviations
| CDR | = | complementarity determining region |
| ER | = | endoplasmic reticulum |
| GFP | = | green fluorescent protein |
| IgG | = | immunoglobulin |
| G MBP | = | maltose binding protein |
| NusA | = | N utilization substance A |
Introduction
Various strategies are used to study the functions of newly discovered proteins, including gene-targeted knockout animals, targeted gene disruption in mammalian cells and knockdown techniques such as siRNA, shRNA, miRNA, CRISPR and TALEN. In addition to these methods, which are active on the nucleotide level, the use of dominant negative mutants or inhibitor molecules were the most widely used methods to interfere directly with protein function. Another method with fast growing relevance that avoids some of the problems encountered with the above technologies is the use of intracellular antibodies (intrabodies). The molecules can be selected and designed to be very specific to the target, which are advantages of the technology. Because they can be thoroughly characterized for specificity in biochemical assays before use in cells, they avoid off-target effects known from nucleotide-based methods, and they can be designed to selectively target splice variants, different isoforms or even one post-translational modification of a protein. Furthermore, the function of target proteins can be knocked down in specific cellular compartments exclusively, while their function remains intact in other cellular compartments, as recently demonstrated by the knockdown of Sec61 in endosomes while maintaining its function in the ER.Citation1 Although intrabodies are typically seen as an experimental tool to reveal the function of proteins by interfering with their function, this approach is also reported to have therapeutic potential against viral infectionsCitation2-6 brain diseasesCitation7-10 or cancer.Citation11-14 Here, the various protein knockdown strategies mediated by intrabodies () are reviewed.
Figure 1. The intrabody approach for the generation of protein interference phenotypes. Antibody fragments containing the antigen binding domains (V regions) specific for a particular protein can be selected from antibody phage display libraries or other in vitro selection systems like bacterial, yeast, mammalian or ribosomal display libraries. Alternatively, the variable region genes can be obtained from hybridoma antibodies by PCR with consensus primers, RACE or PCR with adaptor ligated cDNA. Antibody fragments, typically in scFv format, are then cloned into a specific targeting vector allowing expression of the intrabody in the nucleus, cytoplasm or ER.
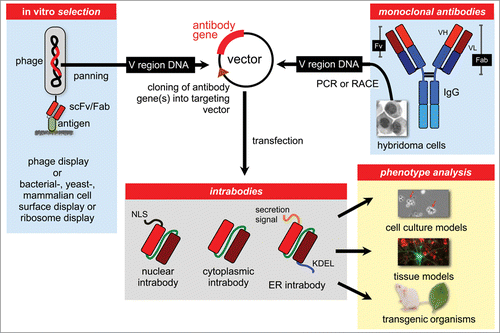
Types of Intrabodies
Intrabodies are antibodies that bind intracellularly to their antigen after being produced in the same cell,Citation15-21 in contrast to antibodies delivered to a living cell from the outside.Citation22-24 Intracellular antibodies can be subdivided into 2 main subgroups according to their mechanism of action. These are: 1) cytosolic intrabodies (cyto-intrabodies) and 2) endoplasmic reticulum (ER) retained intrabodies (ER intrabodies). While cyto-intrabodies block targets in a neutralizing fashion in the cytosol, ER intrabodies knock down proteins in the secretory pathway even without the requirement for the antibody to be neutralizing. By addition of a suitable signal peptide, intrabodies can also be targeted to the nucleus or mitochondria ().Citation15,18, 21
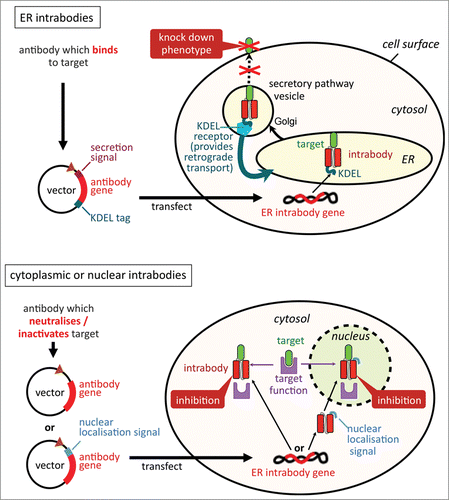
Figure 2. Differences between cytoplasmic/nuclear intrabodies and ER intrabodies. Via their retention signal KDEL, ER intrabodies (A) retain antigens passing the ER by binding to them. As antibodies are naturally produced in the ER, no particular selection for special folding/stability properties is required. In contrast, cytoplasmic/nuclear intrabodies (B) need to fold correctly in the reducing milieu of the cytoplasm. Further, they need to be tested and screened to identify antibodies which are capable, in addition to binding, to neutralise or inactivate their target's activity in the cytoplasmic biochemical milieu.
As antibodies usually are produced in an oxidizing biochemical milieu and folded with the help of ER based chaperones, only a fraction of the repertoire of naïve IgGs can be correctly folded in the reducing environment of the cytoplasm, which does not allow the formation of disulfide bridges.Citation25,26 However, there are numerous examples where intrabodies function against cytosolic targets including examples against EGFR,Citation11 BCR-ABL,Citation13 NSP5 from rotavirus,Citation27 Clostridium botulinum neurotoxin proteases,Citation28,29 core antigen of HBV,Citation6 Bax,Citation30 HIV-1 Vif,Citation2 Etk-kinaseCitation12 or hungtingtin protein.Citation7,8 It has also been shown that cyto-intrabodies can trace cellular components in living cells.Citation31,32
Various approaches have been described for the selection and generation of cyto-intrabodies: 1) use of the intracellular antibody capture technology (IACT);Citation33 2) construction of single domain intrabodies;Citation12,34-36 3) expression of intrabody fusion proteins;Citation37-42 4) complementarity-determining region (CDR) grafting or introduction of synthetic CDRs into suitable preselected frameworks;Citation43-46 and 5) selection of single-chain variable fragments (scFv) without disulfide bonds.Citation7,47 If the functional expression of a particular antibody in the cytoplasm fails, introduction of externally produced antibodies into the cytosol has been proposed using methods such as protein transfection (profection), peptides as protein transduction domains, fusion to targeting proteins or the use of translocation sequences and endosome escape domains.Citation23,48, 49 However, it has been difficult to achieve endosomal escape and to reach the cytoplasm with most of these methods.Citation24
In contrast to cyto-intrabodies, antibodies targeted to the ER are made in their native environment (), thus can be expected to be correctly folded with intact disulfide bridges forming in the oxidizing environment.Citation50 ER intrabodies work by just retaining antigens that pass the secretory pathway. Typically, these are cell-surface molecules, secreted molecules, intravesicular receptors or Golgi-located glycosyltransferases (ref. 3).
ER intrabodies are targeted to the lumen of the ER by a secretory signal peptide, and fusion of the retention sequence KDEL or SEKDEL to their C-terminus prevents their secretion together with the antigen bound to it. The KDEL receptor substrate leaves the ER, is transported to the cis-Golgi apparatus where it binds to the ERD1 and ERD2 receptors which are then recycled back to the ER via COPI-coated vesicles.Citation21,51,52 The ER intrabody-antigen complex may then be degraded via an ER-associated degradation (ERAD) pathway that is either proteasome-dependent or proteasome-independent.Citation53-55
Nuts and Bolts: How to Make Intrabodies
Intrabodies can be generated by cloning the respective cDNA from an existing hybridoma cloneCitation56,57 or more conveniently, new scFvs/Fabs can be selected from in vitro display techniques such as phage display (),Citation58,59 which provide the necessary gene encoding the antibody from the onset and allow a more detailed predesign of antibody fine specificity.Citation60 In addition bacterial-, yeast-, mammalian cell surface display and ribosome display can be employed.Citation61-64 However, the most commonly used in vitro display system for selection of specific antibodies is phage display.Citation59,60,65-67 In a procedure called panning (affinity selection), recombinant antibody phages are selected by incubation of the antibody phage repertoire with the antigen. This process is repeated several times leading to enriched antibody repertoires comprising specific antigen binders to almost any possible target. To date, in vitro assembled recombinant human antibody libraries have already yielded thousands of novel recombinant antibody fragments.Citation58,66,68,69 Antibo-dy phage repertoires can be divided into 4 different types according to the origin of their frameworks and CDRs: immune, naïve, synthetic and semi-synthetic antibody libraries.Citation70 Immune antibody libraries are constructed from antibody genes of B cells from immunized animals or infected patients by PCR, using IgG primers for the heavy and light chain. Nonimmune (also named naïve or universal) libraries are built up from natural, unimmunized, rearranged V genes (e.g., from the IgM B-cell pool.Citation71 Synthetic antibody libraries are constructed entirely in vitro using oligonucleotides that introduce areas of complete or tailored degeneracy into the CDRs of one or more V genes.Citation72,73 Semi-synthetic libraries combine natural and synthetic gene fragments encoding different parts of the scFv or Fab molecule.Citation74
Conversion of scFvs from in vitro selection methods into an ER intrabody only requires a single cloning step into the ER-targeting vector, e.g., pCMV/myc/ER to add the CMV promoter, a secretory leader, the ER retention sequence and a myc tag.Citation56 Additional reporter functions, like green fluorescent protein (GFP), can also be added by using bicistronic vector designs.Citation75
From hybridoma cells, the variable VH and VL domains can be amplified from the hybridoma cDNA using 1) PCR amplification with consensus primers,Citation57,76-78 2) rapid amplification of cDNA-ends (RACECitation79), and 3) PCR amplification using adaptor-ligated antibody cDNA.Citation80 RACE and adaptor-ligated antibody cDNA prevent mismatches that might occur if VH and VL are amplified by consensus primers. VH and VL can be assembled by overlapping PCR introducing a linker taken from phage sequences (Gly4Ser)3Citation56 or human proteins,Citation59 and the assembled scFv will be cloned into a targeting vector to transport scFvs into specific cell compartments.Citation81
The most commonly used format for intrabodies is the scFv, which consists of the H- and L-chain variable antibody domain (VH and VL) held together by a short, flexible linker sequence (frequently (Gly4Ser)3),Citation82 to avoid the need for separate expression and assembly of the 2 antibody chains of a full IgG or Fab molecule.Citation83,84 Alternatively, the Fab format comprising additionally the C1 domain of the heavy chain and the constant region of the light chain has been used. Recently, a new possible format for intrabodies, the scFab, has been described.Citation85 The scFab format promises easier subcloning of available Fab genes into the intracellular expression vector, but it remains to be seen whether this provides any advantage over the well-established scFv format. In addition to scFv and Fab, bispecific formats (for review, see ref. Citation86) have been used as intrabodies. A bispecific Tie-2 x VEGFR-2 antibody targeted to the ER demonstrated an extended half-life compared to the monospecific antibody counterparts.Citation87,88 A bispecific transmembrane intrabody has been developed as a special format to simultaneously recognize intra- and extracellular epitopes of the epidermal growth factor,Citation89 combining the distinct features of the related monospecific antibodies, i.e., inhibition of autophosphorylation and ligand binding. Another intrabody format particularly suitable for cytoplasmic expression are single domain antibodies derived from camelsCitation36 or consisting of one human VH domainCitation34,90 or human VL domain.Citation7,8,12,91 These single domain antibodies often have advantageous properties, e.g., high stability; good solubility; ease of library cloning and selection; high expression yield in E.coli and yeast.
The intrabody gene is expressed inside the target cell after transfection with an expression plasmid or viral transduction with a recombinant virus. With respect to the vector technologies used to supply the antibody mRNA to the target cell, there seem to be few limitations because, in principle, all the usually functional vector-, promoter- and transfection systems for heterologous expression could be employed. Typically, the choice is aimed at providing optimal intrabody transfection and production levels. Microinjection of hybridoma derived or in vitro transcribed mRNA was used in initial proof of principle studies,Citation92-94 but, due to more delicate RNA handling and very small number of transfected cells, this approach has not been used frequently.
Successful transfection and subsequent intrabody production can be analyzed by immunoblot detection of the produced antibody, but, for the evaluation of correct intrabody/antigen-interaction, co-immunoprecipitation from HEK 293 cell extracts transiently cotransfected with the corresponding antigen and intrabody expression plasmids may be used, or the antigen intrabody complex can be confirmed to be localized in the ER by co-staining with the ER marker calnexin.Citation4,5
Examples for Functional Studies with Cytoplasmic Intrabodies
A quite successful group of cytoplasmic intrabodies has been developed against proteins playing an important role in the brain and for Alzheimer's and Parkinson's disease. Using the intracellular antibody capture technology, antibodies against the microtubule-associated protein TAU found in neurofibrillary lesions of Alzheimer's disease brains has been selected.Citation33 This large panel of anti-TAU intrabodies selected from a naïve human antibody library provides a useful tool to study TAU function in degenerating neurons and brains. Meli and colleaguesCitation10 selected conformation-sensitive intrabodies by IACT against Alzheimer's amyloid-β oligomers starting from a phage display library constructed from mice immunized with a truncated version of human amyloid-β peptide. Some antibodies recognized in vivo-produced amyloid-β peptide “deposits” in histological sections from human Alzheimer's disease brains and significantly inhibited amyloid-β peptide oligomer-induced toxicity. A human scFv against oligomeric α-synuclein that inhibits aggregation and prevents α-synuclein-induced toxicity was also isolated from a naïve antibody phage display library.Citation9 α-Synuclein is a presynaptic neuronal protein that is linked genetically and neuropathologically to Parkinson's disease and other neurodegenerative disorders. Interestingly, an anti-α-synuclein clone selected from the same library by a novel biopanning technique based on atomic force microscopy showed an opposite effect compared with the scFv mentioned above. It bound to intracellular aggregates of misfolded ataxin-3 and a pathological fragment of huntingtin and accelerated aggregation of these 2 molecules and increased cytotoxicity.Citation95 Intrabodies have furthermore been selected against the natural precursor of nerve growth factor (proNGF).Citation96 These antibodies can be used for the analysis of intracellular trafficking and signaling of proNGF in living cells. Intrabodies against gephyrin playing a role in clustering receptors have also been selected.Citation97 This strategy could be very useful because the gephyrin knockout mice lead to a lethal phenotype.
Other intrabodies are directed against kinases, for example against the cytoplasmic protein kinase Syk,Citation98 or the respective domains of human epidermal growth factor receptorCitation11 or against the BCR-ABL oncogenic protein.Citation13 Blocking the function of the latter protein may neutralize the oncogenicity of BCR-ABL, and could be used as a potential therapeutic agent in Philadelphia chromosome-positive leukemias. An efficient antibody-caspase 3-mediated cell killing system based on antibody-caspase 3 fusion has been developed.Citation99 In the future, this approach may be used to kill tumor cells expressing tumor-specific proteins. Intrabodies against intracellular kinase domains of cell surface receptors and intracellular kinases (in particular against phosphorylation sites) can also be useful to decipher signaling pathways.Citation11,12, 98-100 Furthermore, oncogenic transcription factors may be attractive targets to inhibit the transcription of oncogenic genes. An example is a functional intrabody against the yeast transcription factor GCN4.Citation46
Another intrabody selected from a naïve antibody phage display library neutralized aggrecanase-2, a factor crucial in the development of osteoarthritis, and dose-dependently improved disease progression in an osteoarthritis mouse model.Citation101 A cytoplasmic intrabody used to investigate the role of the rotavirus non-structural protein NSP5 in the virus replication cycle has been described.Citation27 It was demonstrated that NSP5 is an essential element for the assembly of functional viroplasms. These intrabodies were selected from mice that had been immunized with recombinant GST-NSP5. Intrabodies have also been employed as a strategy to target rabies.Citation102
Selection and Engineering of Intrabodies Suitable for Cytoplasmic Expression
In contrast to ER intrabodies, the prerequisite for a specific protein knockdown by a cytoplasmatic intrabody is that the antigen is neutralized/inactivated through the antibody binding (). As said above, any cytoplasmic intrabody approach will be compromised by failure of the antibody to fold correctly.Citation25,26 Consequently, specialized assays or selection conditions have been proposed to increase the success rates. While the selection of cytoplasmic intrabodies is still often a time-consuming trial and error approach and no reliable standard procedure has emerged, 5 different approaches to generate suitable antibodies are described in detail here and in : 1) In vivo selection of functional intrabodies in eukaryotes such as yeast and in prokaryotes such as E.coli (antigen-dependent and independent); 2) generation of antibody fusion proteins for improving cytosolic stability; 3) use of special frameworks for improving cytosolic stability (e.g., by grafting CDRs or introduction of synthetic CDRs in stable antibody frameworks); 4) use of single domain antibodies for improved cytosolic stability; and 5) selection of disulfide bond free stable intrabodies.
Table 1. Strategies for the selection of functional cytoplasmic intrabodies
Table 2. ER intrabodies applied
Approach 1, intrabody selection strategies, in eukaryotes. An antigen-independent selection method termed “Quality Control” was developed to identify intrabodies that are soluble in the cytoplasm of yeast and mammalian cells.Citation103 For selection in yeast, the scFvs were fused to a transcriptional activation domain and a peptide derived from Gal11P binding to the transcription factor Gal4 (1–100) fragment. Soluble expression of the scFv mediates the specific interaction of peptide Gal 11P with the DNA-bound Gal4 (1–100) fragment and then transcription of lacZ and HIS3 reporter genes can be activated, whereas insoluble scFvs are expected to result in non-functionality of the whole fusion protein and thus no activation of reporter genes occurs. Selection in mammalian cells, based on a similar principle, was implemented by employing scFv-VP16 fusions that bind to the DNA binding domain Gal4. Using the “Quality Control” approach, growth of colonies under selection conditions, indicating scFv clones that are stable and soluble in the cytosol, was below 1% among screened clones, which gives a rough initial estimate of the fraction of antibodies that can be expected to fold correctly in the cytoplasm.Citation103
Yeast two-hybrid technology-based IACT is an antibody-dependent method employed to select suitable intrabodies from scFvs preselected by phage display. The pre-enriched antibody fragment library is fused to the VP16 transcription activation domain. Antigen-specific functional intrabodies are selected after co-transfection of the scFv-VP16 library with antigen coding sequence fused to the LexA DNA binding domain in yeast cells. Complex formation of the antigen and an antigen-specific scFv in the yeast cytoplasm leads to activation of yeast chromosomal reporter genes.Citation33 A single pot library of intracellular antibodies (SPLINT) was generated by amplification of natural V regions from non-immunized mice and intrabodies selected against different antigens.Citation104 A further modification of the original IACT technique was established to select for intrabodies that are able to interrupt protein-protein interactions.Citation105 Concerning the IACT selection strategy, it is important that the source for intrabody selection (particularly naïve, immunized mouse antibody or naïve human antibody phage display repertoires have been employed) must have a very large diversity, because functional cytoplasmic intrabodies were described to be a very small subset of the total antibody repertoire.
Approach 1, intrabody selection strategies, in prokaryotes. Another in vivo selection strategy is the selection of functional cytoplasmic intrabodies fused to a selection marker in E.coli using the twin-arginine translocation (TAT) machinery. This strategy is referred to as intrabody selection after Tat export into the cytoplasm (ISELATE).Citation106 Selection is provided by fusion of an N-terminal Tat-specific signal and C-terminal TEM1 β-lactamase to the coding region of the scFv. With the TAT signal providing the transport of folded proteins, only non-aggregated (therefore presumably correctly folded) intrabodies can be transported from the cytoplasm to the periplasm of E.coli where the fused ß-lactamase provides ampicillin resistance, hence positive selection of the respective clones. Recently, a variant of this selection approach for the isolation of correctly folded membrane-anchored scFvs was developed.Citation107 Karlsson et al found that correctly folded scFvs fused to the Tat signal peptide are transported from the cytoplasm to the periplasm of E.coli, but remain N-terminally anchored with the Tat signal peptide to the inner membrane. Selection is performed with antibody expression plasmid transfected spheroplasts and anti-FLAG antibody, which can only recognize the FLAG epitope fused to the C-terminal end of the antibody if the N-terminal signal peptide is anchored in the inner membrane of E.coli and the scFv localized in the periplasm. Affinity maturation of selected scFvs using error prone PCR resulted in scFv clones expressed in the cytoplasm with higher affinity than the original clones.
A mixed strategy that combined elements described above was also developed in E.coli. In this case, the scFv is N-terminally fused to the Tat signal peptide ssTorA and coexpressed with the antigen C-terminally fused to the β-lactamase gene. It was observed that only those binding pairs that are correctly folded and functional in the cytoplasm could be exported to the periplasm and give rise to antibiotic resistant cells.Citation108 Notably, intrabodies with high stability and high antigen-binding affinity could be selected from resistant bacterial clones on high concentrations of the antibiotic carbenicillin.Citation109
Approach 2, antibody fusion proteins. Several publications proposed the addition of non-intrabody domains to enhance folding or stability of cytoplasmic intrabodies. One approach is to use GFP to improve folding of proteins, including recombinant antibody fragments, in E.coli and mammalian cells.Citation37,39,40 It was shown that GFP variants fold differentially in prokaryotic and eukaryotic cells.Citation110 Recently, scFv-GFP fusions were used to screen for intrabody solubility in mammalian cells by fluorescence activated cell sorting (FACS).Citation39 Mammalian cells were transfected with GFP fusions by retroviral gene transfer, the GFP positive cells sorted and expression of the scFv-GFP fusions analyzed. The GFP signal correlated with soluble expression levels of the scFvs in the cytoplasm; however, only 2 different scFvs were tested. A GFP-tagged cytoplasmically expressed scFv was applied for in vivo labeling of tyrosinated α-tubulin and measurement of microtubule dynamics.Citation37 It remains to be proven whether this strategy is broadly applicable.
In addition to GFP, a Cκ domain,Citation111,112 maltose binding protein (MBP),Citation41 N utilization substance A (NusA) Citation113 or Fc domainCitation38,42 have been proposed as fusion partners, although few successful examples have been presented. Different labs have focused on the inhibition of p53 and activation of mutated p53 inside tumor cells.Citation111,112,114 The anti-p53 intrabodies derived from hybridomas showed different stability inside the cytoplasm, as expected. One of the scFvs targeted to the nucleus or cytoplasm was expressed well as a fusion with the Cκ domain.Citation111 Another scFv showed very low expression inside the cytoplasm after fusion with a Cκ domain.Citation112 Interestingly, 2 intrabodies recognizing p53 mutants that were expressed in the nucleus and cytoplasm restored transcriptional activity of p53 mutants by themselves,Citation114 which demonstrates that solubility depended on the properties of the intrabody rather than on the Cκ domain. Because degradation of proteins can be linked to the presence of regions rich in proline, glutamic acid, serine and threonine (PEST sequences),Citation115 the low content of potential PEST sequences might explain the stability of some intrabodies such as the 2 anti-p53 mutated intrabodies.Citation114 However, the fusion of a proteasome-targeting PEST motif to intrabodies recognizing Huntingtin or α-synuclein-mediated increase of intrabody stability and degradation of the corresponding antigen,Citation116,117 demonstrating that no simple PEST sequence motif-related algorithm will predict the intrabody efficacy.
MBP has extensively been used to enhance folding of intrabodies inside the cytoplasm of E.coli and mammalian cells.Citation41 It was found that the folding of some passenger proteins fused to MBP could be mediated by endogenous chaperones in vivo.Citation118 It was proposed that the solubility-enhancing activity of MBP is mediated by its open conformation and it is assumed that the ligand-binding cleft is involved in the mechanism.Citation119 However, the molecular mechanism of the process is not known, and some reports of improved folding may have been mistakenly based on the improved overall solubility of the fusion.
Approach 3, CDR grafting or introduction of synthetic CDRs in stable frameworks. Antigen-binding loops can be transplanted from one antibody to a different antibody framework, a procedure called CDR loop grafting.Citation120 While the technology gained historical relevance by being used to de-immunize (“humanize”) therapeutic antibodies, it also can be used to transfer a given specificity to a preselected V region framework with optimized production / folding / stability properties. A stable framework-engineered stabilized version of an anti-GCN4 antibody was cytoplasmically expressed as an scFv intrabody in yeast.Citation46 This variant inhibited the activity of β-galactosidase expressed from a GCN4-dependent reporter gene compared to the unfolded original scFv. Another approach used the scFv(F8), which showed very high in vitro stability and functional folding in both the prokaryotic and eukaryotic cytoplasm.Citation43 Several groups of residues in the CDRs important for antigen binding of the poorly stable anti-hen egg lysozyme (HEL) scFv(D1.3) were grafted into the framework of scFv(F8). Five different variants were constructed, and 4 of those could be expressed in the cytoplasm of E.coli and bind to the antigen after purification. The donor framework of scFv(F8) seems to be able to tolerate extensive CDR substitutions without loss of stability in the cytoplasm.Citation45 A synthetic antibody library based on the human framework of anti-β galactosidase scFv13R4 was expressed in the E.coli, yeast and mammalian cell cytoplasm. Randomized CDR 3 loops were introduced in the framework by PCR and a number of functional cytoplasmic intrabodies against different antigens selected, including an intrabody that identified a protein of yet unknown function involved in mast cell degranulation.Citation121 Another approach used amino acid randomization at 4 and 7 different positions in the CDR2 and CDR3 loop of the single domain VH of anti-lysozyme antibody HEL4.Citation44 The library was used for selection of antibodies with different specificities on both purified lysozyme and whole cells.
Approach 4, single domain antibodies. Single domain antibodies are composed of only one V region, which could be either a variable domain of the heavy or light chain. They can be produced from conventional human IgGs (VHs and VLs), from camelid heavy-chain IgGs (VHHs) and from cartilaginous fish IgNARs (VNARs). As for scFvs, libraries of single domain antibodies can be constructed from naïve, synthetic, semi-synthetic, transgenic animals or immunized sources and clone selection is typically done by phage panning or yeast cell surface display. Despite providing only half of the binding surface of an antibody, camelid VH single domain antibodies have demonstrated high affinity for many cognate antigens, high solubility and aggregation resistance. The camelid single domain antibodies are selected from camelid VHH naïve or immune libraries. Several camelid cytoplasmic single domain antibodies have been successfully generated, for example against F-actin capping protein CapG,Citation14 β2-adrenergic receptor,Citation100 Clostridium botulinum neurotoxin (BoNT) proteases,Citation28,29 core antigen of HBV (HBcAg),Citation6 15-acetyldeoxynivalenol,Citation122 caspase-3,Citation123 Bax,Citation30 and nuclear polyA-binding protein.Citation124 A nanobody recognizing the F-actin capping protein CapG was generated after immunization of a llama.Citation14 It was demonstrated that expression of the intrabody in breast cancer cells restrained cell migration and lung metastasis in a xenograft tumor mouse model. Anti-β2-adrenergic receptor single domain intrabodies from the Camelid family inhibiting G protein activation, G protein-coupled receptor kinase-mediated receptor phosphorylation and β-arrestin recruitment have been published.Citation100 An intrabody against caspase-3 was isolated from a naïve phage display llama antibody repertoire.Citation123 Single-domain antibodies against the proapoptotic Bax protein have been selected.Citation30 These anti-Bax VH intrabodies could be used as tools for studying the role of Bax in oxidative-stress-induced apoptosis and for developing novel therapeutics for the degenerative diseases involving oxidative stress.
Yeast surface display has also been used to select camelid VHHs as immune inhibitors of Clostridium botulinum neurotoxin (BoNT) proteases. VHHs were selected from an immunized llama single domain repertoire that remained functional when expressed within neurons and bound to BoNT proteases with high affinity.Citation29 VHHs from a non-immune llama single-domain library have been isolated.Citation28 The X-ray crystal structure of the most potent intrabody in complex with the protease was solved and the structure revealed that the VHH binds in the α-exosite of the enzyme, far from the active site for catalysis. Camelid VHH domain antibodies selected from a non-immune llama single-domain phage display library prevented oculopharyngeal muscular dystrophy associated aggregation of nuclear poly(A)-binding protein.Citation124 Notably, intrabodies also reduce the presence of already existing aggregates. VHH intrabodies against the hepatitis B core antigen (HBcAg) were selected from a single domain phage display library constructed from llamas immunized with noninfectious HBV particles.Citation6 Intrabodies targeted to the nucleus affected HBcAg expression and trafficking in HBV-transfected hepatocytes. Camelid VHH domains selected from a hyper-immunized phagemid library were selected against toxin 15-acetyldeoxynivale.Citation122 Expression of the corresponding intrabodies in yeast resulted in significant resistance to the toxin. The authors suggested that VHH expression in plants may lead to enhanced tolerance to mycotoxins. A camelized rabbit – derived VH single-domain recognizing HIV-1 Vif was constructed starting from an original rabbit scFv.Citation2 A set of 3 mutants of the derived VH single domain antibody with gradual increasing camelization was performed. There was a strong correlation between the improvement in protein solubility in mammalian cells and the gradual increasing camelization. The intrabodies neutralized HIV-1 infectivity. Anti-ß catenin llama single domain intrabodies that inhibit Wnt signaling were recently reported, providing a tool for further study of the Wnt pathway.Citation125
In contrast to the VH domains of camels and llamas, human heavy chain variable domains and human light chain variable domains are more prone to dimerization and aggregation and exhibit poor solubility.Citation35 Nevertheless, several human single domain antibodies have been selected, for example against Etk kinase,Citation12 huntingtin proteinCitation7,8 and B cell super-antigen protein L.91 As for scFvs, a Tat signal peptide and β-lactamase-based strategy was used to isolate human VH domains from a human germ-line VH library, with increased level of thermodynamic stability, reversible folding and soluble expression in E.coli.Citation90 In another approach, aggregation-resistant human heavy chain variable domains were selected on phage by heat denaturation.Citation126 A large human VH domain antibody repertoire was constructed by combinatorial assembly of CDR building blocks from a smaller repertoire comprising a high frequency of aggregation-resistant antibody domains.Citation34 Barthelmy and colleaguesCitation127 evolved and analyzed stable human VH domains from a phage display antibody library. By building libraries comprising the framework of humanized anti-HER2 scFv and different diverse synthetic VH CDR3 loops, they selected the most stably expressed VH domains secreted in E.coli and then introduced mutations that increased the hydrophilicity of the former light chain interface by replacing exposed hydrophobic residues with structurally compatible hydrophilic substitutions. The stability of many in vitro evolved VH domains seemed to be essentially independent from the CDR3 sequence and instead derived from mutations introduced in the second step.
A repertoire of stable human VL domains was isolated after panning with a human naïve VL library with B cell super-antigen protein L. Isolated clones exhibited improved reversibility of thermal unfolding after purification from the periplasm of E.coli.Citation91 Nevertheless, it remains to be demonstrated whether the isolated variable domains can also be stably expressed in the cytoplasm as stable functional intrabodies. An α-hungtingtin single domain antibody comprising a human light chain variable domain (VL) was selected from an original nonfunctional scFv.Citation8 Affinity maturation of the original scFv was performed using error-prone PCR and yeast surface display, and it was shown that the paratope was localized in the VL. However, this selection approach is only applicable when the binding energy of the scFv is provided predominantly by only one of the 2 V domains. A human VL against Etk kinase was selected from a large single domain phage display library, derived from a single human framework of light chain.Citation12 When expressed in Src-transformed cells, the single-domain antibodies interacted with endogenous Etk in the cytoplasm and efficiently blocked its kinase activity, leading to partial inhibition of cellular transformation.
Approach 5, cyto-intrabodies without disulfide bridges. Many stabilizing and destabilizing mutations in the framework or CDRs have been described.Citation128,129 In this context, stable cysteine-free scFvs have been selected and expressed in functional form in the E.coli cytoplasm starting from the levan binding antibody ABPC48. Affinity maturation was performed using DNA-shuffling and random mutagenesis.Citation47 An engineered single domain VL antibody without disulfide-bridge and with high affinity preventing aggregation of huntingtin was selected after affinity maturation of the VL domain of an original anti-huntingtin scFv using error-prone PCR and yeast surface display.Citation7 These intrabodies may have therapeutic potential for the treatment of hungtingtin disease. However, no naïve library strategy has yet emerged from this interesting approach, limiting this method to the tedious optimization of individual antibody clones.
Targeting the “Outside” from the “Inside”: The ER Intrabody Approach
The use of various signal sequences attached to antibody fragments allows its targeting specifically to different intracellular compartments. ER targeting in particular allows interference with the function of the members of the membrane and the plasma proteome (secretome), relying on a different mechanism than cytoplasmic antibodies. Hence, generation of suitable ER intrabodies is much easier because they do not require special folding or stability features, and, in particular, no neutralizing activity toward the antigen is needed. Reasonable antigen binding is sufficient (). This eliminates the necessity for any specialized selection strategy and allows immediate utilization of the vast and quickly growing resource of recombinant research antibodies currently generated.Citation130
Reports of ER intrabodies published so far have shown protein knockdown in vitro and recently in vivo in a mouse model. As a step toward in vivo use, ER intrabodies have been applied in primary tissue explants (Zhang, Korte and Dübel, unpublished data) and against the oncogenic receptors VEGFR-2/KDR and Tie-2 in xenograft mouse tumor models.Citation87,88,131 In addition ER intrabodies specific to amyloid-ß demonstrated an inhibition of amyloid-ß accumulation in an Alzheimer's disease mouse model.Citation132 ER intrabodies have been applied for various purposes ().
Intrabodies have also been used in plants to interfere with cell functions and plant cell-pathogen interactions.Citation133 The first transgenic ER intrabody mouse that constitutively expresses an α-VCAM1 intrabody, resulting in a clear phenotype (aberrant distribution of immature B cells in blood and bone marrow), has recently been described.Citation134 In the future, by using inducible or cell- and tissue specific promoters, this important approach will facilitate the functional analysis of proteins in specific subpopulations of cells or tissues, or with different time kinetics. Moreover, this study hinted at possible quantitative effects, which would allow regulation of knockdown strength.
A therapeutic application of intrabodies would require transfection systems allowing gene therapy, typically viral or non-viral systems.Citation135 Despite growing numbers of clinical studies using gene therapy, some issues are yet unresolved, like risks from insertional mutagenesisCitation136 or low gene transfer efficiency of nonviral vectors.Citation137 The construction of retroviruses with low safety riskCitation138 and the development of transductional and transcriptional targeting for cell-specific gene transferCitation139 are new promising approaches. Furthermore, gene therapy with mRNA might be possible in the future.Citation140
Parameters Influencing the Efficiency of ER Intrabodies
ER antibodies work by retaining their antigen in the ER. For this simple approach, and despite publication of many successful cases, our systematic understanding of the many parameters involved in a successful knockdown by intrabodies is still very limited. The influence of target and antibody expression levels, antibody affinity and epitope structures is still largely unexplored. For instance, a study by Beerli et alCitation141 reported efficient target knockdown by only one of 2 ER intrabodies with nanomolar affinity,Citation141-144 which suggests that high affinity is not sufficient as a predictive criterion for successful ER intrabody-mediated knockdown. This is further supported by a report on the lack of correlation between the affinity and antineoplastic effect of anti-erbB2 targeting ER intrabodies.Citation145 Different knockdown efficiencies not correlated to monovalent affinities have also been reported by others.Citation75 An anti-erbB2 antibody has been reported to bind to its target if transported to the cell surface but not within the cellCitation146 and the reasons for this are not fully understood. The exact reasons for these differences are not yet fully clear, but it can be assumed that individual differences in the epitopes play a key role. Alternatively, the local biochemical milieus may influence individual epitope/idiotype combinations. Study of these parameters in the future is needed, as the results will allow pre-selection of the intrabodies accordingly. If, for instance, the different pH in the Golgi does not allow binding of the ER intrabody to its target in this compartment, in vitro antibody selection strategies such as phage display could easily be adapted by carrying out the selection procedure under buffer conditions that mimic those in the Golgi.
In spite of numerous examples for the successful application of ER intrabodies that can be found in literature (, ref. 3), the majority of these reports mainly focus on the analysis of phenotypical effects and on study of a particular target. Questions concerning the intrinsic properties of the ER intrabody technology and the underlying principles that determine success or failure of ER intrabody-mediated knockdowns have not been satisfactorily addressed so far. For wider and more systematic application of the ER intrabody technology, further insight into the parameters influencing the success of the method itself is needed. A more detailed characterization of the ER intrabody knockdown process will allow elimination of potential off-target effects when interpreting results and identification of specific properties of ER intrabody-mediated knockdowns. Opening new ways of analysis, in addition to the already known unique advantages of ER intrabodies over other methods, is critical.
Table 3. A. Reports of ER intrabody mediated knockdown
Table 3. B. ER intrabody-mediated knockdown: methodological aspects
In the following, we review ER intrabody-mediated knockdown studies with a particular focus on methodological aspects (). The first example of in vivo efficacy of ER intrabodies has recently been described,Citation134 but ER intrabody-mediated knockdowns have so far been employed in vitroCitation3 in a large variety of setups. ER intrabody mediated knockdowns have been performed either in cell lines with constitutiveCitation75,147 or inducibleCitation148 endogenous expression of the target or in cell lines that had been either transientlyCitation149 or stablyCitation141,149, 150 transfected with the target. Correspondingly, transfection of ER intrabodies into these cell lines has been performed transiently,Citation75,149 inducibly stableCitation151 or constitutively stable.Citation151-153 A significant quantitative biochemical analysis of the involved parameters is lacking in most studies published so far. Further, the information on the knockdown process has come from very diverse assays usually optimized for one target, which prevents a systematic analysis of these datasets to obtain general conclusions.
Hence, it is currently difficult to attribute knockdown efficiencies to the biochemical properties of the respective intrabody, e.g., affinity, because knockdown efficiency may also be influenced by the particular expression levels of the ER intrabody and its antigen. Individual developments of transient expression levels over time will also surely affect the knockdown efficiency and a respective time dependence of knockdown efficiency after transient transfection of ER intrabodies has been observed.Citation149
Despite the lack of a systematic analysis of these important parameters, we can nevertheless gain some valuable insights from some examples. For instance, the analysis of 3 different ER intrabodies transiently expressed in the same cell line against the same target allowed some interesting observations.Citation75 The ER intrabodies in this study were analyzed for affinity, expression levels and their ability to bind a linear or conformational epitope. Interestingly, the ER intrabody with the lowest affinity but the highest expression levels gave rise to the highest knockdown efficiency,Citation75 underlining once more the need for further understanding of the knockdown process in a quantitative manner. Moreover, while all antibodies in this study recognized the native target on the cell surface, the ER intrabody with the highest knockdown efficiency also efficiently recognized a linear epitope.Citation75 Recognition of a linear epitope of an unfolded or only partly folded protein, possibly already while the protein is still in the process of translation into the ER, can be imagined to efficiently interfere with the folding process and in this way accelerate degradation of this protein. A systematic characterization of ER intrabody epitopes, particularly of ER intrabodies that are known to accelerate degradation of the target, will in the future be required to evaluate this hypothesis. If the recognition of linear epitopes by ER retained antibodies can indeed promote accelerated degradation of target proteins, this mechanism may in the future be harnessed by selectively generating antibodies against short linear peptide sequences within the target protein instead of whole proteins, and would also allow an important acceleration of the method since the necessity for antigen protein production – the single most retarding process in today's antibody generating pipelines – can be completely avoided.
An important question to understand intrabody generated phenotypes and to avoid incorrect conclusions due to unspecific and off-target effects is for the fate of the intrabody/target protein complex. Only few studies are available so far which tried to assess these questions. It was shown that the ER intrabodies targeting ß-amyloid precursor protein (APP) and the anti-TLR2 ER intrabody are degraded by the proteasome (Ref. Citation154, Böldicke and Burgdorf, unpublished). Although ER intrabodies have been reported to cause accelerated degradation of a target protein,Citation155 there are also reports on accumulation of the target within the cell, although it is depleted from the surface.Citation150,151 The determining factors that are responsible for accelerated degradation, accumulation or even unchanged intracellular target levels are still not clear. However, no detectable ER stress response (unfolded protein response) was observed following substantial overexpression of an anti-p75NTR ER intrabody, confirming the specificity of this particular approachCitation75 and recommending measurement of UPR as a good control allowing for rating the relevance of any future intrabody approaches. Another observation, with possible relevance to the question of the post-translational fate of intrabodies and their antigens, is the lack of target protein glycosylation upon expression of an ER intrabody as a result of the retention in the ER.Citation148,150,153
To compare the results of different ER intrabody-mediated knockdowns, the choice of methods used for detecting knockdowns is critical. The heterogeneity of analysis methods used so far is therefore an obstacle for correlating antibody properties with knockdown efficiency. Methods for detection of ER intrabody-mediated knockdowns ranged from proof on the biochemical level by cell surface stainings followed by flow cytometry to a variety of functional assays including entirely qualitative analysis (see ). While flow cytometric analysis of cell surface stainings are preferable to other methods because they provide direct biochemical evidence for a potential membrane expression knockdown, cell surface stainings have also not been done in a standardized way. In some reports, the same antibody clone that was used as an ER intrabody was also used for the detection of the target on the cell surface,Citation148 which bears the risk of artifacts due to ER intrabodies that still mask the target protein's epitope on the cell surface after being released by cell lysis. Although retention by the KDEL receptor has been described to be very efficient (retention of a 10-fold molar excess of substrate may be possibleCitation156), masking of target proteins on the cell surface due to secretion of ER intrabodies must be excluded. In order to avoid this artifact, cell surface stainings have therefore increasingly been performed with detection antibodies that have been mapped to bind to a different epitope than the ER intrabody.Citation75,134, 149,151
In conclusion, although there are already initial findings that suggest a link between particular antibody properties and knockdown efficiency, a more systematic analysis of these links is required to allow better a priori predictions, and thus to allow the selection of optimal antibodies for future intrabody knockdown approaches.
The ER Intrabody Approach in Comparison to other Knockdown Methods
The cell surface and its receptors provide an interface for communication between the individual cell and its environment, and is therefore a crucial element of cellular decision processes, determining the organization and function of cells, tissues and organs. Complementary to cell surface receptors, secreted factors control cellular decisions by transducing signals via transmembrane receptors to the inside of the cell. Because both the soluble extracellular factors as well as cell surface receptors pass the secretory pathway, the ER intrabody technology can be employed to systematically target these crucial control elements at the protein level.
There have been other attempts to target these proteins at the post-translational level by pharmacological inhibition of receptor tyrosine kinases,Citation157 blocking cell surface receptors or secreted factors extracellularly with antibodiesCitation158,159 or even by targeting the intracellular part of cell surface receptors or signaling molecules downstream of the cell surface receptors via cytosolic intrabodiesCitation11,23 or phosphopeptide mimetics.Citation160,161 Most of these approaches have very narrow application ranges and require tedious individual developments for every case. For instance, not all of the cell surface receptors are receptor tyrosine kinases, and therefore not all of them are accessible to the respective pharmacological agents, and pharmacological inhibitors are only available for some of the receptor tyrosine kinases. The same is true for phosphopeptide mimetics: inhibitory and at the same time cell permeable drugs are only available for a very small number of the signaling molecules, allowing only specialized applications.
Another drawback of pharmacological agents for protein inhibition is the dependence of their effectiveness on biodistribution and bioavailability and the potential side effects that may occur due to lack of specificity, especially as tyrosine kinase inhibitors are generally less specific than antibodies,Citation162,163 which for example, has been shown for protein kinase inhibitors and inhibitory oligonucleotides of TLRs. It was shown that the compounds KT5720, Rottlerin and quercetin inhibited many different protein kinases.Citation164 Inhibitory CpG comprising oligonucleotides developed to inhibit the function of TLR9 inhibit TLR9 signaling, but also bind signal transducer and activator of transcription 1 and 4 (STAT1 and STAT4) and interact with TLRs 3, 7 and 8.Citation165,166 Finally, there is no systematic way to generate them for any possible protein, very much in contrast to antibodies, which can be rationally and systematically generated to almost any target within a few weeks.Citation167
Dominant negative mutants (for review see Ref. Citation168) interfere with protein function by competing with the wildtype, and usually require detailed knowledge of the role of individual amino acids in the protein function. In addition, its dependence on vast overexpression is prone to cause unspecific effects and cell stress. Intrabody approaches do not require any knowledge of the target protein, but even may be used to reveal phenotypes of yet completely unknown proteins, because they can be generated from the genomic sequence information of unknown open reading frames (ORFs), without requiring the antigen for their generation, e.g., by the well-established method of phage display based on synthetic peptides.Citation169
The application range for antibody-based targeting of the cytosolic part of kinase receptors and of the corresponding signaling molecules downstream of cell surface receptors similarly suffers from limitations. As described above, antibodies that can fold correctly if expressed in the cytosol are not easily obtainedCitation19,23, 25 and, although the delivery of antibodies as proteins to the cytosol has been demonstrated in vitro,Citation24 protein delivery from the outside still remains a challenge. ER intrabodies, in contrast, are generally applicable both in vitro and in vivo and can be generated with high specificity against any protein of the secretome, provided that the target protein or a sufficient fragment of it (e.g., synthetic peptide) are available for the selection procedure.Citation60,130,169,170 In vivo studies by ER intrabody gene delivery include knockdown in xenograft tumor models and in an Alzheimer's disease mouse model.Citation88,131,132 The successful in vivo protein knockdown in transgenic intrabody mice by ER intrabodies has opened new opportunities to study genetically lethal phenotypes.Citation134 If the hypothesis can be confirmed that the strength of the in vivo knockdown can be influenced by the strength of an exogenous promoter, different or inducible promoters in mice will certainly facilitate the establishment of novel disease-related preventive and therapeutic mouse models. The potential for intrabody expression restricted to specific tissues by appropriate promoters will yield new insights into the in vivo function of new proteins in subpopulations of cells. Although blocking cell surface receptors by secretion of antibodies to the cellular environment is broadly applicable to cell culture and has also been performed in vivo in transgenic mice,Citation159 this type of interference at the protein level is little defined because the location of the effect in vivo can only poorly be controlled. Since secreted antibodies can be transported to different locations in a transgenic mouse, a secreted antagonistic antibody may not only have an autocrine and paracrine, but also an endocrine effect, which renders tissue specific knockdowns at the protein level impossible by this method.
The most commonly used techniques to knock down proteins of the secretome are so far not those taking action at the post-translational level, but methods that take effect at the RNA or DNA level. RNA interference (RNAi) has become a widely used standard method for knocking down proteins as it is generally applicable provided that the sequence of the target mRNA is known. Only a short RNA sequence (short interfering RNA, siRNA) that can induce degradation of all the homologous cellular mRNA as part of an RNA induced silencing complex (RISC) is required.Citation171,172 Generating siRNA technically was less challenging and less time-consuming than ER intrabody generation by the hybridoma technology, but this situation is currently changed due to the availability of automated miniaturized in vitro antibody selection methods, which provide the antibody gene in the correct format (scFv) for immediate subcloning.
Consequently, the increasing availability of antibody genes and efforts to generate databases with detailed descriptions for affinity reagentsCitation60,169, 173,174 can in the future provide a well-annotated resource of already existing antibody genes that are ready to use as ER intrabodies. For these “ready to use” antibody genes, which are available in the thousands already, generating a knockdown with the ER intrabody will not require any more time than is required for an RNAi-based approach. Although confirmation by a larger data set is needed, a claimed advantage of the ER intrabody technology over RNAi is the potentially longer half-life of the protein-based ER intrabodies compared to RNA-molecules.Citation175 Another inherent property of ER intrabodies that represents an advantage over RNAi concerns specificity, which can be thoroughly assessed biochemically by a multitude of assays before applying an antibody intracellularly, while specificity and off-target effects of RNAi are often more difficult to predict.
RNAi, which is thought to have its origins in nature from an antiviral defense mechanism,Citation176-178 has been found to cause several off-target effects, including an interferon responseCitation179-183 and aberrant expression of up to more than 1000 genes as found by Persengiev et al.Citation181 Off-target silencing of siRNA/shRNA results from imperfect pairing of siRNA strands with sequence motifs that reside primarily in 3′ UTR regions of cellular mRNA. siRNA and miRNA share the same silencing machinery and the magnitude of regulation of siRNA off-target transcripts is similar to that of micro RNAs.Citation184 Furthermore, siRNA recognize TLR3, TLR7 and TLR8 resulting in secretion of interferon-α and synthesis of pro-inflammatory cytokines.Citation185 RNAi and the ER intrabody technology both have broad application areas as they both can be applied in vitro and in vivo. Applications in vivo have already been performed both for RNAi and the ER intrabody technology in the form of in vivo deliveryCitation88,131,186 and in the form of transgenic mice.Citation134,187 However, therapeutic applications are hampered by the hurdle posed by the need for efficient in vivo delivery of nucleic acids, and are still subject to safety concerns for both RNAi and the ER intrabody technology.Citation175,188-190 Consequently, applications have primarily focused on use of the technologies as research tools. The same is true for the classic knockout strategies at the DNA level.
Genetic knockouts in mice have greatly contributed to a better understanding of human disease, and, as a consequence, can yield new therapeutic approaches. However, in spite of substantial efforts aimed at cataloging the mammalian genome, e.g., the International Mouse Phenotyping Consortium (www.mousephenotype.org), genetic knockouts do not always allow complete analysis of essential genes because approximately 30% of genetic knockouts are embryonically lethal.Citation191 If a complete knockout of a gene is lethal early in development, alternative strategies for functional knockdown are required to study functions of the gene in later developmental stages and adults. ER intrabodies offer a solution. Although the genetic knockout of the cell adhesion molecule VCAM1 leads to early embryonic death, mice with an anti-VCAM1 ER intrabody exhibited depletion of VCAM1 at the cell surface, but were nevertheless viable.Citation134 This demonstrates the potential of the ER intrabody technology as an alternative strategy for studying the phenotype of embryonically lethal knockouts in adult mice.
In conclusion, the inherent properties of ER intrabodies implicate some limitations, but also specific strengths that can serve to compensate shortcomings of the other established technologies for functional studies of the secretome.
The Promising Future of Intrabody Approaches
Although intrabody approaches have been known for more than a quarter of a century, and the above examples are very encouraging, this approach is not as widely used for cell biological research as could be expected given the possible benefits. In particular, the technology had a very slow start. What is the reason for that? The answer is not simple, and comprises a multitude of factors. The first intrabody studies used microinjection of mRNA, a cumbersome and inefficient method of limited specificity, resulting in a first decade of very rare applications. In the early nineties, the method gained some momentum with the advent of essential technological elements like the single chain format and recombinant antibody technologies. Typical applications followed the cytoplasmic intrabody approach. In light of the quite substantial requirements of getting an antibody combining both functional neutralization of its target and correct folding in reducing milieu, it can be assumed that those candidates were not easily identified from the collection of available hybridomas. In the early days of intrabody technology, a substantial number of intrabody trials may have failed due to the limited availability of suitable monoclonal antibodies and the limited knowledge about the biochemical requirements. Further, generating recombinant scFv antibodies from hybridoma, and particular expressing it in E. coli for functional validation, was a technology not widespread among the typical cell biology lab of the time. Quite a number of scFvs generated from hybridoma showed very poor production yields in E. coli,Citation192 generating substantial problems for the necessary biochemical characterizations. Another problem, which originated from the use of early hybridoma cell lines as sources for the isolation of antibody DNA, was the frequently observed expression of multiple antibody mRNAs in these cells due to the aneuploid/deregulated state of the hybridoma cells, lacking allelic exclusion and allowing accumulation of mutations.Citation193-196 This mandated substantial experimental efforts to confirm that the predominant V region PCR bands found from any hybridoma indeed encoded the V region subset providing the specificity identified in the supernatant.Citation197 These factors contributed to give the intrabody technology a mixed reputation among the typical possible users who were not acquainted with recombinant antibody technology. Nevertheless, successful projects using hybridoma cells as a starting point accumulated over time and provided a slow but steady development of the technology.
Only in the last decade have recombinant in vitro antibody selections from gene repertoires been utilized in substantial numbers, allowing much quicker access to the antibody and providing the V region encoding DNA without any additional experimental effort. This also substantially increases the chances of getting antibodies with the required complex property combination using appropriately tuned libraries and panning strategies from the start. The technology to generate research antibodies from phage display in large numbers is now robust and reliable,Citation65-67, 69,169 and international research consortia like the “Affinomics” initiative (www.affinomics.org) in the EU and similar initiatives in the US have already generated several thousands of antibodies to hundreds of new potential intrabody targets together with their V region genes, providing right away a vast resource of scFv DNA for future intrabody approaches. Studies on introducing antibodies from the outside to interfere with intracellular functions show the limitations of this approach,Citation24 so the quick availability and fast generation of recombinant antibodies for intrabody approaches may motivate more researchers to try this technology for their functional analysis. Protein functions of bioinformatically predicted ORFs of so far unknown function could be evaluated in respect of resulting knockout phenotypes, even without the availability of the resulting protein, because antibodies can efficiently be generated by phage display to chemically synthetized peptides based on the ORF sequence. In the future, this strategy allows a completely new systematic approach to identify protein functions of yet uncharacterized members of the proteome, particularly straightforward for the membrane proteome where most binders may be able to generate a phenotype after just a single subcloning step into an ER intrabody vector. Further, new methods for the rapid generation of transgenic organisms, like CRISPR/Cas9 or TALEN, are maturing, and will further facilitate the experiments required to screen for a phenotype in vivo.
The application of the intrabody technology, in particular the most facile ER intrabody approach, can thus be expected to grow much more rapidly in the near future than in its first 2 decades.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Michael Hust for providing graphic elements for the figures.
Funding
We are grateful for support by the EU consortium “Affinomics.”
References
- Zehner M, Marschall AL, Bos E, Schloetel J-G, Kreer C, Fehrenschild D, Limmer A, Ossendorp F, Lang T, Koster AJ, et al. Endosomal Sec61 mediates antigen translocation in the cytosol for cross-presentation. Immunity 2015; 42:850-63; PMID:25979419; http://dx.doi.org/10.1016/j.immuni.2015.04.008
- Aires da Silva F, Santa-Marta M, Freitas-Vieira A, Mascarenhas P, Barahona I, Moniz-Pereira J, Gabuzda D, Goncalves J. Camelized rabbit-derived VH single-domain intrabodies against Vif strongly neutralize HIV-1 infectivity. J Mol Biol 2004; 340:525-42; PMID:15210352; http://dx.doi.org/10.1016/j.jmb.2004.04.062
- Böldicke T. Blocking translocation of cell surface molecules from the ER to the cell surface by intracellular antibodies targeted to the ER. J Cell Mol Med 2007; 11:54-70; PMID:17367501; http://dx.doi.org/10.1111/j.1582-4934.2007.00002.x
- Kirschning CJ, Dreher S, Maass B, Fichte S, Schade J, Koster M, Noack A, Lindenmaier W, Wagner H, Böldicke T. Generation of anti-TLR2 intrabody mediating inhibition of macrophage surface TLR2 expression and TLR2-driven cell activation. BMC Biotechnol 2010; 10:31; PMID:20388199; http://dx.doi.org/10.1186/1472-6750-10-31
- Reimer E, Somplatzki S, Zegenhagen D, Hanel S, Fels A, Bollhorst T, Hovest LG, Bauer S, Kirschning CJ, Boldicke T. Molecular cloning and characterization of a novel anti-TLR9 intrabody. Cell Mol Biol Lett 2013; 18:433-46; PMID:23893288; http://dx.doi.org/10.2478/s11658-013-0098-8
- Serruys B, Van Houtte F, Farhoudi-Moghadam A, Leroux-Roels G, Vanlandschoot P. Production, characterization and in vitro testing of HBcAg-specific VHH intrabodies. J Gen Virol 2010; 91:643-52; PMID:19889923; http://dx.doi.org/10.1099/vir.0.016063-0
- Colby DW, Chu Y, Cassady JP, Duennwald M, Zazulak H, Webster JM, Messer A, Lindquist S, Ingram VM, Wittrup KD. Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody. Proc Natl Acad Sci U S A 2004; 101:17616-21; PMID:15598740; http://dx.doi.org/10.1073/pnas.0408134101
- Colby DW, Garg P, Holden T, Chao G, Webster JM, Messer A, Ingram VM, Wittrup KD. Development of a human light chain variable domain (V(L)) intracellular antibody specific for the amino terminus of huntingtin via yeast surface display. J Mol Biol 2004; 342:901-12; PMID:15342245; http://dx.doi.org/10.1016/j.jmb.2004.07.054
- Emadi S, Barkhordarian H, Wang MS, Schulz P, Sierks MR. Isolation of a human single chain antibody fragment against oligomeric α-synuclein that inhibits aggregation and prevents α-synuclein-induced toxicity. J Mol Biol 2007; 368:1132-44; PMID:17391701; http://dx.doi.org/10.1016/j.jmb.2007.02.089
- Meli G, Visintin M, Cannistraci I, Cattaneo A. Direct in vivo intracellular selection of conformation-sensitive antibody domains targeting Alzheimer amyloid-β oligomers. J Mol Biol 2009; 387:584-606; PMID:19361429; http://dx.doi.org/10.1016/j.jmb.2009.01.061
- Hyland S, Beerli RR, Barbas CF, Hynes NE, Wels W. Generation and functional characterization of intracellular antibodies interacting with the kinase domain of human EGF receptor. Oncogene 2003; 22:1557-67; PMID:12629519; http://dx.doi.org/10.1038/sj.onc.1206299
- Paz K, Brennan LA, Iacolina M, Doody J, Hadari YR, Zhu Z. Human single-domain neutralizing intrabodies directed against Etk kinase: a novel approach to impair cellular transformation. Mol Cancer Ther 2005; 4:1801-9; PMID:16276002; http://dx.doi.org/10.1158/1535-7163.MCT-05-0174
- Tse E, Lobato MN, Forster A, Tanaka T, Chung GT, Rabbitts TH. Intracellular antibody capture technology: application to selection of intracellular antibodies recognising the BCR-ABL oncogenic protein. J Mol Biol 2002; 317:85-94; PMID:11916380; http://dx.doi.org/10.1006/jmbi.2002.5403
- Van Impe K, Bethuyne J, Cool S, Impens F, Ruano-Gallego D, De Wever O, Vanloo B, Van Troys M, Lambein K, Boucherie C, et al. A nanobody targeting the F-actin capping protein CapG restrains breast cancer metastasis. Breast Cancer Res 2013; 15:R116; PMID:24330716; http://dx.doi.org/10.1186/bcr3585
- Biocca S, Di Luzio A, Werge T, Cattaneo A. Intracellular immunization: Expression of antibody domains in the cytoplasm and in the nucleus of mammalian cells. Cytotechnology 1991; 5:49-50; PMID:AMBIGUOUS; http://dx.doi.org/10.1007/BF00736806
- Biocca S, Neuberger MS, Cattaneo A. Expression and targeting of intracellular antibodies in mammalian cells. EMBO J 1990; 9:101-8; PMID:2153072
- Carlson JR. A new means of inducibly inactivating a cellular protein. Mol Cell Biol 1988; 8:2638-46; PMID:3136320
- Lobato MN, Rabbitts TH. Intracellular antibodies and challenges facing their use as therapeutic agents. Trends Mol Med 2003; 9:390-6; PMID:13129705; http://dx.doi.org/10.1016/S1471-4914(03)00163-1
- Stocks M. Intrabodies as drug discovery tools and therapeutics. Curr Opin Chem Biol 2005; 9:359-65; PMID:15979379; http://dx.doi.org/10.1016/j.cbpa.2005.06.003
- Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P. Transgenic plants expressing a functional single-chain Fv antibody are specifically protected from virus attack. Nature 1993; 366:469-72; PMID:8247156; http://dx.doi.org/10.1038/366469a0
- Wheeler YY, Chen SY, Sane DC. Intrabody and intrakine strategies for molecular therapy. Mol Ther 2003; 8:355-66; PMID:12946308; http://dx.doi.org/10.1016/S1525-0016(03)00183-7
- Freund G, Sibler AP, Desplancq D, Oulad-Abdelghani M, Vigneron M, Gannon J, Van Regenmortel MH, Weiss E. Targeting endogenous nuclear antigens by electrotransfer of monoclonal antibodies in living cells. mAbs 2013; 5:518-22; PMID:23765067; http://dx.doi.org/10.4161/mabs.25084
- Marschall AL, Frenzel A, Schirrmann T, Schüngel M, Dübel S. Targeting antibodies to the cytoplasm. mAbs 2011; 3:3-16; PMID:21099369; http://dx.doi.org/10.4161/mabs.3.1.14110
- Marschall AL, Zhang C, Frenzel A, Schirrmann T, Hust M, Perez F, Dübel S. Delivery of antibodies to the cytosol: debunking the myths. mAbs 2014; 6:943-56; PMID:24848507; http://dx.doi.org/10.4161/mabs.29268
- Biocca S, Ruberti F, Tafani M, Pierandrei-Amaldi P, Cattaneo A. Redox state of single chain Fv fragments targeted to the endoplasmic reticulum, cytosol and mitochondria. Biotechnology (N Y) 1995; 13:1110-5; PMID:9636285; http://dx.doi.org/10.1038/nbt1095-1110
- Wörn A, Plückthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol 2001; 305:989-1010; PMID:11162109; http://dx.doi.org/10.1006/jmbi.2000.4265
- Vascotto F, Campagna M, Visintin M, Cattaneo A, Burrone OR. Effects of intrabodies specific for rotavirus NSP5 during the virus replicative cycle. J Gen Virol 2004; 85:3285-90; PMID:15483242; http://dx.doi.org/10.1099/vir.0.80075-0
- Dong J, Thompson AA, Fan Y, Lou J, Conrad F, Ho M, Pires-Alves M, Wilson BA, Stevens RC, Marks JD. A single-domain llama antibody potently inhibits the enzymatic activity of botulinum neurotoxin by binding to the non-catalytic α-exosite binding region. J Mol Biol 2010; 397:1106-18; PMID:20138889; http://dx.doi.org/10.1016/j.jmb.2010.01.070
- Tremblay JM, Kuo CL, Abeijon C, Sepulveda J, Oyler G, Hu X, Jin MM, Shoemaker CB. Camelid single domain antibodies (VHHs) as neuronal cell intrabody binding agents and inhibitors of Clostridium botulinum neurotoxin (BoNT) proteases. Toxicon 2010; 56:990-8; PMID:20637220; http://dx.doi.org/10.1016/j.toxicon.2010.07.003
- Gueorguieva D, Li S, Walsh N, Mukerji A, Tanha J, Pandey S. Identification of single-domain, Bax-specific intrabodies that confer resistance to mammalian cells against oxidative-stress-induced apoptosis. FASEB J 2006; 20:2636-8; PMID:17060401; http://dx.doi.org/10.1096/fj.06-6306fje
- Kaiser PD, Maier J, Traenkle B, Emele F, Rothbauer U. Recent progress in generating intracellular functional antibody fragments to target and trace cellular components in living cells. Biochim Biophys Acta 2014; 1844:1933-42; PMID:24792387; http://dx.doi.org/10.1016/j.bbapap.2014.04.019
- Rinaldi AS, Freund G, Desplancq D, Sibler AP, Baltzinger M, Rochel N, Mely Y, Didier P, Weiss E. The use of fluorescent intrabodies to detect endogenous gankyrin in living cancer cells. Exp Cell Res 2013; 319:838-49; PMID:23353833; http://dx.doi.org/10.1016/j.yexcr.2013.01.011
- Visintin M, Settanni G, Maritan A, Graziosi S, Marks JD, Cattaneo A. The intracellular antibody capture technology (IACT): towards a consensus sequence for intracellular antibodies. J Mol Biol 2002; 317:73-83; PMID:11916379; http://dx.doi.org/10.1006/jmbi.2002.5392
- Christ D, Famm K, Winter G. Repertoires of aggregation-resistant human antibody domains. Protein Eng Des Sel 2007; 20:413-6; http://dx.doi.org/10.1093/protein/gzm037
- Kim DY, Hussack G, Kandalaft H, Tanha J. Mutational approaches to improve the biophysical properties of human single-domain antibodies. Biochim Biophys Acta 2014; 1844:1983-2001; PMID:25065345; http://dx.doi.org/10.1016/j.bbapap.2014.07.008
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem 2013; 82:775-97; PMID:23495938; http://dx.doi.org/10.1146/annurev-biochem-063011-092449
- Cassimeris L, Guglielmi L, Denis V, Larroque C, Martineau P. Specific in vivo labeling of tyrosinated α-tubulin and measurement of microtubule dynamics using a GFP tagged, cytoplasmically expressed recombinant antibody. PloS one 2013; 8:e59812; PMID:23555790; http://dx.doi.org/10.1371/journal.pone.0059812
- Czajkowsky DM, Hu J, Shao Z, Pleass RJ. Fc-fusion proteins: new developments and future perspectives. EMBO Mol Med 2012; 4:1015-28; PMID:22837174; http://dx.doi.org/10.1002/emmm.201201379
- Guglielmi L, Denis V, Vezzio-Vie N, Bec N, Dariavach P, Larroque C, Martineau P. Selection for intrabody solubility in mammalian cells using GFP fusions. Protein Eng Des Sel 2011; 24:873-81; http://dx.doi.org/10.1093/protein/gzr049
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 2006; 24:79-88; PMID:16369541; http://dx.doi.org/10.1038/nbt1172
- Shaki-Loewenstein S, Zfania R, Hyland S, Wels WS, Benhar I. A universal strategy for stable intracellular antibodies. J Immunol Methods 2005; 303:19-39; PMID:16045924; http://dx.doi.org/10.1016/j.jim.2005.05.004
- Strube RW, Chen SY. Enhanced intracellular stability of sFv-Fc fusion intrabodies. Methods 2004; 34:179-83; PMID:15312671; http://dx.doi.org/10.1016/j.ymeth.2004.04.003
- Donini M, Morea V, Desiderio A, Pashkoulov D, Villani ME, Tramontano A, Benvenuto E. Engineering stable cytoplasmic intrabodies with designed specificity. J Mol Biol 2003; 330:323-32; PMID:12823971; http://dx.doi.org/10.1016/S0022-2836(03)00530-8
- Mandrup OA, Friis NA, Lykkemark S, Just J, Kristensen P. A novel heavy domain antibody library with functionally optimized complementarity determining regions. PloS One 2013; 8:e76834; PMID:24116173; http://dx.doi.org/10.1371/journal.pone.0076834
- Philibert P, Stoessel A, Wang W, Sibler AP, Bec N, Larroque C, Saven JG, Courtete J, Weiss E, Martineau P. A focused antibody library for selecting scFvs expressed at high levels in the cytoplasm. BMC Biotechnol 2007; 7:81; PMID:18034894; http://dx.doi.org/10.1186/1472-6750-7-81
- Wörn A, Auf der Maur A, Escher D, Honegger A, Barberis A, Plückthun A. Correlation between in vitro stability and in vivo performance of anti-GCN4 intrabodies as cytoplasmic inhibitors. J Biol Chem 2000; 275:2795-803; PMID:10644744; http://dx.doi.org/10.1074/jbc.275.4.2795
- Proba K, Wörn A, Honegger A, Plückthun A. Antibody scFv fragments without disulfide bonds made by molecular evolution. J Mol Biol 1998; 275:245-53; PMID:9466907; http://dx.doi.org/10.1006/jmbi.1997.1457
- Kakimoto S, Tanabe T, Azuma H, Nagasaki T. Enhanced internalization and endosomal escape of dual-functionalized poly(ethyleneimine)s polyplex with diphtheria toxin T and R domains. Biomed Pharmacother 2010; 64:296-301; PMID:20347568; http://dx.doi.org/10.1016/j.biopha.2009.06.017
- Qian Z, LaRochelle JR, Jiang B, Lian W, Hard RL, Selner NG, Luechapanichkul R, Barrios AM, Pei D. Early endosomal escape of a cyclic cell-penetrating peptide allows effective cytosolic cargo delivery. Biochemistry 2014; 53:4034-46; PMID:24896852; http://dx.doi.org/10.1021/bi5004102
- van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol 2005; 40:191-228; PMID:16126486; http://dx.doi.org/10.1080/10409230591008161
- Hardwick KG, Lewis MJ, Semenza J, Dean N, Pelham HR. ERD1, a yeast gene required for the retention of luminal endoplasmic reticulum proteins, affects glycoprotein processing in the Golgi apparatus. EMBO J 1990; 9:623-30; PMID:2178921
- Lewis MJ, Pelham HR. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell 1992; 68:353-64; PMID:1310258; http://dx.doi.org/10.1016/0092-8674(92)90476-S
- Donoso G, Herzog V, Schmitz A. Misfolded BiP is degraded by a proteasome-independent endoplasmic-reticulum-associated degradation pathway. Biochem J 2005; 387:897-903; PMID:15610068; http://dx.doi.org/10.1042/BJ20041312
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol 2005; 7:766-72; PMID:16056268; http://dx.doi.org/10.1038/ncb0805-766
- Schmitz A, Schneider A, Kummer MP, Herzog V. Endoplasmic reticulum-localized amyloid β-peptide is degraded in the cytosol by two distinct degradation pathways. Traffic 2004; 5:89-101; PMID:14690498; http://dx.doi.org/10.1111/j.1600-0854.2004.00159.x
- Böldicke T, Somplatzki S, Sergeev G, Mueller PP. Functional inhibition of transitory proteins by intrabody-mediated retention in the endoplasmatic reticulum. Methods 2012; 56:338-50; PMID:22037249; http://dx.doi.org/10.1016/j.ymeth.2011.10.008
- Dübel S, Breitling F, Fuchs P, Zewe M, Gotter S, Welschof M, Moldenhauer G, Little M. Isolation of IgG antibody Fv-DNA from various mouse and rat hybridoma cell lines using the polymerase chain reaction with a simple set of primers. J Immunol Methods 1994; 175:89-95; PMID:7930642; http://dx.doi.org/10.1016/0022-1759(94)90334-4
- Hust M, Frenzel A, Tomszak F, Kügler J, Dübel S. Antibody Phage Display. Handbook of Therapeutic Antibodies, 2nd ed. Weinheim, Germany: Wiley-VCH; 2014; 43-76.
- Breitling F, Dübel S, Seehaus T, Klewinghaus I, Little M. A surface expression vector for antibody screening. Gene 1991; 104:147-53; PMID:1916287; http://dx.doi.org/10.1016/0378-1119(91)90244-6
- Bradbury AR, Sidhu S, Dubel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 2011; 29:245-54; PMID:21390033; http://dx.doi.org/10.1038/nbt.1791
- Beerli RR, Bauer M, Buser RB, Gwerder M, Muntwiler S, Maurer P, Saudan P, Bachmann MF. Isolation of human monoclonal antibodies by mammalian cell display. Proc Natl Acad Sci U S A 2008; 105:14336-41; PMID:18812621; http://dx.doi.org/10.1073/pnas.0805942105
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 1997; 15:553-7; PMID:9181578; http://dx.doi.org/10.1038/nbt0697-553
- Samuelson P, Gunneriusson E, Nygren PA, Stahl S. Display of proteins on bacteria. J Biotechnol 2002; 96:129-54; PMID:12039531; http://dx.doi.org/10.1016/S0168-1656(02)00043-3
- Zhao XL, Chen WQ, Yang ZH, Li JM, Zhang SJ, Tian LF. Selection and affinity maturation of human antibodies against rabies virus from a scFv gene library using ribosome display. J Biotechnol 2009; 144:253-8; PMID:19818816; http://dx.doi.org/10.1016/j.jbiotec.2009.09.022
- Colwill K, Renewable Protein Binder Working G, Graslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods 2011; 8:551-8; PMID:21572409; http://dx.doi.org/10.1038/nmeth.1607
- Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple S, Clarke KF, Conquer JS, Crofts AM, Crowther SR, Dyson MR, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol 2007; 8:R254; PMID:18047641; http://dx.doi.org/10.1186/gb-2007-8-11-r254
- Sidhu SS. Antibodies for all: The case for genome-wide affinity reagents. FEBS Lett 2012; 586:2778-9; PMID:22664378; http://dx.doi.org/10.1016/j.febslet.2012.05.044
- Hust M, Frenzel A, Schirrmann T, Dübel S. Selection of recombinant antibodies from antibody gene libraries. Methods Mol Biol 2014; 1101:305-20; PMID:24233787; http://dx.doi.org/10.1007/978-1-62703-721-1_14
- Mersmann M, Meier D, Mersmann J, Helmsing S, Nilsson P, Gräslund S, Colwill K, Hust M, Dübel S. Towards proteome scale antibody selections using phage display. N Biotechnol 2010; 27:118-28; PMID:19883803; http://dx.doi.org/10.1016/j.nbt.2009.10.007
- Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23:1105-16; PMID:16151404; http://dx.doi.org/10.1038/nbt1126
- Hust M, Frenzel A, Meyer T, Schirrmann T, Dübel S. Construction of human naive antibody gene libraries. Methods Mol Biol 2012; 907:85-107; PMID:22907347; http://dx.doi.org/10.1007/978-1-61779-974-7_5
- Hayashi N, Welschof M, Zewe M, Braunagel M, Dübel S, Breitling F, Little M. Simultaneous mutagenesis of antibody CDR regions by overlap extension and PCR. Biotechniques 1994; 17:310, 2, 4-5.
- Rothe C, Urlinger S, Lohning C, Prassler J, Stark Y, Jager U, Hubner B, Bardroff M, Pradel I, Boss M, et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J Mol Biol 2008; 376:1182-200; PMID:18191144; http://dx.doi.org/10.1016/j.jmb.2007.12.018
- Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, Rem L, Frans N, Daukandt M, Pieters H, et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat Biotechnol 2005; 23:344-8; PMID:15723048; http://dx.doi.org/10.1038/nbt1067
- Zhang C, Helmsing S, Zagrebelsky M, Schirrmann T, Marschall AL, Schüngel M, Korte M, Hust M, Dübel S. Suppression of p75 neurotrophin receptor surface expression with intrabodies influences Bcl-xL mRNA expression and neurite outgrowth in PC12 cells. PloS One 2012; 7:e30684; PMID:22292018; http://dx.doi.org/10.1371/journal.pone.0030684
- Nam CH, Moutel S, Teillaud JL. Generation of murine scFv intrabodies from B-cell hybridomas. Methods Mol Biol 2002; 193:301-27; PMID:12325520
- Pope AR, Embleton MJ, Mernaugh R. Construction and use of antibody gene repertoires. In: Mc Cafferty J, Hoogenboom HR, Chiswell DJ (Eds.), Antibody engineering: A practical approach. New York, NY: Oxford University Press; 1996. p. 1–40.
- Toleikis L, Frenzel A. Cloning single-chain antibody fragments (ScFv) from hyrbidoma cells. Methods Mol Biol 2012; 907:59-71; PMID:22907345; http://dx.doi.org/10.1007/978-1-61779-974-7_3
- Ruberti F, Cattaneo A, Bradbury A. The use of the RACE method to clone hybridoma cDNA when V region primers fail. J Immunol Methods 1994; 173:33-9; PMID:8034983; http://dx.doi.org/10.1016/0022-1759(94)90280-1
- Ladiges W, Osman GE. Molecular characterization of immunoglobulin genes. In: Howard GC, Bethell DR (Eds.), Basic Methods in Antibody Production and Characterization. Boca Raton, FL: CRC Press Ltd; 2000:169-91.
- Persic L, Righi M, Roberts A, Hoogenboom HR, Cattaneo A, Bradbury A. Targeting vectors for intracellular immunisation. Gene 1997; 187:1-8; PMID:9073060; http://dx.doi.org/10.1016/S0378-1119(96)00627-0
- Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A 1988; 85:5879-83; PMID:3045807; http://dx.doi.org/10.1073/pnas.85.16.5879
- Levin R, Mhashilkar AM, Dorfman T, Bukovsky A, Zani C, Bagley J, Hinkula J, Niedrig M, Albert J, Wahren B, et al. Inhibition of early and late events of the HIV-1 replication cycle by cytoplasmic Fab intrabodies against the matrix protein, p17. Mol Med 1997; 3:96-110; PMID:9085253
- Reinman M, Jantti J, Alfthan K, Keranen S, Soderlund H, Takkinen K. Functional inactivation of the conserved Sem1p in yeast by intrabodies. Yeast 2003; 20:1071-84; PMID:12961755; http://dx.doi.org/10.1002/yea.1027
- Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, Kirsch MI, Meier D, Dübel S. Single chain Fab (scFab) fragment. BMC Biotechnol 2007; 7:14; PMID:17346344; http://dx.doi.org/10.1186/1472-6750-7-14
- Kontermann RE. Dual targeting strategies with bispecific antibodies. mAbs 2012; 4:182-97; PMID:22453100; http://dx.doi.org/10.4161/mabs.4.2.19000
- Jendreyko N, Popkov M, Beerli RR, Chung J, McGavern DB, Rader C, Barbas CF, 3rd. Intradiabodies, bispecific, tetravalent antibodies for the simultaneous functional knockout of two cell surface receptors. J Biol Chem 2003; 278:47812-9; PMID:12947084; http://dx.doi.org/10.1074/jbc.M307002200
- Jendreyko N, Popkov M, Rader C, Barbas CF, 3rd. Phenotypic knockout of VEGF-R2 and Tie-2 with an intradiabody reduces tumor growth and angiogenesis in vivo. Proc Natl Acad Sci U S A 2005; 102:8293-8; PMID:15928093; http://dx.doi.org/10.1073/pnas.0503168102
- Müller N, Hartmann C, Genssler S, Koch J, Kinner A, Grez M, Wels WS. A bispecific transmembrane antibody simultaneously targeting intra- and extracellular epitopes of the epidermal growth factor receptor inhibits receptor activation and tumor cell growth. Int J Cancer 2014; 134:2547-59; PMID:24243620; http://dx.doi.org/10.1002/ijc.28585
- Kim DS, Song HN, Nam HJ, Kim SG, Park YS, Park JC, Woo EJ, Lim HK. Directed evolution of human heavy chain variable domain (VH) using in vivo protein fitness filter. PloS One 2014; 9:e98178; PMID:24892548; http://dx.doi.org/10.1371/journal.pone.0098178
- Kim DY, To R, Kandalaft H, Ding W, van Faassen H, Luo Y, Schrag JD, St-Amant N, Hefford M, Hirama T, et al. Antibody light chain variable domains and their biophysically improved versions for human immunotherapy. mAbs 2014; 6:219-35; PMID:24423624; http://dx.doi.org/10.4161/mabs.26844
- Biocca S, Pierandrei-Amaldi P, Cattaneo A. Intracellular expression of anti-p21ras single chain Fv fragments inhibits meiotic maturation of xenopus oocytes. Biochem Biophys Res Commun 1993; 197:422-7; PMID:8267576; http://dx.doi.org/10.1006/bbrc.1993.2496
- Burke B, Warren G. Microinjection of mRNA coding for an anti-Golgi antibody inhibits intracellular transport of a viral membrane protein. Cell 1984; 36:847-56; PMID:6323023; http://dx.doi.org/10.1016/0092-8674(84)90034-5
- Valle G, Jones EA, Colman A. Anti-ovalbumin monoclonal antibodies interact with their antigen in internal membranes of Xenopus oocytes. Nature 1982; 300:71-4; PMID:7133132; http://dx.doi.org/10.1038/300071a0
- Kvam E, Nannenga BL, Wang MS, Jia Z, Sierks MR, Messer A. Conformational targeting of fibrillar polyglutamine proteins in live cells escalates aggregation and cytotoxicity. PloS one 2009; 4:e5727; PMID:19492089; http://dx.doi.org/10.1371/journal.pone.0005727
- Paoletti F, Malerba F, Konarev PV, Visintin M, Scardigli R, Fasulo L, Lamba D, Svergun DI, Cattaneo A. Direct intracellular selection and biochemical characterization of a recombinant anti-proNGF single chain antibody fragment. Arch Biochem Biophys 2012; 522:26-36; PMID:22516657; http://dx.doi.org/10.1016/j.abb.2012.04.003
- Zacchi P, Dreosti E, Visintin M, Moretto-Zita M, Marchionni I, Cannistraci I, Kasap Z, Betz H, Cattaneo A, Cherubini E. Gephyrin selective intrabodies as a new strategy for studying inhibitory receptor clustering. J Mol Neurosci 2008; 34:141-8; PMID:18008186; http://dx.doi.org/10.1007/s12031-007-9018-6
- Dauvillier S, Merida P, Visintin M, Cattaneo A, Bonnerot C, Dariavach P. Intracellular single-chain variable fragments directed to the Src homology 2 domains of Syk partially inhibit Fc epsilon RI signaling in the RBL-2H3 cell line. J Immunol 2002; 169:2274-83; http://dx.doi.org/10.4049/jimmunol.169.5.2274
- Tse E, Rabbitts TH. Intracellular antibody-caspase-mediated cell killing: an approach for application in cancer therapy. Proc Natl Acad Sci U S A 2000; 97:12266-71; PMID:11050246; http://dx.doi.org/10.1073/pnas.97.22.12266
- Staus DP, Wingler LM, Strachan RT, Rasmussen SG, Pardon E, Ahn S, Steyaert J, Kobilka BK, Lefkowitz RJ. Regulation of beta2-adrenergic receptor function by conformationally selective single-domain intrabodies. Mol Pharmacol 2014; 85:472-81; PMID:24319111; http://dx.doi.org/10.1124/mol.113.089516
- Chiusaroli R, Visentini M, Galimberti C, Casseler C, Mennuni L, Covaceuszach S, Lanza M, Ugolini G, Caselli G, Rovati LC, et al. Targeting of ADAMTS5s ancillary domain with the recombinant mAb CRB0017 ameliorates disease progression in a spontaneous murine model of osteoarthritis. Osteoarthritis Cartilage 2013; 21:1807-10; PMID:23954517; http://dx.doi.org/10.1016/j.joca.2013.08.015
- Liu Y, Sun L, Yu P, Li A, Li C, Tang Q, Li D, Liang M. Viral suppression function of intracellular antibody against C-terminal domain of rabies virus phosphoprotein. Acta Biochim Biophys Sin 2015; http://dx.doi.org/10.1093/abbs/gmv060
- Auf der Maur A, Tissot K, Barberis A. Antigen-independent selection of intracellular stable antibody frameworks. Methods 2004; 34:215-24; PMID:15312674; http://dx.doi.org/10.1016/j.ymeth.2004.04.004
- Visintin M, Meli GA, Cannistraci I, Cattaneo A. Intracellular antibodies for proteomics. J Immunol Methods 2004; 290:135-53; PMID:15261577; http://dx.doi.org/10.1016/j.jim.2004.04.014
- Visintin M, Melchionna T, Cannistraci I, Cattaneo A. In vivo selection of intrabodies specifically targeting protein-protein interactions: a general platform for an “undruggable” class of disease targets. J Biotechnol 2008; 135:1-15; PMID:18395925; http://dx.doi.org/10.1016/j.jbiotec.2008.02.012
- Fisher AC, DeLisa MP. Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery. J Mol Biol 2009; 385:299-311; PMID:18992254; http://dx.doi.org/10.1016/j.jmb.2008.10.051
- Karlsson AJ, Lim HK, Xu H, Rocco MA, Bratkowski MA, Ke A, DeLisa MP. Engineering antibody fitness and function using membrane-anchored display of correctly folded proteins. J Mol Biol 2012; 416:94-107; PMID:22197376; http://dx.doi.org/10.1016/j.jmb.2011.12.021
- Waraho D, DeLisa MP. Versatile selection technology for intracellular protein-protein interactions mediated by a unique bacterial hitchhiker transport mechanism. Proc Natl Acad Sci U S A 2009; 106:3692-7; PMID:19234130; http://dx.doi.org/10.1073/pnas.0704048106
- Waraho-Zhmayev D, Meksiriporn B, Portnoff AD, DeLisa M. Optimizing recombinant antibodies for intracellular function using hitchhiker-mediated survival selection. Protein Eng Des Sel 2014; 27: 351-358; PMID:25225416; http://dx.doi.org/10.1093/protein/gzu038
- Sacchetti A, Cappetti V, Marra P, Dell'Arciprete R, El Sewedy T, Crescenzi C, Alberti S. Green fluorescent protein variants fold differentially in prokaryotic and eukaryotic cells. J Cell Biochem Suppl 2001; Suppl 36:117-28; PMID:11455577; http://dx.doi.org/10.1002/jcb.1091
- Cohen PA, Mani JC, Lane DP. Characterization of a new intrabody directed against the N-terminal region of human p53. Oncogene 1998; 17:2445-56; PMID:9824155; http://dx.doi.org/10.1038/sj.onc.1202190
- Jannot CB, Hynes NE. Characterization of scFv-421, a single-chain antibody targeted to p53. Biochem Biophys Res Commun 1997; 230:242-6; PMID:9016757; http://dx.doi.org/10.1006/bbrc.1996.5930
- Zheng L, Baumann U, Reymond JL. Production of a functional catalytic antibody ScFv-NusA fusion protein in bacterial cytoplasm. J Biochem 2003; 133:577-81; PMID:12801908; http://dx.doi.org/10.1093/jb/mvg074
- Caron de Fromentel C, Gruel N, Venot C, Debussche L, Conseiller E, Dureuil C, Teillaud JL, Tocque B, Bracco L. Restoration of transcriptional activity of p53 mutants in human tumour cells by intracellular expression of anti-p53 single chain Fv fragments. Oncogene 1999; 18:551-7; PMID:9927212; http://dx.doi.org/10.1038/sj.onc.1202338
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 1986; 234:364-8; PMID:2876518; http://dx.doi.org/10.1126/science.2876518
- Joshi SN, Butler DC, Messer A. Fusion to a highly charged proteasomal retargeting sequence increases soluble cytoplasmic expression and efficacy of diverse anti-synuclein intrabodies. mAbs 2012; 4:686-93; PMID:22929188; http://dx.doi.org/10.4161/mabs.21696
- Butler DC, Messer A. Bifunctional anti-huntingtin proteasome-directed intrabodies mediate efficient degradation of mutant huntingtin exon 1 protein fragments. PloS One 2011; 6:e29199; PMID:22216210; http://dx.doi.org/10.1371/journal.pone.0029199
- Raran-Kurussi S, Waugh DS. The ability to enhance the solubility of its fusion partners is an intrinsic property of maltose-binding protein but their folding is either spontaneous or chaperone-mediated. PloS One 2012; 7:e49589; PMID:23166722; http://dx.doi.org/10.1371/journal.pone.0049589
- Nallamsetty S, Waugh DS. Mutations that alter the equilibrium between open and closed conformations of Escherichia coli maltose-binding protein impede its ability to enhance the solubility of passenger proteins. Biochem Biophys Res Commun 2007; 364:639-44; PMID:17964542; http://dx.doi.org/10.1016/j.bbrc.2007.10.060
- Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 1986; 321:522-5; PMID:3713831; http://dx.doi.org/10.1038/321522a0
- Mazuc E, Guglielmi L, Bec N, Parez V, Hahn CS, Mollevi C, Parrinello H, Desvignes JP, Larroque C, Jupp R, et al. In-cell intrabody selection from a diverse human library identifies C12orf4 protein as a new player in rodent mast cell degranulation. PloS One 2014; 9:e104998; PMID:25122211; http://dx.doi.org/10.1371/journal.pone.0104998
- Doyle PJ, Saeed H, Hermans A, Gleddie SC, Hussack G, Arbabi-Ghahroudi M, Seguin C, Savard ME, Mackenzie CR, Hall JC. Intracellular expression of a single domain antibody reduces cytotoxicity of 15-acetyldeoxynivalenol in yeast. J Biol Chem 2009; 284:35029-39; PMID:19783651; http://dx.doi.org/10.1074/jbc.M109.045047
- McGonigal K, Tanha J, Palazov E, Li S, Gueorguieva-Owens D, Pandey S. Isolation and functional characterization of single domain antibody modulators of Caspase-3 and apoptosis. Appl Biochem Biotechnol 2009; 157:226-36; PMID:18553063; http://dx.doi.org/10.1007/s12010-008-8266-4
- Verheesen P, de Kluijver A, van Koningsbruggen S, de Brij M, de Haard HJ, van Ommen GJ, van der Maarel SM, Verrips CT. Prevention of oculopharyngeal muscular dystrophy-associated aggregation of nuclear polyA-binding protein with a single-domain intracellular antibody. Hum Mol Genet 2006; 15:105-11; PMID:16319127; http://dx.doi.org/10.1093/hmg/ddi432
- Newnham LE, Wright MJ, Holdsworth G, Kostarelos K, Robinson MK, Rabbitts TH, Lawson AD. Functional inhibition of β-catenin-mediatedWnt signaling by intracellular VHHantibodies. mAbs 2015; 7:180-91; PMID:25524068; http://dx.doi.org/10.4161/19420862.2015.989023
- Jespers L, Schon O, Famm K, Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat Biotechnol 2004; 22:1161-5; PMID:15300256; http://dx.doi.org/10.1038/nbt1000
- Barthelemy PA, Raab H, Appleton BA, Bond CJ, Wu P, Wiesmann C, Sidhu SS. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J Biol Chem 2008; 283:3639-54; PMID:18045863; http://dx.doi.org/10.1074/jbc.M708536200
- Yasui H, Ito W, Kurosawa Y. Effects of substitutions of amino acids on the thermal stability of the Fv fragments of antibodies. FEBS Lett 1994; 353:143-6; PMID:7926039; http://dx.doi.org/10.1016/0014-5793(94)01027-7
- Jager M, Plückthun A. Folding and assembly of an antibody Fv fragment, a heterodimer stabilized by antigen. J Mol Biol 1999; 285:2005-19; PMID:9925781; http://dx.doi.org/10.1006/jmbi.1998.2425
- Dübel S, Stoevesandt O, Taussig MJ, Hust M. Generating recombinant antibodies to the complete human proteome. Trends Biotechnol 2010; 28:333-9; PMID:20538360; http://dx.doi.org/10.1016/j.tibtech.2010.05.001
- Popkov M, Jendreyko N, McGavern DB, Rader C, Barbas CF, 3rd. Targeting tumor angiogenesis with adenovirus-delivered anti-Tie-2 intrabody. Cancer Res 2005; 65:972-81; PMID:15705898
- Sudol KL, Mastrangelo MA, Narrow WC, Frazer ME, Levites YR, Golde TE, Federoff HJ, Bowers WJ. Generating differentially targeted amyloid-β specific intrabodies as a passive vaccination strategy for Alzheimer disease. Mol Ther 2009; 17:2031-40; PMID:19638957; http://dx.doi.org/10.1038/mt.2009.174
- Gahrtz M, Conrad U. Immunomodulation of plant function by in vitro selected single-chain Fv intrabodies. Methods Mol Biol 2009; 483:289-312; PMID:19183906; http://dx.doi.org/10.1007/978-1-59745-407-0_17
- Marschall AL, Single FN, Schlarmann K, Bosio A, Strebe N, van den Heuvel J, Frenzel A, Dübel S. Functional knock down of VCAM1 in mice mediated by endoplasmatic reticulum retained intrabodies. mAbs 2014; 6:1394-401.
- Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet 2011; 12:316-28; PMID:21468099; http://dx.doi.org/10.1038/nrg2971
- Knight S, Collins M, Takeuchi Y. Insertional mutagenesis by retroviral vectors: current concepts and methods of analysis. Curr Gene Ther 2013; 13:211-27; PMID:23590635; http://dx.doi.org/10.2174/1566523211313030006
- Jin L, Zeng X, Liu M, Deng Y, He N. Current progress in gene delivery technology based on chemical methods and nano-carriers. Theranostics 2014; 4:240-55; PMID:24505233; http://dx.doi.org/10.7150/thno.6914
- Papayannakos C, Daniel R. Understanding lentiviral vector chromatin targeting: working to reduce insertional mutagenic potential for gene therapy. Gene Ther 2013; 20:581-8; PMID:23171920; http://dx.doi.org/10.1038/gt.2012.88
- Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet 2007; 8:573-87; PMID:17607305; http://dx.doi.org/10.1038/nrg2141
- Kallen KJ, Thess A. A development that may evolve into a revolution in medicine: mRNA as the basis for novel, nucleotide-based vaccines and drugs. Ther Adv Vaccines 2014; 2:10-31; PMID:24757523; http://dx.doi.org/10.1177/2051013613508729
- Beerli RR, Wels W, Hynes NE. Inhibition of signaling from Type 1 receptor tyrosine kinases via intracellular expression of single-chain antibodies. Breast Cancer Res Treat 1996; 38:11-7; PMID:8825118; http://dx.doi.org/10.1007/BF01803779
- Harwerth IM, Wels W, Schlegel J, Muller M, Hynes NE. Monoclonal antibodies directed to the erbB-2 receptor inhibit in vivo tumour cell growth. British J Cancer 1993; 68:1140-5; PMID:7903153; http://dx.doi.org/10.1038/bjc.1993.494
- Sato JD, Kawamoto T, Le AD, Mendelsohn J, Polikoff J, Sato GH. Biological effects in vitro of monoclonal antibodies to human epidermal growth factor receptors. Mol Biol Med 1983; 1:511-29; PMID:6094961
- Wels W, Beerli R, Hellmann P, Schmidt M, Marte BM, Kornilova ES, Hekele A, Mendelsohn J, Groner B, Hynes NE. EGF receptor and p185erbB-2-specific single-chain antibody toxins differ in their cell-killing activity on tumor cells expressing both receptor proteins. Int J Cancer 1995; 60:137-44; PMID:7814146; http://dx.doi.org/10.1002/ijc.2910600120
- Arafat W, Gomez-Navarro J, Xiang J, Siegal GP, Alvarez RD, Curiel DT. Antineoplastic effect of anti-erbB-2 intrabody is not correlated with scFv affinity for its target. Cancer Gene Ther 2000; 7:1250-6; PMID:11023197; http://dx.doi.org/10.1038/sj.cgt.7700228
- Grim JE, Siegal GP, Alvarez RD, Curiel DT. Intracellular expression of the anti-erbB-2 sFv N29 fails to accomplish efficient target modulation. Bioch Biophys Res Commun 1998; 250:699-703; PMID:9784409; http://dx.doi.org/10.1006/bbrc.1998.9391
- Graus-Porta D, Beerli RR, Hynes NE. Single-chain antibody-mediated intracellular retention of ErbB-2 impairs Neu differentiation factor and epidermal growth factor signaling. Mol Cell Biol 1995; 15:1182-91; PMID:7532277
- Richardson JH, Sodroski JG, Waldmann TA, Marasco WA. Phenotypic knockout of the high-affinity human interleukin 2 receptor by intracellular single-chain antibodies against the α subunit of the receptor. Proc Natl Acad Sci U S A 1995; 92:3137-41; PMID:7724529; http://dx.doi.org/10.1073/pnas.92.8.3137
- Strebe N, Guse A, Schüngel M, Schirrmann T, Hafner M, Jostock T, Hust M, Müller W, Dübel S. Functional knockdown of VCAM-1 at the posttranslational level with ER retained antibodies. J Immunol Methods 2009; 341:30-40; PMID:19038261; http://dx.doi.org/10.1016/j.jim.2008.10.012
- Beerli RR, Wels W, Hynes NE. Intracellular expression of single chain antibodies reverts ErbB-2 transformation. J Biol Chem 1994; 269:23931-6; PMID:7929040
- Richardson JH, Waldmann TA, Sodroski JG, Marasco WA. Inducible knockout of the interleukin-2 receptor α chain: expression of the high-affinity IL-2 receptor is not required for the in vitro growth of HTLV-I-transformed cell lines. Virology 1997; 237:209-16; PMID:9356333; http://dx.doi.org/10.1006/viro.1997.8779
- Yuan Q, Strauch KL, Lobb RR, Hemler ME. Intracellular single-chain antibody inhibits integrin VLA-4 maturation and function. Biochem J 1996; 318 ( Pt 2):591-6; PMID:8809051
- Böldicke T, Weber H, Mueller PP, Barleon B, Bernal M. Novel highly efficient intrabody mediates complete inhibition of cell surface expression of the human vascular endothelial growth factor receptor-2 (VEGFR-2/KDR). J Immunol Methods 2005; 300:146-59; PMID:15946674; http://dx.doi.org/10.1016/j.jim.2005.03.007
- Paganetti P, Calanca V, Galli C, Stefani M, Molinari M. β-site specific intrabodies to decrease and prevent generation of Alzheimer Abeta peptide. J Cell Biol 2005; 168:863-8; PMID:15767460; http://dx.doi.org/10.1083/jcb.200410047
- Beerli RR, Wels W, Hynes NE. Autocrine inhibition of the epidermal growth factor receptor by intracellular expression of a single-chain antibody. Biochem Biophys Res Commun 1994; 204:666-72; PMID:7980527; http://dx.doi.org/10.1006/bbrc.1994.2511
- Pelham HR. The dynamic organisation of the secretory pathway. Cell Struct Funct 1996; 21:413-9; PMID:9118249; http://dx.doi.org/10.1247/csf.21.413
- Chen X, Ye H, Kuruvilla R, Ramanan N, Scangos KW, Zhang C, Johnson NM, England PM, Shokat KM, Ginty DD. A chemical-genetic approach to studying neurotrophin signaling. Neuron 2005; 46:13-21; PMID:15820690; http://dx.doi.org/10.1016/j.neuron.2005.03.009
- Ruberti F, Capsoni S, Comparini A, Di Daniel E, Franzot J, Gonfloni S, Rossi G, Berardi N, Cattaneo A. Phenotypic knockout of nerve growth factor in adult transgenic mice reveals severe deficits in basal forebrain cholinergic neurons, cell death in the spleen, and skeletal muscle dystrophy. J Neurosci 2000; 20:2589-601; PMID:10729339
- Capsoni S, Tiveron C, Vignone D, Amato G, Cattaneo A. Dissecting the involvement of tropomyosin-related kinase A and p75 neurotrophin receptor signaling in NGF deficit-induced neurodegeneration. Proc Natl Acad Sci U S A 2010; 107:12299-304; PMID:20566851; http://dx.doi.org/10.1073/pnas.1007181107
- Shakespeare WC. SH2 domain inhibition: a problem solved? Curr Opin Chem Biol 2001; 5:409-15; PMID:11470604; http://dx.doi.org/10.1016/S1367-5931(00)00222-2
- Mandal PK, Gao F, Lu Z, Ren Z, Ramesh R, Birtwistle JS, Kaluarachchi KK, Chen X, Bast RC, Jr., Liao WS, et al. Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3. J Med Chem 2011; 54:3549-63; PMID:21486047; http://dx.doi.org/10.1021/jm2000882
- Hojjat-Farsangi M. Small-Molecule Inhibitors of the Receptor Tyrosine Kinases: Promising Tools for Targeted Cancer Therapies. Int J Mol Sci 2014; 15:13768-801; PMID:25110867; http://dx.doi.org/10.3390/ijms150813768
- Liu S, Kurzrock R. Toxicity of targeted therapy: Implications for response and impact of genetic polymorphisms. Cancer Treat Rev 2014; 40:883-91; PMID:24867380; http://dx.doi.org/10.1016/j.ctrv.2014.05.003
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 2000; 351:95-105; PMID:10998351; http://dx.doi.org/10.1042/0264-6021:3510095
- Graham KL, Lee LY, Higgins JP, Steinman L, Utz PJ, Ho PP. Treatment with a toll-like receptor inhibitory GpG oligonucleotide delays and attenuates lupus nephritis in NZB/W mice. Autoimmunity 2010; 43:140-55; PMID:19845477; http://dx.doi.org/10.3109/08916930903229239
- Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol 2006; 6:823-35; PMID:17063184; http://dx.doi.org/10.1038/nri1957
- Braden BC, Goldbaum FA, Chen BX, Kirschner AN, Wilson SR, Erlanger BF. X-ray crystal structure of an anti-Buckminsterfullerene antibody fab fragment: biomolecular recognition of C(60). Proc Natl Acad Sci U S A 2000; 97:12193-7; PMID:11035793; http://dx.doi.org/10.1073/pnas.210396197
- Sheppard D. Dominant negative mutants: tools for the study of protein function in vitro and in vivo. Am J Respir Cell Mol Biol 1994; 11:1-6; PMID:8018332; http://dx.doi.org/10.1165/ajrcmb.11.1.8018332
- Hust M, Meyer T, Voedisch B, Rülker T, Thie H, El-Ghezal A, Kirsch MI, Schütte M, Helmsing S, Meier D, et al. A human scFv antibody generation pipeline for proteome research. J Biotechnol 2011; 152:159-70; PMID:20883731; http://dx.doi.org/10.1016/j.jbiotec.2010.09.945
- Geyer CR, McCafferty J, Dubel S, Bradbury AR, Sidhu SS. Recombinant antibodies and in vitro selection technologies. Methods Mol Biol 2012; 901:11-32; PMID:22723092; http://dx.doi.org/10.1007/978-1-61779-931-0_2
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411:494-8; PMID:11373684; http://dx.doi.org/10.1038/35078107
- Schmidt FR. About the nature of RNA interference. Appl Microbiol Biotechnol 2005; 67:429-35; PMID:15703909; http://dx.doi.org/10.1007/s00253-004-1882-1
- Cottingham K. Antibodypedia seeks to answer the question: “how good is that antibody?.” J Proteome Res 2008; 7:4213.
- Bourbeillon J, Orchard S, Benhar I, Borrebaeck C, de Daruvar A, Dubel S, Frank R, Gibson F, Gloriam D, Haslam N, et al. Minimum information about a protein affinity reagent (MIAPAR). Nat Biotechnol 2010; 28:650-3; PMID:20622827; http://dx.doi.org/10.1038/nbt0710-650
- Cao T, Heng BC. Intracellular antibodies (intrabodies) versus RNA interference for therapeutic applications. Ann Clin Lab Sci 2005; 35:227-9; PMID:16081577
- Wang QC, Nie QH, Feng ZH. RNA interference: antiviral weapon and beyond. World J Gastroenterol 2003; 9:1657-61; PMID:12918096
- Montgomery MK. RNA interference: historical overview and significance. Methods Mol Biol 2004; 265:3-21; PMID:15103066
- Zhou R, Rana TM. RNA-based mechanisms regulating host-virus interactions. Immunol Rev 2013; 253:97-111; PMID:23550641; http://dx.doi.org/10.1111/imr.12053
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 2003; 5:834-9; PMID:12942087; http://dx.doi.org/10.1038/ncb1038
- Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs? Trends Genet 2004; 20:521-4; PMID:15475108; http://dx.doi.org/10.1016/j.tig.2004.08.006
- Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA (New York, NY) 2004; 10:12-8; http://dx.doi.org/10.1261/rna5160904
- Sledz CA, Williams BR. RNA interference and double-stranded-RNA-activated pathways. Biochem Soc Trans 2004; 32:952-6; PMID:15506933; http://dx.doi.org/10.1042/BST0320952
- Snove O, Jr., Holen T. Many commonly used siRNAs risk off-target activity. Biochem Biophys Res Commun 2004; 319:256-63; PMID:15158470; http://dx.doi.org/10.1016/j.bbrc.2004.04.175
- Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov 2010; 9:57-67; PMID:20043028; http://dx.doi.org/10.1038/nrd3010
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol 2005; 23:457-62; PMID:15778705; http://dx.doi.org/10.1038/nbt1081
- de Fougerolles AR. Delivery vehicles for small interfering RNA in vivo. Hum Gene Ther 2008; 19:125-32; PMID:18257677; http://dx.doi.org/10.1089/hum.2008.928
- Kleinhammer A, Wurst W, Kuhn R. Constitutive and conditional RNAi transgenesis in mice. Methods 2011; 53:430-6; PMID:21184828; http://dx.doi.org/10.1016/j.ymeth.2010.12.015
- Paroo Z, Corey DR. Challenges for RNAi in vivo. Trends Biotechnol 2004; 22:390-4; PMID:15283982; http://dx.doi.org/10.1016/j.tibtech.2004.06.004
- Cejka D, Losert D, Wacheck V. Short interfering RNA (siRNA): tool or therapeutic? Clin Sci (Lond) 2006; 110:47-58; PMID:16336204; http://dx.doi.org/10.1042/CS20050162
- Wirth T, Parker N, Yla-Herttuala S. History of gene therapy. Gene 2013; 525:162-9; PMID:23618815; http://dx.doi.org/10.1016/j.gene.2013.03.137
- Dickerson JE, Zhu A, Robertson DL, Hentges KE. Defining the role of essential genes in human disease. PloS One 2011; 6:e27368; PMID:22096564; http://dx.doi.org/10.1371/journal.pone.0027368
- Li JY, Sugimura K, Boado RJ, Lee HJ, Zhang C, Duebel S, Pardridge WM. Genetically engineered brain drug delivery vectors: cloning, expression and in vivo application of an anti-transferrin receptor single chain antibody-streptavidin fusion gene and protein. Protein Eng 1999; 12:787-96; PMID:10506289; http://dx.doi.org/10.1093/protein/12.9.787
- Blatt NB, Bill RM, Glick GD. Characterization of a unique anti-DNA hybridoma. Hybridoma 1998; 17:33-9; PMID:9523235; http://dx.doi.org/10.1089/hyb.1998.17.33
- Bradbury A. Antibody Reproducibility challenges: The Solution lies in the sequence. Nature 2015; in press.
- Toleikis L, Broders O, Dübel S. Cloning single chain antibody fragments (scFv) from hybridoma clones. In Decker J, Reischl U (Eds.), Molecular Diagnosis of Infectious Diseases, 2nd ed. Totowa, NY: Humana Press Inc; 2004:447-58.
- Zack DJ, Wong AL, Stempniak M, Weisbart RH. Two kappa immunoglobulin light chains are secreted by an anti-DNA hybridoma: implications for isotypic exclusion. Mol Immunol 1995; 32:1345-53; PMID:8643104; http://dx.doi.org/10.1016/0161-5890(95)00112-3
- Strebe N, Breitling F, Moosmayer D, Brocks D, Dübel S. Cloning of Variable domains from mouse hybridoma by PCR. In Antibody Engineering (Vol. 1; 2nd ed.). Berlin: Springer Protocols; 2010:3-14.
- Wheeler YY, Kute TE, Willingham MC, Chen SY, Sane DC. Intrabody-based strategies for inhibition of vascular endothelial growth factor receptor-2: effects on apoptosis, cell growth, and angiogenesis. FASEB J 2003; 17:1733-5; PMID:12958192
- Alvarez RD, Barnes MN, Gomez-Navarro J, Wang M, Strong TV, Arafat W, Arani RB, Johnson MR, Roberts BL, Siegal GP, et al. A cancer gene therapy approach utilizing an anti-erbB-2 single-chain antibody-encoding adenovirus (AD21): a phase I trial. Clin Cancer Res 2000; 6:3081-7; PMID:10955787
- Deshane J, Siegal GP, Wang M, Wright M, Bucy RP, Alvarez RD, Curiel DT. Transductional efficacy and safety of an intraperitoneally delivered adenovirus encoding an anti-erbB-2 intracellular single-chain antibody for ovarian cancer gene therapy. Gynecol Oncol 1997; 64:378-85; PMID:9062138; http://dx.doi.org/10.1006/gyno.1996.4566
- Grim J, Deshane J, Siegal GP, Alvarez RD, DiFiore P, Curiel DT. The level of erbB2 expression predicts sensitivity to the cytotoxic effects of an intracellular anti-erbB2 sFv. J Mol Med 1998; 76:451-8; PMID:9625302; http://dx.doi.org/10.1007/s001090050237
- Wright M, Grim J, Deshane J, Kim M, Strong TV, Siegal GP, Curiel DT. An intracellular anti-erbB-2 single-chain antibody is specifically cytotoxic to human breast carcinoma cells overexpressing erbB-2. Gene Ther 1997; 4:317-22; PMID:9176517; http://dx.doi.org/10.1038/sj.gt.3300372
- Jannot CB, Beerli RR, Mason S, Gullick WJ, Hynes NE. Intracellular expression of a single-chain antibody directed to the EGFR leads to growth inhibition of tumor cells. Oncogene 1996; 13:275-82; PMID:8710366
- Wang W, Zhou J, Xu L, Zhen Y. Antineoplastic effect of intracellular expression of a single-chain antibody directed against type IV collagenase. J Environ Pathol Toxicol Oncol 2000; 19:61-8; PMID:10905509
- Sangboonruang S, Thammasit P, Intasai N, Kasinrerk W, Tayapiwatana C, Tragoolpua K. EMMPRIN reduction via scFv-M6-1B9 intrabody affects alpha3beta1-integrin and MCT1 functions and results in suppression of progressive phenotype in the colorectal cancer cell line Caco-2. Cancer Gene Ther 2014; 21:246-55; PMID:24924201; http://dx.doi.org/10.1038/cgt.2014.24
- Thammasit P, Sangboonruang S, Suwanpairoj S, Khamaikawin W, Intasai N, Kasinrerk W, Tayapiwatana C, Tragoolpua K. Intracellular Acidosis Promotes Mitochondrial Apoptosis Pathway: Role of EMMPRIN Down-regulation via Specific Single-chain Fv Intrabody. J Cancer 2015; 6:276-86; PMID:25663946; http://dx.doi.org/10.7150/jca.10879
- Figini M, Ferri R, Mezzanzanica D, Bagnoli M, Luison E, Miotti S, Canevari S. Reversion of transformed phenotype in ovarian cancer cells by intracellular expression of anti folate receptor antibodies. Gene Ther 2003; 10:1018-25; PMID:12776159; http://dx.doi.org/10.1038/sj.gt.3301962
- Guillaume-Rousselet N, Jean D, Frade R. Cloning and characterization of anti-cathepsin L single chain variable fragment whose expression inhibits procathepsin L secretion in human melanoma cells. Biochem J 2002; 367:219-27; PMID:12241546; http://dx.doi.org/10.1042/BJ20020350
- Accardi L, Paolini F, Mandarino A, Percario Z, Di Bonito P, Di Carlo V, Affabris E, Giorgi C, Amici C, Venuti A. In vivo antitumor effect of an intracellular single-chain antibody fragment against the E7 oncoprotein of human papillomavirus 16. Int J Cancer 2014; 134:2742-7; PMID:24226851; http://dx.doi.org/10.1002/ijc.28604
- Rondon IJ, Marasco WA. Intracellular antibodies (intrabodies) for gene therapy of infectious diseases. Annu Rev Microbiol 1997; 51:257-83; PMID:9343351; http://dx.doi.org/10.1146/annurev.micro.51.1.257
- Poznansky MC, Foxall R, Mhashilkar A, Coker R, Jones S, Ramstedt U, Marasco W. Inhibition of human immunodeficiency virus replication and growth advantage of CD4+ T cells from HIV-infected individuals that express intracellular antibodies against HIV-1 gp120 or Tat. Hum Gene Ther 1998; 9:487-96; PMID:9525310; http://dx.doi.org/10.1089/hum.1998.9.4-487
- Zhou P, Goldstein S, Devadas K, Tewari D, Notkins AL. Cells transfected with a non-neutralizing antibody gene are resistant to HIV infection: targeting the endoplasmic reticulum and trans-Golgi network. J Immunol 1998; 160:1489-96.
- Walsh R, Nuttall S, Revill P, Colledge D, Cabuang L, Soppe S, Dolezal O, Griffiths K, Bartholomeusz A, Locarnini S. Targeting the hepatitis B virus precore antigen with a novel IgNAR single variable domain intrabody. Virology 2011; 411:132-41; PMID:21239030; http://dx.doi.org/10.1016/j.virol.2010.12.034
- Liao W, Strube RW, Milne RW, Chen SY, Chan L. Cloning of apoB intrabodies: specific knockdown of apoB in HepG2 cells. Biochem Biophys Res Commun 2008; 373:235-40; PMID:18558087; http://dx.doi.org/10.1016/j.bbrc.2008.06.020
- Heintges T, zu Putlitz J, Wands JR. Characterization and binding of intracellular antibody fragments to the hepatitis C virus core protein. Biochem Biophys Res Commun 1999; 263:410-8; PMID:10491307; http://dx.doi.org/10.1006/bbrc.1999.1350
- Blazek D, Celer V, Navratilova I, Skladal P. Generation and characterization of single-chain antibody fragments specific against transmembrane envelope glycoprotein gp46 of maedi-visna virus. J Virol Methods 2004; 115:83-92; PMID:14656464; http://dx.doi.org/10.1016/j.jviromet.2003.09.020
- Steinberger P, Andris-Widhopf J, Buhler B, Torbett BE, Barbas CF, 3rd. Functional deletion of the CCR5 receptor by intracellular immunization produces cells that are refractory to CCR5-dependent HIV-1 infection and cell fusion. Proc Natl Acad Sci U S A 2000; 97:805-10; PMID:10639161; http://dx.doi.org/10.1073/pnas.97.2.805
- Swan CH, Buhler B, Steinberger P, Tschan MP, Barbas CF, 3rd, Torbett BE. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery. Gene Ther 2006; 13:1480-92; PMID:16738691; http://dx.doi.org/10.1038/sj.gt.3302801
- Cordelier P, Kulkowsky JW, Ko C, Matskevitch AA, McKee HJ, Rossi JJ, Bouhamdan M, Pomerantz RJ, Kari G, Strayer DS. Protecting from R5-tropic HIV: individual and combined effectiveness of a hammerhead ribozyme and a single-chain Fv antibody that targets CCR5. Gene Ther 2004; 11:1627-37; PMID:15295615; http://dx.doi.org/10.1038/sj.gt.3302329
- BouHamdan M, Strayer DS, Wei D, Mukhtar M, Duan LX, Hoxie J, Pomerantz RJ. Inhibition of HIV-1 infection by down-regulation of the CXCR4 co-receptor using an intracellular single chain variable fragment against CXCR4. Gene Ther 2001; 8:408-18; PMID:11313818; http://dx.doi.org/10.1038/sj.gt.3301411
- Mukhtar M, Acheampong E, Khan MA, Bouhamdan M, Pomerantz RJ. Down-modulation of the CXCR4 co-receptor by intracellular expression of a single chain variable fragment (SFv) inhibits HIV-1 entry into primary human brain microvascular endothelial cells and post-mitotic neurons. Brain Res Mol Brain Res 2005; 135:48-57; PMID:15857668; http://dx.doi.org/10.1016/j.molbrainres.2004.11.015
- Beyer F, Doebis C, Busch A, Ritter T, Mhashilkar A, Marasco WM, Laube H, Volk HD, Seifert M. Decline of surface MHC I by adenoviral gene transfer of anti-MHC I intrabodies in human endothelial cells-new perspectives for the generation of universal donor cells for tissue transplantation. J Gene Med 2004; 6:616-23; PMID:15170732; http://dx.doi.org/10.1002/jgm.548
- Busch A, Marasco WA, Doebis C, Volk HD, Seifert M. MHC class I manipulation on cell surfaces by gene transfer of anti-MHC class I intrabodies-a tool for decreased immunogenicity of allogeneic tissue and cell transplants. Methods 2004; 34:240-9; PMID:15312677; http://dx.doi.org/10.1016/j.ymeth.2004.03.017
- Mhashilkar AM, Doebis C, Seifert M, Busch A, Zani C, Soo Hoo J, Nagy M, Ritter T, Volk HD, Marasco WA. Intrabody-mediated phenotypic knockout of major histocompatibility complex class I expression in human and monkey cell lines and in primary human keratinocytes. Gene Ther 2002; 9:307-19; PMID:11938450; http://dx.doi.org/10.1038/sj.gt.3301656
- Koistinen P, Pulli T, Uitto VJ, Nissinen L, Hyypia T, Heino J. Depletion of alphaV integrins from osteosarcoma cells by intracellular antibody expression induces bone differentiation marker genes and suppresses gelatinase (MMP-2) synthesis. Matrix Biol 1999; 18:239-51; PMID:10429943; http://dx.doi.org/10.1016/S0945-053X(99)00022-0
- Koistinen P, Heino J. The selective regulation of α Vbeta 1 integrin expression is based on the hierarchical formation of α V-containing heterodimers. J Biol Chem 2002; 277:24835-41; PMID:11997396; http://dx.doi.org/10.1074/jbc.M203149200
- Koistinen P, Ahonen M, Kahari VM, Heino J. alphaV integrin promotes in vitro and in vivo survival of cells in metastatic melanoma. Int J Cancer 2004; 112:61-70; PMID:15305376; http://dx.doi.org/10.1002/ijc.20377
- Richardson JH, Hofmann W, Sodroski JG, Marasco WA. Intrabody-mediated knockout of the high-affinity IL-2 receptor in primary human T cells using a bicistronic lentivirus vector. Gene Ther 1998; 5:635-44; PMID:9797868; http://dx.doi.org/10.1038/sj.gt.3300644
- Tragoolpua K, Intasai N, Kasinrerk W, Mai S, Yuan Y, Tayapiwatana C. Generation of functional scFv intrabody to abate the expression of CD147 surface molecule of 293A cells. BMC Biotechnol 2008; 8:5; PMID:18226275; http://dx.doi.org/10.1186/1472-6750-8-5
- Kovaleva M, Bussmeyer I, Rabe B, Grotzinger J, Sudarman E, Eichler J, Conrad U, Rose-John S, Scheller J. Abrogation of viral interleukin-6 (vIL-6)-induced signaling by intracellular retention and neutralization of vIL-6 with an anti-vIL-6 single-chain antibody selected by phage display. J Virol 2006; 80:8510-20; PMID:16912301; http://dx.doi.org/10.1128/JVI.00420-06
- Cardinale A, Filesi I, Vetrugno V, Pocchiari M, Sy MS, Biocca S. Trapping prion protein in the endoplasmic reticulum impairs PrPC maturation and prevents PrPSc accumulation. J Biol Chem 2005; 280:685-94; PMID:15513919; http://dx.doi.org/10.1074/jbc.M407360200
- Vetrugno V, Cardinale A, Filesi I, Mattei S, Sy MS, Pocchiari M, Biocca S. KDEL-tagged anti-prion intrabodies impair PrP lysosomal degradation and inhibit scrapie infectivity. Biochem Biophys Res Commun 2005; 338:1791-7; PMID:16288721; http://dx.doi.org/10.1016/j.bbrc.2005.10.146
- Peng JL, Wu S, Zhao XP, Wang M, Li WH, Shen X, Liu J, Lei P, Zhu HF, Shen GX. Downregulation of transferrin receptor surface expression by intracellular antibody. Biochem Biophys Res Commun 2007; 354:864-71; PMID:17266924; http://dx.doi.org/10.1016/j.bbrc.2007.01.052
- Intasai N, Tragoolpua K, Pingmuang P, Khunkaewla P, Moonsom S, Kasinrerk W, Lieber A, Tayapiwatana C. Potent inhibition of OKT3-induced T cell proliferation and suppression of CD147 cell surface expression in HeLa cells by scFv-M6-1B9. Immunobiology 2009; 214:410–21.
- Meli G, Lecci A, Manca A, Krako N, Albertini V, Benussi L, Ghidoni R, Cattaneo A. Conformational targeting of intracellular Abeta oligomers demonstrates their pathological oligomerization inside the endoplasmic reticulum. Nat Commun 2014; 5:3867; PMID:24861166; http://dx.doi.org/10.1038/ncomms4867
- Steinberger P, Sutton JK, Rader C, Elia M, Barbas CF, 3rd. Generation and characterization of a recombinant human CCR5-specific antibody. A phage display approach for rabbit antibody humanization. J Biol Chem 2000; 275:36073-8; PMID:10969070; http://dx.doi.org/10.1074/jbc.M002765200
