ABSTRACT
Antibodies with pH-dependent binding to both target antigens and neonatal Fc receptor (FcRn) provide an alternative tool to conventional neutralizing antibodies, particularly for therapies where reduction in antigen level is challenging due to high target burden. However, the requirements for optimal binding kinetic framework and extent of pH dependence for these antibodies to maximize target clearance from circulation are not well understood. We have identified a series of naturally-occurring high affinity antibodies with pH-dependent target binding properties. By in vivo studies in cynomolgus monkeys, we show that pH-dependent binding to the target alone is not sufficient for effective target removal from circulation, but requires Fc mutations that increase antibody binding to FcRn. Affinity-enhanced pH-dependent FcRn binding that is double-digit nM at pH 7.4 and single-digit nM at pH 6 achieved maximal target reduction when combined with similar target binding affinities in reverse pH directions. Sustained target clearance below the baseline level was achieved 3 weeks after single-dose administration at 1.5 mg/kg. Using the experimentally derived mechanistic model, we demonstrate the essential kinetic interplay between target turnover and antibody pH-dependent binding during the FcRn recycling, and identify the key components for achieving maximal target clearance. These results bridge the demand for improved patient dosing convenience with the “know-how” of therapeutic modality by design.
Introduction
The ability of antibodies to target their cognate antigens with high affinity and specificity has enabled them to become one of the most successful therapeutic drug classes in the treatment of many human diseases.Citation1-3 Despite the demonstrated clinical success, the effective neutralization of moderately and highly-abundant target antigens to achieve therapeutic efficacy while providing dosing convenience to patients remains a challenge.Citation4,Citation5 Highly-abundant target antigens are known to be difficult to antagonize with antibodies (e.g., omalizumab against IgE, eculizumab against complement component C5) because of the high doses or frequent injections required for complete target suppression.Citation6,Citation7 Moderately abundant soluble antigens with high turnover rates can also present substantial challenges, in part due to the reduced clearance of the antibody-target complex relative to the target alone.Citation8-10 This can lead to a substantial increase in total antigen (free plus complex) concentrations over the baseline level, which in turn requires high or frequent antibody dosing for antigen neutralization (e.g., IL-6, hepcidin, IL-13).Citation11-13 With therapeutic market pressures for greater patient compliance and treatment cost-effectiveness, there is a continuous demand for new therapeutic entities that enable dose reduction, sustained clinical efficacy, and improved overall convenience to the patient.
In recent years, antibodies with pH-dependent antigen binding characteristics aimed at improving their pharmacokinetic (PK) properties were introduced.Citation14 In comparison to conventional antibodies, a pH-dependent target binding antibody, also referred to as recycling antibody, readily binds to antigens at neutral pH, but dissociate from them once internalized into the acidic endosome (). This allows the antigen-free antibody to be recycled back to the cell surface by neonatal Fc receptor (FcRn), while the dissociated antigen is trafficked to the lysosome for degradation. By repeating this cycle of antigen binding in plasma and dissociating in endosomes, the half-life of pH-dependent antibodies is extended, leading to increased target exposure and greater antigen clearance, thus enabling lower therapeutic dosage or dosing frequency. Examples of this approach have been described in which monoclonal antibodies (mAbs) with engineered pH-dependent target binding properties generated by incorporating pH sensitive histidine residues in the variable domain have reduced target-mediated clearance and prolonged pharmacodynamic (PD) effects relative to their wild-type counterparts in vivo.Citation14-17
Figure 1. Schematic representation of FcRn trafficking mechanisms. mAb, FcRn and target are represented by yellow/blue ribbons, gray ribbons and closed red circles, respectively. mAb-target complexes are taken up by either (a) fluid phase pinocytosis (kFPE) or (b) FcRn mediated endocytosis (kRME). Enhanced pH 6.0 FcRn binding shuttles mAb from late endosome and lysosomal degradation pathways (kLE and kLys) toward recycling pathways (kRcy). Dissociated targets in early endosome proceed toward degradation pathways in lysosome. Salvaged mAbs (c) having weaker pH 7.4 FcRn affinity are either released to extracellular fluid or undergo futile cycling back to early endosomes. mAb bound to FcRn on cell surface (d) can also bind free target and undergo FcRn mediated endocytosis. Intracellular trafficking parameters are summarized in Supplementary Table 1.
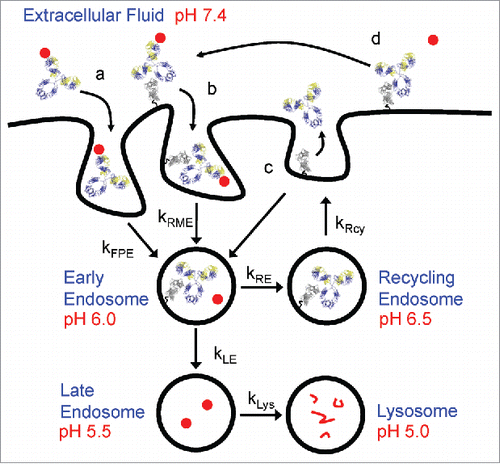
To further enhance the efficiency of antibody recycling and antigen degradation, an improved modality called sweeping antibody, which combines pH-dependent target binding with enhanced FcRn binding were subsequently developed.Citation18,Citation19 In contrast to conventional antibodies whose uptake rate into the cell is limited by nonspecific fluid-phase pinocytosis, a sweeping antibody with engineered Fc regions is capable of binding to FcRn at neutral pH on the cell surface.Citation20,Citation21 As a result of increased FcRn binding, the uptake rate of antibody-antigen complexes into the cell is accelerated, thereby allowing greater amounts of soluble antigens to be degraded compared with a recycling antibody. A study of an Fc-engineered sweeping mAb showed that soluble antigen concentrations were reduced by as much as 1000-fold compared with its conventional counterpart, postulating this antigen-sweeping mode of action could hold superior therapeutic potential over traditional antibody therapies.Citation18
Although these novel approaches offer advantages over conventional antibodies for the neutralization of soluble antigens with moderate to high target burden, the therapeutic modality framework for achieving sustained in vivo target clearance has not been established. This framework comprises complicated intracellular trafficking pathways between the antibody, antigen, and FcRn in response to pH changes, and is dependent on the kinetic interplay between target turnover, antibody-antigen and antibody-FcRn interactions concurrently ( and Supplementary Table 1).
Here, we investigate the molecular design framework encompassing the ideal kinetic balance for sweeping antibodies to maximize target clearance in circulation and suitable target profiles for sweeping antibodies. We identified 5 mouse-derived antibodies with natural pH-dependent binding characteristics against 2 soluble antigens with different binding affinities at neutral and acidic pH environments through biosensor screening, and paired each onto 3 human IgG1 Fc variants of increasing FcRn-binding affinities. Through studies in cynomolgus (cyno) monkeys, we show that accumulation of total antigen in plasma is reduced concomitant with an increase in FcRn binding at both neutral and acidic pH, to the extent that the total antigen concentration in plasma is maintained below the baseline level up to 21 d from a single-dose administration at 1.5 mg/kg. We further establish a mechanistic PK/PD model to elucidate the mode of action of these sweeping antibodies and identify the key requirements of the molecular design framework to advance the therapeutic concept for clinical applications.
Results
Antibodies with pH-dependent antigen binding properties are naturally occurring
Individual connective tissue growth factor (CTGF) and proprotein convertase subtilisin/kexin type 9 (PCSK9) immunization campaigns were conducted in mice to generate antibodies with high affinities. In seeking antibodies with inherent binding affinity differences at neutral and acidic pH, a screening surface plasmon resonance (SPR) assay was modified such that the antigen dissociation rate constant was obtained separately for pH 7.4 and pH 6.0 solutions while the association rate constant was measured at pH 7.4 (refer to Materials and Methods). This assay was designed to mimic the dynamic changes in pH that the antibody-antigen complex experiences during the FcRn-mediated recycling. Among the 4038 IgG clones that were screened, 124 IgG clones exhibited faster dissociation rates, or weaker apparent affinities, at pH 6.0 with > 2 ratios of dissociation rates between pH 6.0 and pH 7.4. These IgG clones were all in the range of pM to single-digit nM target binding affinities at neutral pH. This translates into a discovery rate of 3.1% for in vivo-derived IgGs with natural pH-dependent antigen binding profiles. The identification of pH-dependent anti-CTGF antibodies were performed the same way (data not shown).
Five IgG clones, 3 against cyno PCSK9 and 2 against cyno CTGF were selected for the in vivo study. As shown by SPR analyses, these mAbs exhibit single-digit nM binding affinity at pH 7.4 and weakened affinity in the double-digit nM range at pH 6.0 (). While these mAbs bind with similar affinity at neutral pH, they demonstrate various degrees of pH-dependence as a result of faster dissociation at pH 6.0. The fold shift ratio between the 2 pH solutions ranges from as low as ∼3 for mAb B, ∼5 for mAb D and mAb E, and ∼8 for mAb C, respectively, to as high as ∼33 for mAb A (). Visual comparison of the corresponding SPR sensorgrams shows that the dissociation phase is faster at pH 6.0 versus at pH 7.4 for all the mAbs, in agreement with the kinetic rate constants obtained from the 1:1 model fit (). Among the 5 mAbs, mAb A exhibits the fastest dissociation from the antigen at pH 6.0, as illustrated by the rapid drop of signal to the baseline as soon as the antibody-antigen complex is exposed to pH 6.0 (a). As a result of this rapid dissociation, the apparent affinity for mAb A at pH 6.0 is the weakest among all with the largest ratio of pH-dependency.
Figure 2. SPR sensorgrams of pH-dependent antigen binding antibodies binding to cognate antigens and 1:1 kinetic model fit overlays. (a-e) Binding measurements of 10-min antigen association and 45-min dissociation at pH 7.4 (left panels) and pH 6.0 (right panels) for anti-PCSK9 mAb A (a), mAb B (b), and mAb C (c), and anti-CTGF mAb D (d) and mAb E (e), respectively. The colored lines represent the binding response signals at different antigen concentrations (100 nM, red; 50 nM, teal; 25 nM, blue; 12.5 nM, green; and 6.25 nM, pink), and the overlaid black lines represent the fitted curves.
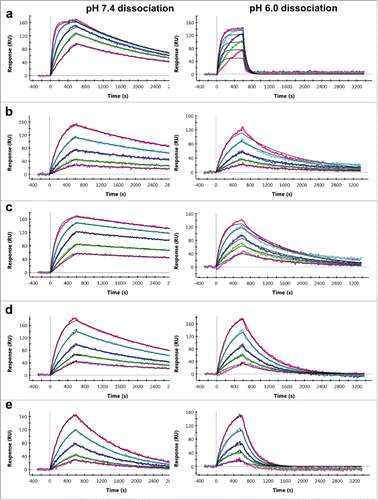
Table 1. pH-dependent binding kinetics and affinities of anti-CTGF and anti-PCSK9 mAbs selected for in vivo studies.
Engineered Fc variants increase FcRn binding at both neutral and acidic pH
In earlier studies, mutated variants of human IgG1 containing the triple mutations M252Y/S254T/T256E (YTE), M252Y/N286E/N434Y (YEY), and M252Y/V308P/N434Y (YPY) were generated with the goal of increasing binding to FcRn.Citation22,Citation23 Through recombinant engineering, the variable region sequences of the 5 identified pH-dependent mAbs were constructed onto each of these IgG1 Fc variants. From bio-layer interferometry (BLI) biosensor analyses, these Fc variants also bind in a pH-dependent manner with significant binding improvement to both human and cyno FcRn by 10s to100s of fold relative to the wild-type (WT) molecule (). The rank order from the greatest to the least FcRn binding improvement is YPY > YEY > YTE. The binding affinities reach as high as single-digit nM affinity in YEY and YPY variants at pH 6.0 with double-digit nM affinity at pH 7.4. Even though there is a binding difference between the human and cyno FcRn (as expected), the rank order of the Fc variants in increasing the binding affinity is the same for both species. As shown by the representative binding profiles and the fitted curves (Supplementary Fig. 1), the improvement in the Fc binding to FcRn can be visualized by saturation at lower FcRn concentrations. While the majority of the affinities was obtained from equilibrium analyses due to the fast on and fast off binding profiles, it was possible to fit the pH 6.0 YEY and YPY binding curves to the standard 1:1 kinetic binding model due to increased affinities for FcRn. In addition, the determined affinities at both pH conditions are comparable and within errors of the biosensor technique for each Fc variant group regardless of differences in the variable region and target specificity.Citation24
Table 2. pH-dependent binding affinities of antibody Fc variants to human and cyno FcRn.
Enhanced FcRn binding affinity increases PCSK9 and CTGF clearance
To evaluate the PK/PD effects of pH-dependent antigen binding antibodies with altered FcRn affinities in vivo, in vivo efficacies of the 3 anti-PCSK9 and 2 anti-CTGF mAbs were tested in cyno monkeys. Mean pre-dose levels of PCSK9 and CTGF were 1.9 nM (95% CI: 1.6–2.3, n = 36) and 1.3 nM (95% CI: 1.1–1.6, n = 33), respectively. Assuming constant rates of target synthesis, the cumulative amount of PCSK9 and CTGF produced over 10 d was estimated to be ∼5 and ∼40 nmol/kg, respectively. Based on these amounts, a 1.5 mg/kg mAb dose (10 nmol/kg) was selected to evaluate the sweeping mechanism to reduce total plasma levels of these targets. The time profiles for changes in the plasma concentrations of antibody (), percent changes in the circulating antigen level relative to pre-dose (), as well as the associated PK/PD parameters (Supplementary Table 2), were obtained.
Figure 3. Plasma or serum concentration-time profiles of pH-dependent antigen binding antibodies in WT, YTE, YEY, and YPY Fc variants in cynomolgus monkeys. The anti-PCSK9 mAb A (a), mAb B (b), and mAb C (c), and anti-CTGF mAb D (d) and mAb E (e) were administered intravenously at single dose of 1.5 mg/kg. Each data point represents the mean ± s.d. (n = 3 each). For the YEY and YPY variants, only data points up to 10 or 14 d are shown due to significant drop in measurable concentrations.
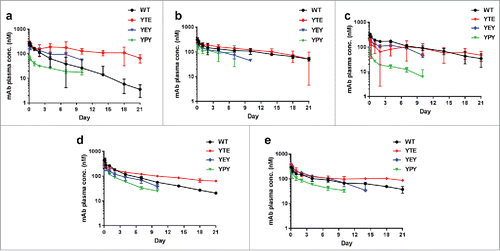
Following an intravenous dose, total (free plus bound) systemic clearance of WT anti-PCSK9 and anti-CTGF mAbs was in the expected range for humanized mAbs displaying linear clearance in monkeys, suggesting that the clearance of the mAb-target molecular complex was comparable to that of the mAb alone ( and Supplementary Table 2). Clearance of YTE variants was equal to or up to 5 times slower than their respective WT variants, whereas the apparent clearance of YPY variants was 2 to 11 times faster ( and Supplementary Table 2). YEY variants displayed clearance intermediate between YTE and YPY. Consistent with the formation of anti-drug antibodies, a precipitous drop in mAb concentrations was observed for some variants 10–14 d post-dose (); therefore, the analysis was restricted to data up to and including 10 day.
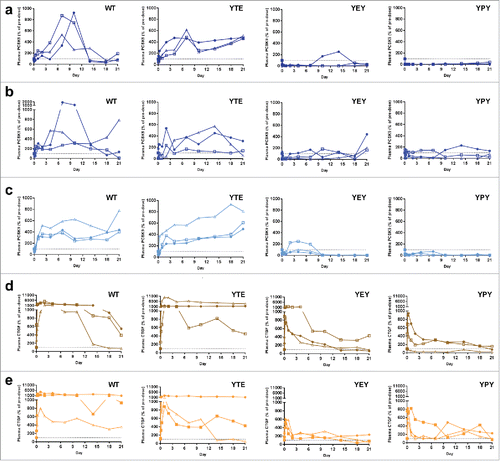
shows that for the WT variants, total (free plus complex) mean PCSK9 and CTGF concentrations (AUC0–10 days) increased up to 6- and 21-fold relative to baseline, respectively. Variants with increased FcRn binding affinity significantly reduced total mean PCSK9 and CTGF levels up 70-fold and 18-fold relative to WT, respectively. The YPY molecules generated the most robust reductions in both total and free target levels. In the case of anti-PCSK9 mAbs, total target levels were reduced 11-fold (mAb A) and 2–3-fold (mAb B and mAb C) lower than baseline levels 10 d post-dose, whereas for the anti-CTGF mAbs (mAb D and mAb E), total target levels were reduced to around baseline levels ( and Supplementary Table 2). Increasing doses up to 10 mg/kg of the YPY variant did not result in further reductions in either total or free CTGF levels (Supplementary Fig. 2). Furthermore, endogenous serum IgG levels were not affected by doses up to 10 mg/kg of the YPY variant (Supplementary Fig. 3), indicating that the lack of dose-dependent changes in CTGF levels were not a result of saturating membrane or endosomal pools of FcRn. Free target levels also displayed FcRn-dependent reductions relative to WT, but are less quantitative due to inherent limitations associated with measuring free target levels for low affinity mAbs (Supplementary Table 2).Citation25 As further evidence for the efficacy of these sweeping mAbs to result in meaningful downstream PD effects, decreases in total and free PCSK9 for the YEY and YPY variants translated to sustained reductions in LDL relative to the WT and YTE variants (Supplementary Fig. 4).
Figure 4. In vivo antigen characterization of pH-dependent antigen binding antibodies with altered FcRn binding affinities in cynomolgus monkeys. The anti-PCSK9 mAb A (a), mAb B (b), and mAb C (c), and anti-CTGF mAb D (d) and mAb E (e) were administered intravenously at single dose of 1.5 mg/kg (n = 3 animals per group, represented by different symbols). The percentage antigen was calculated by dividing the pre-dose antigen concentration with the plasma antigen concentration measured at each time point. The time-course percentage antigen inhibition is plotted for each animal. The dotted line at 100% represents the baseline normalized pre-dose antigen concentration in each animal.
Optimal FcRn and target binding properties identified using a mechanistic model
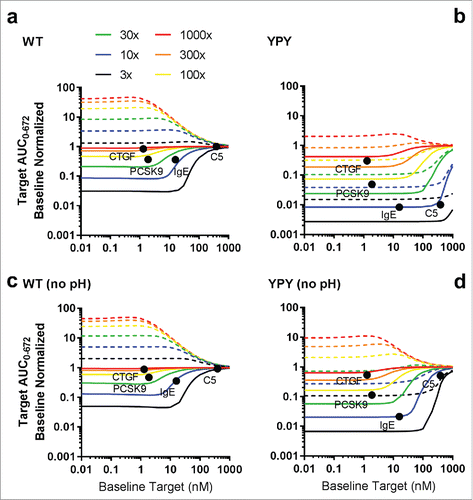
With the exception of pH 6.0 target binding affinities or pH 6.0 target dissociation rates, where a weak correlation with free target levels was observed (R2 0.34, data not shown), in vitro binding or kinetic parameters did not display a correlation with either free or total target levels. To further identify the optimal binding properties for reducing free target levels via the antigen sweeping mechanism, a mechanistic model that incorporated intracellular trafficking, target binding and FcRn binding kinetics was implemented (Supplementary Fig. 5). The model was calibrated using the data described above for anti-PCSK9 and anti-CTGF mAbs (Supplementary Fig. 6–7). All simulations were performed using a 1.5 mg/kg dose, which represents a maximum feasible subcutaneous dose in humans assuming a solubility of 100 mg/mL and 1–2 mL injection volume. For the purpose of this analysis, a greater than 90% reduction in free target levels relative to baseline was considered optimal. In the absence of enhanced FcRn binding affinity, the optimal pH 7.4 binding affinities and pH 6.0 dissociation rates were ≤ 1 nM and ≥ 0.03 s−1, respectively (a). Increased FcRn affinity extended the range of accessible pH 7.4 binding target affinities to at least 10 nM and significantly reduced the requirements for pH-dependent target binding (b-d). In addition to the effect of pH-dependent target binding, the pH 7.4 target association rate was found to play a major role. As the mAb-target association rate increased, the target clearance also increased for both the WT and YPY antibodies (), indicating that an mAb-target association rate of ∼1.0E+5 M−1s−1 and higher was required to maintain a proper balance between encounter complex formation and target depletion. Using the optimal pH-dependent target binding affinity and kinetic rate parameters identified from and , the PK/PD profiles in response to changes in FcRn binding affinity and pH-dependency were generated (Supplementary Fig. 8). The simulations showed that the optimal pH 6.0 and pH 7.4 FcRn binding affinities were between 1–10 nM and 100–1000 nM, respectively. Furthermore, the influence of target abundance and turnover on the efficiency of the antigen sweeping system was explored. The model predicted significant reductions in free and total target levels for YPY mAbs in the presence and absence of modest (10-fold) pH-dependent target binding compared with WT (a and c). Consistent with the in vivo study findings, the model indicated that enhanced FcRn binding affinity can greatly extend the range of tractable targets, including moderately abundant/fast turnover targets (e.g., PCSK9, CTGF), and highly abundant/slow turnover targets (e.g., IgE, C5) (b and d).
Figure 5. Simulations describing the effect of pH-dependent target binding on free target levels in cynomolgus monkey following a single 1.5 mg/kg intravenous dose. The pH 6.0 target affinity was varied from 1 to 1000x weaker than the pH 7.4 target affinity. The target association rate was fixed to 1.0E + 5 M−1s−1. FcRn binding affinity was fixed to the model derived estimates for WT (a), YTE (b), YEY (c) and YPY (d) variants. Target concentration and target clearance were set to 1 nM and 0.0547 L/hr, respectively. Clearance of the target-mAb complex was set equal to the mAb.
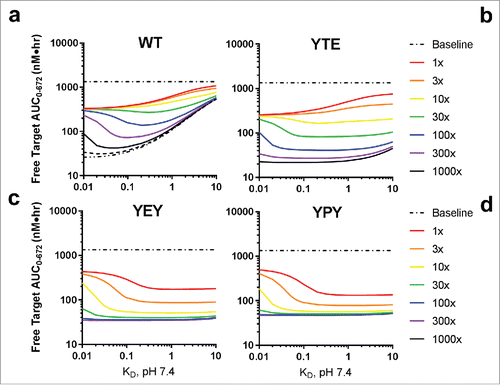
Figure 6. Simulated effect of target association rate on free target levels in cynomolgus monkey following a single 1.5 mg/kg intravenous dose. The target association rate was set to 1.0E + 4 (a), 1.0E + 5 (b) or 1.0E + 6 M−1s−1 (c). The pH 6.0 target affinity was varied from 1 to 10000x weaker than the pH 7.4 target affinity. FcRn binding affinity was fixed to the model derived estimates for WT (left panels) and YPY (right panels) variants. Target concentration and target clearance were set to 1 nM and 0.0547 L/hr, respectively. Clearance of the target-mAb complex was set equal to the mAb.
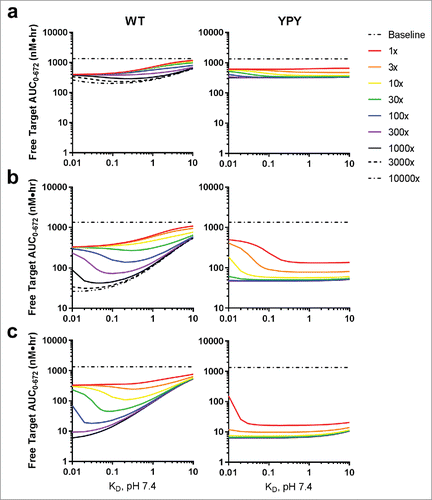
Figure 7. Simulation of baseline-normalized free (solid lines) and total target levels (dashed lines) in cynomolgus monkeys following a single 1.5 mg/kg intravenous dose of pH-dependent (a, b) and non-pH-dependent (c, d) mAbs in both the WT and YPY variants. Target abundance was varied from 0.01 to 1000 nM and target clearance was varied from 3 to 1000 times faster than the clearance of a mAb in monkey in the absence of target-mediated disposition (0.001 L/hr). The closed black circles represent the predicted baseline normalized free target levels for CTGF, PCSK9, IgE and C5. These predicted values were derived by plotting the experimentally measured baseline concentrations against the interpolated target clearances, as defined by the solid lines. Clearance of the target-mAb complex was set equal to the mAb. The pH 6.0 and pH 7.4 target binding affinities were set to 10 nM and 1 nM, respectively, the association rate was fixed to 1.0E + 5 M−1s−1, and the target concentration was 1 nM.
Discussion
Herein, we demonstrated that antibodies with binding affinities comparable to baseline target levels and modest pH dependence can result in significant FcRn-dependent decreases in both total and free target. The concept of a sweeping antibody was first introduced by Igawa et al, where a series of anti-IL-6R antibodies were used to modulate exogenous plasma levels of hIL-6R in wild-type and hFcRn transgenic mice.Citation18 The efficiency of the antigen sweeping mechanism in these models was determined to be highly sensitive to pH-dependent FcRn and pH-dependent target binding. To our knowledge, our study is the first to describe the efficiency of the sweeping mechanism in non-human primates using antibodies with naturally derived pH-dependent target binding properties against endogenous proteins.
Studies published to date with pH-dependent mAbs were based on the modification of variable region sequence on the pre-existing mAb to confer pH-dependent binding.Citation14-16,Citation18 Extensive histidine scanning mutagenesis of residues specifically in the antibody complementarity-determining regions was performed and subsequently characterized. Although this histidine-based engineering approach has demonstrated promising results, it is a labor- and time-intensive process that requires detailed analysis of individual mutants to validate the engineered pH dependence. In some cases, incorporating pH-dependent properties has led to reduced or substantial loss of target binding affinity at neutral pH, which could in turn affect efficacy.Citation16 An alternative de novo approach to isolate pH-dependent antibodies selected by phage display from libraries enriched in histidines was demonstrated.Citation26 Although this process is more efficient and direct compared with the engineering approach, these phage-derived pH-dependent antibodies may display biophysical issues and require other sequence optimization efforts due to known potential challenges associated with antibodies of non-immune origins.Citation27 We therefore chose the approach of in vivo immunization and direct selection of antibodies for pH-dependent target binding properties from immunized animals. Through the use of a modified SPR method to mimic the dynamic pH changes that a mAb-antigen complex encounters during the endosomal recycling process, we successfully identified over a hundred IgG clones exhibiting a variety of single-digit nM or pM neutral pH affinities with greater than 2-fold faster dissociation rates at acidic pH. Although the discovery of these mAbs was rare (3.1% of the binders), we demonstrated that it was feasible to directly identify mAbs with natural pH-dependent binding properties from early candidate screening with the support of high-throughput biosensor technology.Citation24 Additional analyses indicate these pH-dependent mAbs exhibited diversity in both the sequence and epitopes on the antigen with relevance in biologic function (data not shown).
CTGF and PCSK9 were selected for this study because they have similar baseline concentrations; however, the target turnover of CTGF is ∼10 times faster than PCSK9. As such these 2 proteins represent moderate to highly abundant targets with relatively fast turnover, which have historically been difficult to modulate with once-monthly subcutaneous doses. pH-dependent anti-CTGF and anti-PCSK9 antibodies with single-digit nM affinity and WT FcRn affinity resulted in transient modulation of total and free target levels, arising from: 1) accumulation of antibody-target complexes that, in accordance with Le Chatelier's principle, reduced maximum achievable target suppression; and 2) consumption of free antibody binding sites from relatively fast target turnover. Variants with increased FcRn affinity resulted in significant reductions in both total and free target levels, despite having modest pH-dependent target binding. YPY variants generally displayed the most robust and sustained reductions in total target levels, followed by YEY, even though the apparent clearance of these variants was several fold higher. In contrast to previous studies, there did not appear to be a strong correlation between in vitro target binding or kinetic parameters and antibody or target concentration-time profiles.Citation15,Citation28 Our study, however, evaluated a much narrower range of target binding affinities, which may have masked intrinsic differences between molecules.
To further interrogate the potential utility of the antigen sweeping mechanism to enhance target suppression, Haraya et al implemented a modified dynamic antibody-antigen binding target-mediated drug disposition model.Citation29 The model incorporated empirically-derived composite parameters that represented uptake clearance and differential recycling of antibody in the antibody-target complex. Although this model was able to capture the observed phenotype for the sweeping mechanism, the capacity of such a model to address pertinent questions related to a priori identification of optimal antibody properties was confounded by the model's simplicity. In light of these limitations, we implemented an experimentally calibrated physiologically-based mechanistic model that explicitly captured the dynamic interplay between pH-dependent target binding, pH-dependent FcRn binding and intracellular trafficking kinetics.
The mechanistic model was able to corroborate previously reported experimental data for pH-dependent antibodies that suggested the pH 7.4/pH 6.0 KD ratio was less important than the pH 7.4 target affinity and pH 6.0 target dissociation rate. For pH-dependent antibodies with WT FcRn affinity, target binding affinities ≤ 1 nM at neutral pH were required to maintain a proper balance between target engagement and disposal. The optimal pH 6.0 dissociation half-life was determined to be less than ≤ 70s and was independent of pH 7.4 binding affinity or target association rate. This value is in excellent accord with the pH 6.0 dissociation half-lives (10 to 78s) reported for pH-dependent PCSK9 antibodies that displayed prolonged PD relative to non-pH-dependent antibodies or antibodies with much slower pH 6.0 dissociation half-lives. For high affinity antibodies, predicted free target levels converged toward an asymptote that reflected the maximum achievable target suppression arising from rate-limited encounter-complex formation, as described by the law of mass action. In line with these observations, increasing the target association rate from the slow binding (104 M−1s−1) to fast binding (106 M−1s−1) resulted in dramatic reductions in free target levels for both pH-dependent and sweeping antibodies.
Engineering or naturally screening for antibodies with such profound differences in pH-dependent binding characteristics, especially for mAbs with sub-nM affinities, may be difficult to achieve, resulting in the preference for antibodies with single-digit nM affinities. Enhanced FcRn binding at neutral pH significantly lessened the pH-dependent target binding criteria required to achieve the same level of target suppression relative to WT variants. Modest pH-dependent target binding contributed an additional 3 to 5-fold improvement in total and free target reductions, compared with YPY variants without pH dependence. YPY variants with modest pH-dependent target binding are predicted to extend the range of accessible target levels and turnover rates from 10 to 100 nM and 0.01 to 0.1 L/hr, compared with the corresponding WT variants. These results suggest that, in addition to PCSK9, a sweeping antibody would be capable of significantly reducing baseline levels of IgE and C5 with a 1.5 mg/kg once monthly subcutaneous dose. In alignment with experimental observations, anti-CTGF antibodies were predicted to fall slightly outside the accessible target space, achieving a maximum 70% reduction of free target levels. While this may be acceptable from a therapeutic standpoint for CTGF, these levels fall outside the desired range of ≥ 90%. Another salient conclusion from the modeling results is the need to properly match the target affinity with target abundance and antibody concentrations. While the sweeping mechanism is predicted to loosen these requirements somewhat compared with conventional antibodies, low abundance/high turnover targets (e.g., cytokines) may require higher affinity antibodies to drive sufficiently rapid uptake clearance.
High affinity FcRn binding at neutral pH has been associated with increased antibody clearance, and may also interfere with the endogenous IgG recycling at sufficiently high doses.Citation30,Citation31 Antagonistic anti-FcRn antibodies, with high affinity binding (1–2 nM) at both neutral and acidic pH, reduced endogenous serum IgG levels up to 70% in a dose-dependent manner following a single 5 mg/kg or 20 mg/kg dose.Citation31 Although YPY variants demonstrated similar binding to FcRn at pH 6.0, the pH 7.4 affinity was 15-fold weaker. A single intravenous dose of the YPY variant did not affect endogenous monkey plasma IgG levels up to 10 mg/kg. Based on modeling and simulations, the optimal pH 6.0 and pH 7.4 FcRn binding affinities were predicted to be around 1 nM and 100 nM, respectively. The observed pH-dependent binding characteristics for the YEY and YPY variants are very close to these optimal values, suggesting that there may be little room for improving the overall efficiency of the sweeping mechanism through modulation of FcRn affinity.
The model described in this study shares the same overarching structure described previously for other physiologically-based pharmacokinetic (PBPK) models, including vascular, endosomal, interstitial fluid and lymphatic compartments.Citation32,Citation33 The major difference between the current model and previously described models resides in how intracellular trafficking was handled in the endosomal compartment. In contrast to previously described PBPK models, the model used in this study instantiates rapid FcRn-mediated internalization (∼3 s) and intracellular trafficking kinetics (∼20 s), consistent with the time-scales observed in vitro by fluorescence microscopy.Citation34-36 Fast intracellular trafficking kinetics coupled with lower copies of FcRn per endothelial cell (105) were required to capture the relatively prolonged distribution phase observed for YPY variants in monkeys. Three transit compartments were used to describe FcRn-mediated intracellular trafficking between the membrane surface (pH 7.4), early endosomes (pH 6.0) and recycling endosomes (pH 6.5). The model also incorporated futile cycling, whereby mAb that was not released from FcRn at pH 7.4 was cycled back to the early endosome.
An alternate pH-dependent endosomal trafficking model has been proposed by Chen and Balthasar.Citation37 This model incorporated a gradual time-dependent reduction in endosomal pH over 10.8 min across 5 transit compartments that varied in pH from 7.4 to 6.0, and did not account for FcRn-mediated internalization from the cell surface. In the terminal pH 6.0 compartment, mAb bound to FcRn was recycled, whereas unbound mAb was degraded. A more conventional modeling framework that incorporated slower intracellular trafficking kinetics (10–20 min) coupled with higher copies of FcRn per cell (106) grossly underestimated the duration of the distribution phase observed for YPY and YEY variants. Interestingly, the model described here predicts that YPY and YEY variants undergo several additional cycles of intracellular trafficking before being released from FcRn at neutral pH, compared with ∼1 cycle for WT and YTE variants. This may explain why YPY and YEY variants are less sensitive to pH-dependent target binding, as high affinity FcRn binding provides several additional opportunities for these antibodies to release their target in the early endosome before dissociating from FcRn at neutral pH.
Consistent with other PBPK models, sensitivity analyses for the current model indicated that the PK profile was most sensitive to lymph flow, endosomal volume, FcRn binding affinity and number of FcRn binding sites per cell (Supplementary Figure 9 and 10). Total target levels for WT variants were most sensitive to endosomal volume; whereas, total target levels for YPY variants were most sensitive to FcRn binding affinity, number of FcRn binding sites per cell and FcRn intracellular trafficking rates (Supplementary Figure 11). The sensitivity of the model to these parameters reflects their importance in capturing the dynamic interplay between target uptake and disposal for sweeping antibodies. The model more accurately captured the accumulation of total target levels for WT mAbs when the apparent endosomal volume was set to fairly large values, i.e., ∼50% of the cell volume or ∼2-fold higher than the maximum estimated endosomal volume of endothelial cells, assuming ∼100 billion capillary endothelial cells per monkey.Citation38 The size of the endosomal compartment partially contributed to the dissociation of antibody-antigen complexes by reducing their effective endosomal concentration. The relatively large apparent endosomal volume implemented in this model is likely an artifact of not accounting for the highly compartmentalized and multivesicular nature of endosomes, and will be the subject of further investigation.
In summary, we demonstrate that naturally occurring high affinity antibodies with pH-dependent target binding properties can be identified through direct screening. These antibodies, coupled with increased FcRn binding, can effectively neutralize targets with high baseline concentration or rapid turnover in vivo at doses that are amenable for once monthly subcutaneous dosing, thereby increasing doing convenience for patients. We also provide valuable insights into the kinetic attributes for the optimal design of sweeping antibodies serving uniquely as a ‘delivery vehicle’ to maximize target clearance. These results collectively provide guidance for the optimal selection, design, and evaluation of such sweeping antibodies in early discovery to facilitate the innovative therapeutic approach, potentially resulting in a shortened timeline to the clinic.
Materials and methods
Generation of anti-PCSK9 and anti-CTGF antibodies
NMRI x C57/BI6 mice were immunized with recombinant PCSK9 or CTGF proteins. Hybridomas were generated by fusion with PAI myeloma cells using standard methods. Antibodies in the cultured supernatant hybridomas were screened with Luminex bead-based assays using respective antigen. Positive binders with MFI > 2000 signal were advanced to screen for antigen binding kinetics at neutral and acidic pH solutions using SPR. Variable gene sequences from antibodies with pH-dependent binding characteristics were recovered from hybridomas using standard methods.
Expression vector containing IgG1 Fc mutations
Each heavy and light chain variable gene sequence was cloned into existing in-house pTT5 expression vectors using standard methods. The following mutations were made in the human IgG1 constant region: Met252 to Tyr, Ser254 to Thr and Thr256 to Glu for the Fc-YTE variant; Met252 to Tyr, Asn286 to Glu, and Asn434 to Tyr for the Fc-YEY variant; and Met252 to Tyr, Val308 to Pro, and Asn434 to Tyr for the Fc-YPY variant. These mutations were introduced into the vector by infusing the gBlocks® gene fragment (Integrated DNA Technologies) with an In-Fusion PCR Cloning Kit (Clontech Laboratories).
Expression, purification and quality verification of mAbs
Chinese hamster ovary (CHO) cells were transfected with expression vectors containing heavy chain and light chain cassettes using Freestyle CHO Expression Medium with 8 mM Glutamax (Invitrogen). The transfected cells were incubated for 10 days, and then the medium was harvested and used to purify the antibodies with the ÄKTA affinity chromatography system and MabSelect Sure resin (GE Healthcare) following standard methods.Citation39 The purified mAbs were formulated in 60 mM sodium acetate (pH 5.0), and their concentrations were determined by adsorption at 280 nm using an extinction coefficient of 1.36 in NanoDrop™ 8000 Spectrophotometer (Thermo Fisher Scientific). The quality and monomer content of the purified antibodies were accessed by analytical techniques, including SDS-Page (NuPAGE, Life Technologies) and UPLC size-exclusion chromatography (ACQUITY, Water Corporations). The endotoxin level was measured by Endosafe®-PTS™ (Charles River Laboratories) and the sequence and glycosylation patterns were confirmed via intact mass analysis by TOF Mass Spectrometry (Agilent 6224 TOF LC/MS, Agilent Technologies).
Surface plasmon resonance analyses of pH-dependent antigen binding kinetics
The binding kinetics of the mAbs to respective antigen was evaluated at 25oC using a ProteOn XPR36 instrument (Bio-Rad Laboratories) equipped with 2 separate buffer inlets. The buffer inlet A is placed in a pH 7.4 PBS-T-EDTA buffer (PBS, 0.005% Tween 20, and 3 mM EDTA) whereas the inlet B is placed in a pH 6.0 citrate buffer (10 mM citrate, 150 mM NaCl, 0.005% Tween 20, and 3 mM EDTA). Recombinant protein A/G was purchased from Thermo Fisher Scientific (catalog # 21186). To couple the protein A/G to the biosensor surfaces, we used an amine coupling kit containing 400 mM EDC, 100 mM N-hydroxysulfosuccinimide (sulfo-NHS), and 1 M ethanolamine-HCl (pH 8.5) (Bio-Rad, catalog # 1762410). The individual flow channels in the GLM sensor chip were activated by injecting a fresh mixture of EDC/sulfo-NHS (400 mM/100 mM), followed by the immobilization of 30 µg/mL protein A/G prepared in pH 4.5 sodium acetate and the blocking of excess reactive esters with 1 M ethanolamine. Each activation, immobilization, and deactivation step was performed for 5 min with 6 parallel injections in the horizontal direction at 30 μl/min. The prepared protein A/G surfaces were subsequently conditioned with 3 18-sec pulses of glycine (pH 1.5) at 100 μl/min in both the horizontal and vertical directions. A group of 6 mAbs diluted at 0.25 µg/mL in PBS-T-EDTA were injected in parallel in the vertical direction for 160 s at 25 μl/min. Following a switch in orientation of the sensor chip and a blank buffer injection of 60 s, the antigen binding kinetics of the mAbs was measured by injecting 5 concentrations of cyno PCSK9 or cyno CTGF (100 nM – 6.25 nM) prepared by 2-fold serial dilution in PBS-T-E-DTA. The binding interactions were monitored in running buffer over a 10-min association period and a 45-min dissociation period at 40 μl/min. After each binding cycle, the protein A/G surface was regenerated with 2 18-sec pulses of glycine (pH 1.5) at 100 μl/min in both the horizontal and vertical directions.
In the evaluation of binding kinetics at pH 7.4, both the antigen association and dissociation steps were measured in pH 7.4 PBS-T-EDTA by using inlet A as the running buffer. To measure the dissociation at pH 6.0, the running buffer was switched from inlet A to inlet B containing the pH 6.0 citrate buffer immediately after the 10-min antigen association period. This buffer switch was introduced to mimic the antibody recycling process in vivo, which presents that following the antigen binding in the pH 7.4 cytoplasmic environment, the subsequent internalization of the antibody-antigen complex via pinocytosis expose the complex to the pH 6 acidic endosomal environment where the antigen dissociates.
The collected binding sensorgrams were double-referenced using channel inter-spots and a parallel in-line buffer blank subtraction by the integrated ProteOn Manager software (v.3.1.0.6). The processed binding curves were then fitted using the Langmuir model describing a 1:1 binding stoichiometry. In all of the analyses, ka is the association rate constant for the antibody-antigen binding reaction, kd is the dissociation rate constant of the antibody-antigen complex, and KD is the equilibrium dissociation constant (i.e., affinity) defined by kd/ka.
BioLayer interferometry analyses of mAbs binding to FcRn
The various antibody Fc variants were investigated at 25oC using an Octet RED384 instrument (ForteBio, Pall Corporation) equipped with 16 anti-human Fab-CH1 biosensor tips. Each mAb prepared at 20 μg/mL in 1 × KB (PBS pH [7.4], 0.02% Tween-20, 0.1% albumin, and 0.05% sodium azide) running buffer was dispensed into a 384-well tilted-bottom microplate at a volume of 90 μl per well for 16 wells. Both the human and cyno FcRn proteins were dialyzed into individual pH 7.4 1 × KB and pH 6.0 citrate buffer and then dispensed into the same microplate at 15 titrating concentration (11500 nM to 0.70 nM) prepared by 2-fold serial dilution in respective buffer. Prior to the binding measurements, the sensor tips were pre-hydrated in 1 × KB for 5 min, followed by 3 cycles of pre-conditioning with 15-sec dips in glycine (pH 1.5), alternating with 15-sec dips in 1 × KB. The sensor tips were then transferred to the mAb-containing wells for a 300-sec loading step. After a 60-sec baseline dip in 1 × KB, the binding kinetics were measured by dipping the mAb-coated sensors into the wells containing human or cyno FcRn at pH 7.4 1 × KB and pH 6.0 citrate buffer simultaneously. The binding interactions were monitored over a 20-min association period and followed by a 10-min dissociation period in new wells containing fresh buffer. The AHC sensor tips were regenerated with 2 18-sec dips in glycine (pH 1.5) between each binding cycle.
The collected data were referenced using a parallel buffer blank subtraction, and the baseline was aligned to the y-axis and smoothed by a Savitzky-Golay filter in the data analysis software (v.9.0.0.4). Equilibrium analysis was performed on weak interactions with fast on- and off- binding profiles, whereas 1:1 Langmuir kinetic model was applied to fit the stronger interactions exhibiting curvature at low FcRn concentrations. For each antibody Fc variant, 2 values of KD values were generated, with one representing the binding affinity of the mAb and FcRn at pH 7.4 and the other at pH 6.0, respectively.
In vivo pharmacokinetic studies
Male cynomolgus monkeys, naïve to previous treatment with biologics, were administered a single intravenous dose (n = 3/dose group). Blood samples were collected before dosing and up to 6 weeks post-dose. Clinical chemistry samples (LDL, HDL, cholesterol and triglycerides) were collected and analyzed for anti-PCSK9 antibodies. In vivo studies and clinical chemistry analysis were conducted at Charles River Laboratory. Studies were approved by IACUC and were in compliance with US Department of Agriculture Animal Welfare Act (9CFR Parts 1, 2, and 3).
Determination of total anti-PCSK9 and anti-CTGF mAbs
Drug concentrations were measured in monkey serum or plasma using enzyme linked immunosorbent assays (ELISA). Goat anti-human IgG, adsorbed against monkey IgG (Novus Cat# NB7487 or MyBioSource Cat# NC0672104) and horseradish peroxidase (HRP)-conjugated goat anti-human IgG, adsorbed against monkey IgG (Southern Biotech Cat# 2049–05 or Bethyl Cat# A80–319P) were used as the capture and detection reagents, respectively. PK parameters were calculated using non-compartmental and 2-compartmental analysis in Phoenix WinNonlin 6.3 (Certara, MD, USA).
Determination of free and total PCSK9
Free PCSK9 serum concentrations were determined using an electrochemiluminescent assay (Meso Scale Discovery, MSD). Diluted serum samples (1:1, in binding buffer, Thermo Fisher) were passed through a Protein G resin-packed column (Thermo Fisher) to separate free PCSK9 from PCSK9-bound antibody complexes. Filtered serum samples were incubated with biotinylated and sulfo-labeled anti-PCSK9 antibody (R&D Systems Cat# AF3888) and then transferred to a MSD Gold streptavidin-coated microplate. Total (free and therapeutic antibody bound) PCSK9 levels were determined by ELISA, using unlabeled capture and biotinylated-detection antibodies that recognized distinct PCSK9 epitopes from the therapeutic mAb. Streptavidin-conjugated HRP (Jackson ImmunoResearch) was used as the secondary detection reagent.
Determination of free and total CTGF
Free and total (free and therapeutic mAb bound) CTGF plasma concentrations were determined using the Gyrolab platform with biotinylated and Alexa Fluor 647-labeled anti-CTGF antibodies, as capture and detection reagents, respectively. Free target levels were measured using an anti-CTGF antibody that was directed to the same CTGF epitope as the therapeutic mAb. Total target levels were measured using anti-CTGF capture and detection antibodies that recognized distinct epitopes from the therapeutic mAb.
Modeling and simulation
A minimalistic PBPK model (Supplementary Figure 5) with explicit representation of target binding, FcRn binding and intracellular trafficking kinetics was used to describe the plasma concentration time-course for total mAb and total target levels.Citation40 The clearance (CL) rate for CTGF was set to the glomerular filtration rate in monkey, 125 mL/h/kg.Citation41 The CL rate for PCSK9 in monkeys, 11 mL/h/kg, was estimated from mechanistic PK studies with high affinity non-pH-dependent anti-PCSK9 antibodies.Citation15,Citation28 Target synthesis rates were varied to achieve the baseline level of CTGF or PCSK9 observed for each cohort of monkeys. Target binding affinities were set to their experimentally determined values. FcRn binding affinities were varied within 2-standard deviation of the experimentally determined values. The number of FcRn molecules per endothelial cell was fixed to 1.0E + 5 copies per cell.Citation42 Intracellular trafficking rate constants were calibrated to match the observed α and β phase half-lives for mAbs displaying linear kinetics in monkey.Citation40 Dissociated target molecules in the early endosome were rapidly sorted to an intra-endosomal compartment, thereby preventing re-association with available mAb binding sites. An additional term was included in the vascular endothelium to account for potential epitope-dependent differences in the adsorptive endocytosis of mAb-target complexes. Model parameters are summarized in Supplementary Table 1.
Author contributions
S.S. conceived the idea and guided the study. D.Y. and C.G. wrote the manuscript. D.Y., C.G., S.S. and R.K. deliberated and finalized the contents of the manuscript. S.S., R.K., D.Y. and S.C. strategized the parameters to test for pH-dependent target binding and FcRn affinity. S.S., R.K., D.Y., S.C., J.A. and S.R. formulated the in vivo study design. M.L. designed the immunogens and immunization strategy for both CTGF and PCSK9. H.X and H.S. contributed to V-gene recovery, sequencing, and construct synthesis. Z.H. contributed to the production and purification of the antibodies. D.Y. conducted pH-dependent SPR screening for the PCSK9 immunization campaign and analyzed the data. I.R. led the project team that identified the pH-dependent antibodies against CTGF. C.K. performed SPR characterization on the anti-CTGF candidates. D.Y. and H.S. developed the pH-dependent BLI assay and analyzed the FcRn binding kinetics. C.G. developed the mechanistic PBPK model and analyzed the simulations. D.Y. and S.C. qualified and prepared the materials for delivery to the CRO. J.A. facilitated the study at the CRO and wrote the PK method section. T.B., M.M. and E.W. conducted bioanalysis on the serum/plasma samples and wrote the corresponding method sections. R.K. and S.S. provided technical guidance, conceptual advice, and oversight throughout the entire process.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary_Materials.pdf
Download PDF (1.1 MB)Acknowledgements
We thank the Biotherapeutics Discovery group for antibody generation and production of the antibodies for the in vivo study. We also thank the PCSK9 and CTGF project teams for allowing us to use the antibodies for the study.
References
- Strebhardt K, Ullrich A. Paul Ehrlich's magic bullet concept: 100 years of progress. Nat Rev Cancer. 2008;8:473-80. doi:10.1038/nrc2394. PMID:18469827
- Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol. 2010;10:301-16. doi:10.1038/nri2761. PMID:20414204
- Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278-87. doi:10.1038/nrc3236. PMID:22437872
- Tabrizi MA, Tseng C-ML, Roskos LK. Elimination mechanisms of therapeutic monoclonal antibodies. Drug Discov Today. 2006;11:81-8. doi:10.1016/S1359-6446(05)03638-X. PMID:16478695
- Kamath AV. Translational pharmacokinetics and pharmacodynamics of monoclonal antibodies. Drug Discov Today Technol. 2016;21–22:75-83. doi:10.1016/j.ddtec.2016.09.004. PMID:27978991
- Lowe PJ, Tannenbaum S, Gautier A, Jimenez P. Relationship between omalizumab pharmacokinetics, IgE pharmacodynamics and symptoms in patients with severe persistent allergic (IgE-mediated) asthma. Br J Clin Pharmacol. 2009;68:61-76. doi:10.1111/j.1365-2125.2009.03401.x. PMID:19660004
- Zareba KM. Eculizumab: A novel therapy for paroxysmal nocturnal hemoglobinuria. Drugs Today. 2007;43:539. doi:10.1358/dot.2007.43.8.1130446. PMID:17925885
- Finkelman FD, Madden KB, Morris SC, Holmes JM, Boiani N, Katona IM, Maliszewski CR. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J Immunol Baltim Md 1950 1993;151:1235-44.
- Rehlaender BN, Cho MJ. Antibodies as carrier proteins. Pharm Res. 1998;15:1652-6. doi:10.1023/A:1011936007457. PMID:9833983
- Davda JP, Hansen RJ. Properties of a general PK/PD model of antibody-ligand interactions for therapeutic antibodies that bind to soluble endogenous targets. mAbs. 2010;2:576-88. doi:10.4161/mabs.2.5.12833. PMID:20676036
- Martin PL, Cornacoff J, Prabhakar U, Lohr T, Treacy G, Sutherland JE, Hersey S, Martin E. Reviews Preclinical Safety and Immune-Modulating Effects of Therapeutic Monoclonal Antibodies to Interleukin-6 and Tumor Necrosis Factor-α in Cynomolgus Macaques. J Immunotoxicol. 2005;1:131-9. doi:10.1080/15476910490894904. PMID:18958646
- Xiao JJ, Krzyzanski W, Wang Y-M, Li H, Rose MJ, Ma M, Wu Y, Hinkle B, Perez-Ruixo JJ. Pharmacokinetics of Anti-hepcidin Monoclonal Antibody Ab 12B9m and Hepcidin in Cynomolgus Monkeys. AAPS J. 2010;12:646-57. doi:10.1208/s12248-010-9222-0. PMID:20737261
- Hodsman P, Ashman C, Cahn A, De Boever E, Locantore N, Serone A, Pouliquen I. A phase 1, randomized, placebo-controlled, dose-escalation study of an anti-IL-13 monoclonal antibody in healthy subjects and mild asthmatics. Br J Clin Pharmacol. 2013;75:118-28. doi:10.1111/j.1365-2125.2012.04334.x. PMID:22616628
- Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, Moriyama C, Watanabe T, Takubo R, Doi Y, et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol. 2010;28:1203-7. doi:10.1038/nbt.1691. PMID:20953198
- Chaparro-Riggers J, Liang H, DeVay RM, Bai L, Sutton JE, Chen W, Geng T, Lindquist K, Casas MG, Boustany LM, et al. Increasing Serum Half-life and Extending Cholesterol Lowering in Vivo by Engineering Antibody with pH-sensitive Binding to PCSK9. J Biol Chem. 2012;287:11090-7. doi:10.1074/jbc.M111.319764. PMID:22294692
- Devanaboyina SC, Lynch SM, Ober RJ, Ram S, Kim D, Puig-Canto A, Breen S, Kasturirangan S, Fowler S, Peng L, et al. The effect of pH dependence of antibody-antigen interactions on subcellular trafficking dynamics. mAbs. 2013;5:851-9. doi:10.4161/mabs.26389. PMID:24492341
- Fukuzawa T, Sampei Z, Haraya K, Ruike Y, Shida-Kawazoe M, Shimizu Y, Gan SW, Irie M, Tsuboi Y, Tai H, et al. Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases. Sci Rep. 2017;7:1080. doi:10.1038/s41598-017-01087-7. PMID:28439081
- Igawa T, Maeda A, Haraya K, Tachibana T, Iwayanagi Y, Mimoto F, Higuchi Y, Ishii S, Tamba S, Hironiwa N, et al. Engineered Monoclonal Antibody with Novel Antigen-Sweeping Activity In Vivo. PLoS ONE. 2013;8:e63236. doi:10.1371/journal.pone.0063236. PMID:23667591
- Igawa T, Mimoto F, Hattori K. pH-dependent antigen-binding antibodies as a novel therapeutic modality. Biochim Biophys Acta BBA - Proteins Proteomics. 2014;1844:1943-50. doi:10.1016/j.bbapap.2014.08.003. PMID:25125373
- Raghavan M, Bonagura VR, Morrison SL, Bjorkman PJ. Analysis of the pH Dependence of the Neonatal Fc Receptor/Immunoglobulin G Interaction Using Antibody and Receptor Variants. Biochemistry (Mosc). 1995;34:14649-57; doi:10.1021/bi00045a005
- Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7:715-25. doi:10.1038/nri2155. PMID:17703228
- Dall'Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J Biol Chem. 2006;281:23514-24. doi:10.1074/jbc.M604292200. PMID:16793771
- Igawa T, Ishii S, Maeda A, Nakai T. Antibodies with modified affinity to fcrn that promote antigen clearance [Internet]. 2016 [cited 2016 Oct 19]; Available from: http://www.google.com/patents/US20160244526.
- Yang D, Singh A, Wu H, Kroe-Barrett R. Comparison of biosensor platforms in the evaluation of high affinity antibody-antigen binding kinetics. Anal Biochem. 2016;508:78-96. doi:10.1016/j.ab.2016.06.024. PMID:27365220
- Lee JW, Kelley M, King LE, Yang J, Salimi-Moosavi H, Tang MT, Lu J-F, Kamerud J, Ahene A, Myler H, et al. Bioanalytical approaches to quantify “total” and “free” therapeutic antibodies and their targets: technical challenges and PK/PD applications over the course of drug development. AAPS J. 2011;13:99-110. doi:10.1208/s12248-011-9251-3. PMID:21240643
- Bonvin P, Venet S, Fontaine G, Ravn U, Gueneau F, Kosco-Vilbois M, Proudfoot AE, Fischer N. De novo isolation of antibodies with pH-dependent binding properties. mAbs. 2015;7:294-302. doi:10.1080/19420862.2015.1006993. PMID:25608219
- Lee E-C, Liang Q, Ali H, Bayliss L, Beasley A, Bloomfield-Gerdes T, Bonoli L, Brown R, Campbell J, Carpenter A, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat Biotechnol. 2014;32:356-63. doi:10.1038/nbt.2825. PMID:24633243
- Henne KR, Ason B, Howard M, Wang W, Sun J, Higbee J, Tang J, Matsuda KC, Xu R, Zhou L, et al. Anti-PCSK9 Antibody Pharmacokinetics and Low-Density Lipoprotein-Cholesterol Pharmacodynamics in Nonhuman Primates Are Antigen Affinity-Dependent and Exhibit Limited Sensitivity to Neonatal Fc Receptor-Binding Enhancement. J Pharmacol Exp Ther. 2015;353:119-31. doi:10.1124/jpet.114.221242. PMID:25653417
- Haraya K, Tachibana T, Iwayanagi Y, Maeda A, Ozeki K, Nezu J, Ishigai M, Igawa T. PK/PD analysis of a novel pH-dependent antigen-binding antibody using a dynamic antibody–antigen binding model. Drug Metab Pharmacokinet. 2016;31:123-32. doi:10.1016/j.dmpk.2015.12.007. PMID:26944099
- Borrok MJ, Wu Y, Beyaz N, Yu X-Q, Oganesyan V, Dall'Acqua WF, Tsui P. pH-dependent Binding Engineering Reveals an FcRn Affinity Threshold That Governs IgG Recycling. J Biol Chem. 2014;290:4282-90. doi:10.1074/jbc.M114.603712. PMID:25538249
- Nixon AE, Chen J, Sexton DJ, Muruganandam A, Bitonti AJ, Dumont J, Viswanathan M, Martik D, Wassaf D, Mezo A. Fully Human Monoclonal Antibody Inhibitors of the Neonatal Fc Receptor Reduce Circulating IgG in Non-Human Primates. Front Immunol. 2015;6. doi:10.3389/fimmu.2015.00176. PMID:25954273
- Garg A, Balthasar JP. Physiologically-based pharmacokinetic (PBPK) model to predict IgG tissue kinetics in wild-type and FcRn-knockout mice. J Pharmacokinet Pharmacodyn. 2007;34:687-709. doi:10.1007/s10928-007-9065-1. PMID:17636457
- Shah DK, Betts AM. Towards a platform PBPK model to characterize the plasma and tissue disposition of monoclonal antibodies in preclinical species and human. J Pharmacokinet Pharmacodyn. 2012;39:67-86. doi:10.1007/s10928-011-9232-2. PMID:22143261
- Prabhat P, Gan Z, Chao J, Ram S, Vaccaro C, Gibbons S, Ober RJ, Ward ES. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc Natl Acad Sci. 2007;104:5889-94. doi:10.1073/pnas.0700337104. PMID:17384151
- Ober RJ, Martinez C, Vaccaro C, Zhou J, Ward ES. Visualizing the Site and Dynamics of IgG Salvage by the MHC Class I-Related Receptor, FcRn. J Immunol. 2004;172:2021-9. doi:10.4049/jimmunol.172.4.2021. PMID:14764666
- Ober RJ, Martinez C, Lai X, Zhou J, Ward ES. Exocytosis of IgG as mediated by the receptor, FcRn: An analysis at the single-molecule level. Proc Natl Acad Sci. 2004;101:11076-81. doi:10.1073/pnas.0402970101. PMID:15258288
- Chen Y, Balthasar JP. Evaluation of a catenary PBPK model for predicting the in vivo disposition of mAbs engineered for high-affinity binding to FcRn. AAPS J. 2012;14:850-9. doi:10.1208/s12248-012-9395-9. PMID:22956476
- Gil J, Silage DA. Morphometry of pinocytotic vesicles in the capillary endothelium of rabbit lungs using automated equipment. Circ Res. 1980;47:384-91. doi:10.1161/01.RES.47.3.384. PMID:7408120
- Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 1999.
- Giragossian C, Vage C, Li J, Pelletier K, Piché-Nicholas N, Rajadhyaksha M, Liras J, Logan A, Calle RA, Weng Y. Mechanistic investigation of the preclinical pharmacokinetics and interspecies scaling of PF-05231023, a fibroblast growth factor 21-antibody protein conjugate. Drug Metab Dispos Biol Fate Chem. 2015;43:803-11. doi:10.1124/dmd.114.061713. PMID:25805881
- Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10:1093-5. doi:10.1023/A:1018943613122. PMID:8378254
- Wiśniewski JR, Ostasiewicz P, Duś K, Zielińska DF, Gnad F, Mann M. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol Syst Biol. 2012;8:611. doi:10.1038/msb.2012.44. PMID:22968445
