ABSTRACT
Protein therapeutic higher order structure (HOS) is a quality attribute that can be assessed to help predict shelf life. To model product shelf-life values, possible sample-dependent pathways of degradation that may affect drug efficacy or safety need to be evaluated. As changes in drug thermal stability over time can be correlated with an increased risk of HOS perturbations, the effect of long-term storage on the product should be measured as a function of temperature. Here, complementary high-resolution mass spectrometry methods for HOS analysis were used to identify storage-dependent changes of biotherapeutics (bevacizumab (Avastin), trastuzumab (Herceptin), rituximab (Rituxan), and the NIST reference material 8671 (NISTmAb)) under accelerated or manufacturer-recommended storage conditions. Collision-induced unfolding ion mobility-mass spectrometry data showed changes in monoclonal antibody folded stability profiles that were consistent with the appearance of a characteristic unfolded population. Orthogonal hydrogen-deuterium exchange-mass spectrometry data revealed that the observed changes in unfolding occurred in parallel to changes in HOS localized to the periphery of the hinge region. Using intact reverse-phase liquid chromatography-mass spectrometry, we identified several mass species indicative of peptide backbone hydrolysis, located between the variable and constant domains of the heavy chain of bevacizumab. Taken together, our data highlighted the capability of these approaches to identify age- or temperature-dependent changes in biotherapeutic HOS.
Introduction
The reported shelf-life of a therapeutic product is generally calculated using data from several sources and combined with statistical modelling to estimate the duration over which a product is expected to retain therapeutic efficacy, purity and safety.Citation1 To correctly model these shelf-lives, several quality attributes (QAs) are identified through quality risk management and experimentation, which may include any physical, chemical or biological properties/characteristics.Citation2 In the US, shelf-life assessment is often performed using a Quality by Design approach that ensures that the knowledge base accounts for the association between the clinical properties and selected QAs of a product, while further accounting for how variations in the manufacturing process may affect the QAs and clinical properties.Citation2–Citation4
Given that process-related changes in these QAs are to be expected during the biotherapeutic production cycle, substantial efforts have been made to identify any attribute that may affect the stability, safety or efficacy of the product.Citation5 Notable product QAs include, but are not limited to, formulation,Citation6,Citation7 impurity-dependent fragmentation/proteolysis,Citation8 glycan profileCitation9,Citation10 and oxygen- or light-dependent protein modifications.Citation11,Citation12 Additionally, the cellular systems, such as Chinese hamster ovary cells,Citation13 used to express these biotherapeutics can cause variations in the active pharmaceutical ingredient QAs between production batches.Citation5,Citation14
Subvisible protein aggregates, given their known association with immune-mediated responses, are a QA with direct safety-related concerns.Citation15,Citation16 Several manufacturer-dependent factors known to enhance the risk of forming these subvisible aggregates include, but are not limited to, filtration,Citation17 agitation,Citation18 freeze-thaw and lyophilization.Citation19–Citation21 Because unfolding of the higher order structure (HOS) of monoclonal antibodies (mAbs) provides a pathway for aggregates to form, the effect of mAb thermal stability as a function of time on the formulated products needs to be evaluated. While higher temperatures lead to a greater risk of unfolding within proteins, including transient unfolding events, reduced thermal stability will further enhance the risk of potential protein denaturation, misfolding and aggregation.Citation22
Substantial efforts have been made by the manufacturers during product development and manufacture cycles to identify the consequences of long-term storage on the stability and safety of the products through the optimization of accessible storage and formulation conditions, using real-time/real-temperature data supported by the use of forced degradation studies.Citation23–Citation25 While ICH guidelines recommend that the expiration date only be calculated based on these real-time and real-condition datasets, accelerated storage studies are still considered extremely valuable despite the non-Arrhenius degradation behaviors associated with their use. Notably, accelerated storage studies represent an important approach for analytical method validation, in addition to the preliminary assessment of manufacturing process changes, such as scale-up and/or formulation changes.Citation23
Product formulation considerations known to affect the long-term stability of biotherapeutic products include both the concentrationCitation7 and type of non-ionic detergent used (e.g., Polysorbate 20 vs Polysorbate 80).Citation26 Notable work by Singh and co-workers, using a variety of analytical methods, indicated that both Polysorbate 20 and Polysorbate 80 exhibited a high affinity for the mAb N-terminal variable (VH/VL)- and constant (C1/CL)- containing antigen-binding fragment (Fab) domain compared to the C-terminal Fc domains. Compared to Polysorbate 20, Polysorbate 80 was shown to exhibit a higher affinity for the Fab domain, and as a consequence may, provide enhanced levels of protection against tertiary structure perturbations within this region of the mAb.Citation26,Citation27
Consistent with above concerns regarding long-term thermal stability on biotherapeutic HOS, we investigated the consequences of accelerated storage conditions (6 months at 25 ± 2°C, 60 ± 5% RH (relative humidity)) on four commercially available mAbs, bevacizumab (Avastin), trastuzumab (Herceptin), rituximab (Rituxan), and the NIST reference material 8671 (NISTmAb).Citation28 To provide additional sample comparison data, two different lots of rituximab were studied, including an in-date (unexpired) lot and an out-of-date (expired) lot with an expiration date several years before the start of the study, which was used as a negative control. Each of these biotherapeutic products exhibits variations in terms of their formulations () and sequences (). To compare the effect of accelerated storage on the HOS of these products, four different mass spectrometric methods, including native ion mobility-mass spectrometry (IM-MS), collision-induced unfolding (CIU), hydrogen-deuterium exchange-mass spectrometry (HDX-MS), and intact protein reverse phase-liquid chromatography-mass spectrometry (RP-LC-MS), were used for analysis, each providing a complementary dataset.
Table 1. Product formulations of mAbs studied.
Table 2. Blast identity sequence similarity score,Citation30 as a percentage for all samples studied.
Results
Native ion mobility-mass spectrometry and collision-induced unfolding of biotherapeutics subjected to accelerated storage conditions
The effects of accelerated storage conditions on the native-like folded protein structure of the biotherapeutics included in this study were examined with gas-phase travelling-wave collision cross sections measured in Nitrogen (TWCCSN2) measurements using native IM-MS.Citation31,Citation32 With optimized instrumental parameters, native IM-MS methods are generally accepted to provide biologically relevant measurements of the rotationally averaged solution phase structure.Citation33,Citation34 Several relevant review articles provide comprehensive information on IM-MS.Citation31,Citation32
Calculated TWCCSN2 values comparing the selected biotherapeutics before and after incubating for six months at the accelerated storage conditions (25 ± 2°C with 60 ± 5% RH) stipulated by the International Council for Harmonisation (ICH) are presented in ; additional data is provided in supporting information (SI) Table 1. Across all samples studied, the TWCCSN2 values were consistent with previously published theoretical cross section values using the trajectory method.Citation36 However, these data were not consistent with previously published data that inferred a degree of gas phase structural compaction occurring.Citation36,Citation37 Therefore, accounting for the errors presented, no change in the rotationally averaged structure of the mAbs studied was observed after the 6-months incubation period at the stated accelerated storage conditions.
Figure 1. Native ion mobility-mass spectrometry analysis of mAbs before and after incubation at the ICH-stipulated accelerated aging conditions. Comparison of TWCCSN2 data for samples before and after accelerated aging at 25°C, 60% relative humidity. Plots of m/z vs drift time (tD) are shown for bevacizumab both before (a) and after (b) accelerated aging. Averaged TWCCSN2 data for all samples are shown in c. Replicate errors shown are 2x the standard deviation. * highlights the +22, nearest-native, charge stateCitation35 used to calculate TWCCSN2 presented in c.
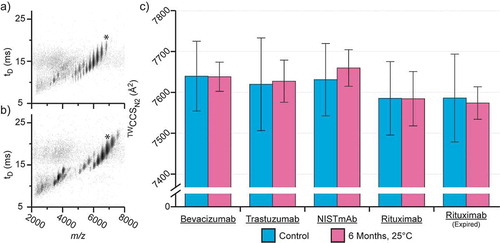
These data suggest that either there is no change in rotationally averaged mAb structure, or highly localized minor structural variations in HOS may be too subtle to resolve using cross-sectional analysis given the lower spatial resolution of the measurement. Thus, orthogonal approaches were applied to examine the impact of accelerated storage conditions on the structural stability of the mAbs studied.
In tandem with the above native IM-MS studies, we further leveraged the power of CIU,Citation34,Citation38–Citation40 which is considered an orthogonal measurement to differential scanning calorimetry,Citation41,Citation42 to measure differences in the gas phase stability of the chosen biotherapeutics before and after sample incubation. CIU leverages the capability of IM-MS to measure changes in structural conformation as a function of gas-phase collisional activation.Citation38 Changes in the structural stability of the gas phase ions can then be quantified against reference datasets using difference plots and scaled deviation score (SDS) values.Citation43
Data comparing the stability of a control against mAbs subjected to accelerated aging conditions (6 months) are presented in , with additional data presented in SI Figures 1 and 2. These data demonstrated that, as a consequence of the storage conditions used, changes in biotherapeutic structural stability were readily observed by CIU-IM-MS. These changes were especially pronounced for the unexpired lots (bevacizumab, trastuzumab, NISTmAb and in-date rituximab), after incubating at the 6-month accelerated storage conditions, with localized SDS unfolding differences all exceeding 300 at 1.44 keV (lab frame energy) compared to the baseline at 0.5 keV (). These observations were further supported by greatly increased root-mean-square deviation (RMSD) difference plot scores (> 6%), indicative of these changes in unfolding (). Consistent with these observations, excluding the expired rituximab lot that showed no difference, samples stored under the manufacturer-recommended conditions (2–8°C) retained these reproducible changes in unfolding (SI Figure 1). However, compared to lots incubated at the ICH-stipulated accelerated storage conditions, samples incubated at the manufacturer-recommended conditions exhibited smaller changes in SDS intensity (≤200, SI Figure 1) at 1.44 keV.
Figure 2. Comparison of collisionally unfolded mAbs before and after incubation at the ICH-stipulated accelerated aging conditions. Control CIU (a) data revealed three dominant unfolding populations (1, 2 and 3) in addition to the native-like conformation (N) for all mAbs studied. Samples subjected to accelerated aging (b) exhibit a similar unfolding profile, with an additional unfolding population (x) observed between the native and previously initial unfolding population (1). Difference plot analysis (c) highlights the extent of the unfolding differences between samples. Plotting the scaled deviation scores (SDS) (d) revealed characteristic spikes consistent with the appearance of a structural unfolding population denoted as ‘x.’ Despite minimal observable changes in the difference plot for the expired rituximab, the SDS plot highlights the consequences of accelerated aging on the sample.
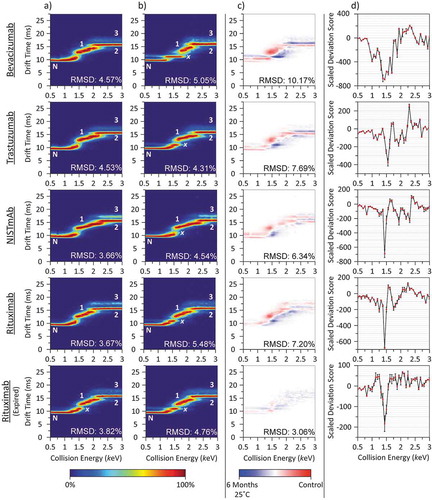
Compared to the unexpired sample lots (bevacizumab, trastuzumab, NISTmAb and in-date rituximab), the expired rituximab lot exhibited smaller changes in SDS intensity under the accelerated storage conditions (−183.6 ± 28.6 at 1.44 keV) (). No changes were observed for those samples that were stored under the manufacturer recommended conditions (2–8°C, SI Figure 1d). These observations are further supported by minimal changes in the global unfolding profile, by difference plot analysis with RMSD values ≤ 3.06% (, and SI Figure 1c).
Closer inspection of the data presented above revealed that the most pronounced changes in SDS, at 1.44 keV (), occurred in tandem with the emergence of a new structural unfolding species identified here as ‘x’. Importantly, this pronounced change in the SDS was found to be consistent across all samples studied (bevacizumab, trastuzumab, NISTmAb, in-date and expired rituximab), and highlighted a possible universal pathway for changes in HOS stability that occurred as a function of time. These data further revealed additional downstream unfolding differences in stability, notably trastuzumab was shown to exhibit a spike in SDS at 2.25 keV consistent with the unfolding transition between intermediate 1 and 2. Given an SDS difference threshold of ±100, changes in the unfolding transition between intermediates 1 and 2 were observed in all but the expired rituximab sample. These data therefore revealed that, while the most prevalent change in stability may be argued to exist between the native state and initial unfolding intermediate, the SDS approach is capable of monitoring more subtle localized changes in biotherapeutic stability across the entirety of the unfolding study.
Importantly, our data highlighted that the structural stability of ‘x’ increased over time at the expense of both the native conformation and proceeding unfolding intermediate (intermediate 1, ). The increased stability of ‘x’ was further observed in tandem with the reduced lifetime of the native-like fold under CIU conditions (). Taken together these observations are attributed to the thermal stability decrease of the biotherapeutics tested over time. The reduced thermal stability of the native conformation may indicate an increased risk of protein misfolding that may in turn act as a pathway to subsequent protein aggregation.Citation44 Furthermore, the existence of such misfolded species could further increase the inherent risk of immunogenic responses, either directly or through an downstream subvisible aggregate formation.Citation45
Hydrogen-deuterium exchange mass spectrometry of biotherapeutics subjected to accelerated storage conditions
While CIU-IM-MS is capable of detecting storage-dependent changes in thermal stability, we were interested in confirming the impact of these conditions on the HOS of mAbs studied using an in-solution labelling approach. HDX-MS was therefore chosen for these studies among all available in-solution labelling approaches, including covalent labelling and fast photochemical oxidation of proteins,Citation46–Citation48 due to both the commercial availability of suitable hardware and well-established methodology. HDX-MS measures mass shifts associated with labile hydrogen-deuterium exchange events as a function of time to determine localized changes in analyte HOS relative to a reference. Areas of increased or lower relative deuterium uptake are indicative of changes in HOS, which reflect the reduced or enhanced protection, respectively, of labile hydrogens in a manner highly correlated with solvent exposure.Citation49,Citation50
Sequence coverage, replicable across all conditions studied, for the respective mAbs were as follows: bevacizumab 86.7% HC/59.8% LC; trastuzumab 89.3% HC/86.4% LC; NISTmAb RM8671 79.1% HC/53.1% LC; and rituximab 84.2% HC/98.1% LC. Coverage maps are shown in SI Figure 3. Regions of poor mAb sequence coverage were primarily located within the VL domain (bevacizumab and NISTmAb RM8671), the inter-chain disulfide bond dense stretch of the heavy chain hinge region and the C-terminal flanking constant (C2) domain that contains the conserved mAb N-linked glycosylation site. Regions of poor sequence coverage represent the limit of the method, as differences in comparator HOS will not be detected in these areas by this technique.
Data comparing the fractional and relative deuterium uptake for control samples, as well as those subjected to accelerated aging, are presented in . The same data for samples subjected to storage at the manufacturer-recommended conditions are included with SI Figure 4. These data demonstrated an important trend among the majority of samples studied: while sporadic, differences (>±1 Da relative difference)Citation50 in deuterium uptake were observed across the length of the mAb, the majority of these changes were clustered within the Fab structural domains (VH, VL, C1, and CL). These observations were further supported by the mirrored changes in deuterium uptake between the C1 domain of the mAb heavy chain and CL of the adjacent light chain. These results are consistent with those performed by Zhang and co-workers, who reported that the most significant changes in deuterium uptake for a bevacizumab sample subjected to thermal stress (3 minutes at 65 or 70°C) were located within the N-terminal Fab domain.Citation54 Compared to our data, the changes in deuterium uptake observed by Zhang and co-workers occurred over a broader region of the Fab domain, including both the variable (VH/VL) and constant domains (C1/CL). While differences in relative deuterium uptake were observed in both the heavy and light chain of the N-terminal variable domain (VH/VL) of our dataset, the differences existing between the dataset here and of Zhang and co-workers are considered to be due to differences in the incubation conditions used. Taken together, these changes highlight potential pharmacodynamic concerns, given the requirement of the Fab domain for in situ antigen binding efficacy.Citation51,Citation52
Figure 3. Hydrogen-deuterium exchange-mass spectrometry analysis of mAbs before and after incubation at the ICH-stipulated accelerated aging conditions. Butterfly line plots comparing deuterium uptake of control samples (top half) and those subjected to 6 months accelerated aging (bottom half), under the various deuterium incubation period studies (a). Relative difference uptake bar charts compare the sum difference in deuteration between the control and accelerated aging samples (a). Colored bars are used to highlight the position of the respective peptides relative to the core domains of the mAb, highlighted on the representative IgG1 structure (b).Citation51,Citation52 Peptides with relative difference values exceeding a significance threshold of ±1 Da are highlighted using the black and red striped bars. Sequence coverage maps for all mAbs studied are shown in the supporting information (SI Figure 3).
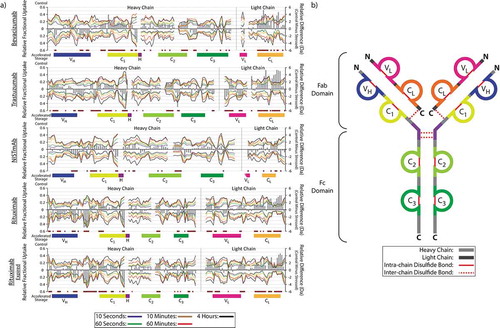
Compared to all other samples studied here, the NISTmAb presented smaller changes in relative deuterium uptake. The observed differences were also more sporadic, and located across the length of both the heavy and light chains. These differences are attributed to its greater thermal stability as observed by CIU-IM-MS ().
Reverse phase accurate mass analysis of biotherapeutics subjected to accelerated storage conditions
To examine if any changes occurred in the modifications, or if any proteolytic cleavages occurred after the ICH-recommended 6-month accelerated storage period, intact mAbs were analyzed by RP-LC-MS. This approach uses minimal sample handling, combined with a shortened reverse-phase chromatography step to permit the rapid intact mass analysis of all biotherapeutic glycoforms, and protein fragments.Citation55 This approach was chosen because the glycan profile has been shown to impact immunotolerance,Citation9 while proteolytic fragments represent previously reported age-dependent impurities.Citation55–Citation57
Intact RP-LC-MS data comparing the fractional abundance of all species observed are presented in . As a function of time, accounting for replicate error, no changes in the glycoform distributions were observed. When comparing the expired and unexpired lots of rituximab both before and after the accelerated storage conditions, we were able to identify differences in the high fractional abundance galactose-containing glycans. While these observations are potentially indicative of the effects of sample aging, we cannot confirm these conclusions given the absence of prior data supporting the glycan distribution of the expired rituximab sample, at an age comparable to that of the unexpired lot (i.e., before its own expiration date).
Figure 4. Reverse phase intact accurate mass analysis of mAbs before and after incubation at the ICH-stipulated accelerated aging conditions. Data highlight the fractional abundance of all observed glycoforms for bevacizumab (a), trastuzumab (b), NISTmAb (c) and both expired and in-date lots of rituximab (d). Masses consistent with a non-biologically relevant 2:2 ratio of heavy chain (HC) and light chain (LC) are denoted proceeding the glycan identification. * denotes one series of a heavy chain clipped variants, observed only in bevacizumab, with the site of the proteolytic cleavage highlighted. Error bars presented are representative of 2x the standard deviation. The glycan nomenclature is consistent with that commonly used to describe immunoglobulin glycoforms.Citation53 Measured masses, including PPM mass errors for all data, are presented in the supplementary information (SI Table 26).
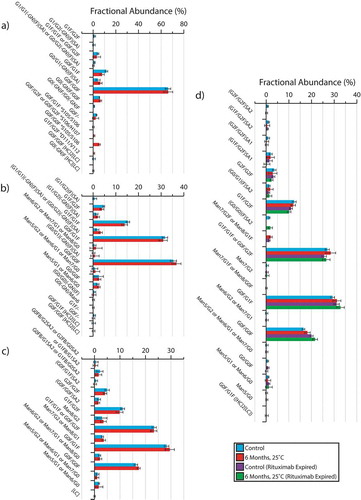
While no in-lot changes in the biotherapeutic glycoforms were identified for any of the samples studied, based on the intact mass analysis of bevacizumab, we were able to identify a number of proteolytic fragments after the 6-month incubation period. Three sites for proteolytic cleavage, all lacking the heavy chain N-terminal domain, were identified. The cleavage sites were all found within the variable (VH) to first constant (C1) interdomain region of the heavy chain, S105/S106, S106/H107, and D111/V112. The S105/S106 cleavage products were found exhibiting two different glycan distributions, G0F/G1F and G0F/G0F, while S106/H107 was identified based on mass alignment with the intact G1F/G1F glycoform. All three of these proteolytic products were identified with mass errors less than 20 ppm (SI Table 2). Comparatively, the D111/V112 (G1F/G2F) proteolytic fragment was identified with a mass error of 75 ppm, and therefore represented a lower confidence mass assignment.
Taken together, these time-dependent clipped variants represent potential product-related concerns, given the potential pharmacodynamics impact of removing the N-terminal antigen binding domain.Citation51 The clustering of these fragments within the interchain VH/C1 domain is of further interest, as it represents a potential hotspot for product degradation similar to previously published data.Citation58 Accounting for the mass accuracy of the method, except glycan and proteolytic light chain/heavy chain clipping variants, no other degradations were observed. This is unsurprising given that degradation events, such as deamidation, may impart only small mass shifts (+1 Da). As these modifications can affect the total product charge, however, ion exchange chromatographyCitation59 or microfluidic capillary electrophoresisCitation60 approaches may be better suited for identifying these subtle changes in mass.
Discussion
The impact of accelerated storage on the HOS and thermal stability of a series of mAb products are reported here as assessed by complementary mass spectrometry techniques. Notably, these data support the application of CIU-IM-MS for revealing storage-dependent changes in structural stability. Such observations highlight possible concerns regarding protein misfolding events, as a function of increased system entropy, which may result in the reduced safety or efficacy of the products. In contrast, native IM-MS approaches revealed no measurable differences in the rotationally averaged cross-sectional structure of the mAb sample set. Considering that several degrees of freedom are exhibited by mAbs both in solutionCitation61,Citation62 and the gas phase,Citation36,Citation37 these results are not surprising given that any minor changes in structure would be readily masked by this inherent structural flexibility. These results are consistent with previous reports presented by Tian and co-workers,Citation63 where IgG subtypes (IgG1, IgG2, IgG3, and IgG4) were observed to be structurally indistinguishable by IM-MS despite observed differences in CIU pathways. The observed differences in CIU were attributed to differences in the respective disulfide bonding patterns of the IgG subtypes, with the extent and pattern of disulfide bonds imparting varying degrees of conformational stability. Based on these critical data, we speculate here that the observed differences in CIU between biotherapeutics subjected to unfolding at 0 and 6 months () may be indicative of disulfide bond rearrangements occurring over the stated time period. Supporting these considerations, data presented by Paborji and co-workersCitation64 identified an increased risk of disulfide bond breakage occurring within a mAb incubated at higher temperatures. Additional reports further support the conclusion that these breakages, along with any sequential bond formation, can affect the stability of mAbs.Citation65–Citation67 However, given the absence of any disulfide mapping approach in our study, such as those previously reported in the literature,Citation68 we cannot conclusively support our hypothesis using CIU-IM-MS data alone.
Complementing our CIU-IM-MS stability analysis, our HDX-IM-MS data revealed the impact of accelerated aging on the HOS of our chosen biotherapeutics. These HDX-IM-MS data indicate that the most notable changes in deuterium uptake occur within the Fab domain (). Similarly, Zhang and co-workers reported the most significant changes in deuterium uptake for a bevacizumab sample subjected to thermal stress were located within the N-terminal Fab domain.Citation54 In their work, the authors were further able to show that the observed changes in deuterium exchange were due to localized unfolding events, which were correlated with increased non-native aggregate states observed by complementary methods,Citation54 and revealed potential sample-related safety or efficacy concerns.
Using intact RP-LC-MS accurate mass analysis, no changes in protein modifications with a fractional abundance above 0.5% were observed. These observations are important given the known effects of modifications, including oxidation, on the thermal stability of proteins.Citation69,Citation70 However, given the overall mass complexity of these biotherapeutics, more subtle changes in intact mass may be masked, and would be better characterized using methods that are capable of separations based on charge (e.g., ion exchange chromatographyCitation59 or microfluidic capillary electrophoresisCitation60). Alternatively, or in combination with the above separation techniques, steps could be made to reduce the mass complexity of the analyte using limited or complete proteolysis with middle-down and bottom-up proteomics approaches.Citation71,Citation72
While no new protein modifications were observed, we were able to identify proteolytic degradation products for bevacizumab. These proteolytic degradation products, centered within the heavy chain VH/C1 inter-domain region, are supportive of a ‘proteolytic hot-spot’ which is consistent with previously published data.Citation58 These proteolytic fragments are of concern when discussing the QA of a biotherapeutic, given the consequences of such proteolysis events on the structure of the antigen binding domain.
Based on our data and the methods used herein, we argue here that the most predominant accelerated aging-dependent QA changes can be attributed to changes in the HOS of the Fab domain (). These changes in HOS can in turn be correlated with the reduced thermal stability of the native mAb conformation, preceding the formation of an increased stability unfolding population that we have identified here as ‘x’ (). Furthermore, our RP-LC-MS studies have revealed a potential hot-spot for proteolysis between the VH and C1 domains of the bevacizumab Fab domain (). Taken together these data highlight potential pharmacodynamics and pharmacokinetics issues given the importance of the Fab domain and HOS stability on the efficacy of these biotherapeutic products.
While the observed changes in HOS and stability appear ubiquitous between the biotherapeutics studied, notable differences between CIU-IM-MS and HDX-MS data are observed when these are compared side by side. Comparing the impact of the accelerated storage conditions used, bevacizumab revealed the most notable change in unfolding, with population ‘x’ exhibiting an increased gas phase lifetime as a function of activation energy (~625 eV) compared to the remaining samples studied (≤315 eV) (). While trastuzumab has a point-of-use concentration (21 mg/ml) and non-ionic detergent (0.1 mg/ml Polysorbate 20) profile comparable to bevacizumab (25 mg/ml, 0.4 mg/ml Polysorbate 20), the former is supplied and stored as a lyophilized product prior to the point of use reconstitution. Under carefully controlled conditions, and given suitably low residual moisture content (<0.5%), lyophilization is known to be capable of increasing storage stability.Citation73
Based on the above considerations, the observed thermal stability differences between the native state of bevacizumab and all other samples studied may be represented by one or both of the following sample dependent variables: 1) formulation dependent effects, including the impact of the non-ionic detergent used and application of lyophilization to increase the long-term storage stability prior to the point of use ();Citation7, Citation6,Citation73 and 2) sequence-dependent effects (), likely localized within the variable domains given an average similarity of 61 ± 15% compared to >98% across the remaining sequence.
In conclusion, our investigation of a series of commercial therapeutic products importantly highlights the consequences of biotherapeutic accelerated storage on the HOS and thermal stability. These data further reveal the capability of native IM-MS, CIU-IM-MS and HDX-IM-MS to examine these differences. Future work is needed to identify the consequences of product formulation differences, notably the effect of detergent concentration, to enable us to more conclusively determine the cause and effect relationships that we observed. These future studies would benefit from the expansion of analysis approaches to include both disulfide bond mapping, and a suitable charge variant analysis approach in lieu of the RP-LC-MS approach used here. Additional considerations for experimental methods that provide aggregate level information, including size-exclusion chromatographyCitation74 and subvisible particle analysis approaches,Citation75 would provide valuable data on the downstream effects of these changes in HOS and stability.
Materials and methods
Five different mAb lots, across four different products, were chosen for this study. Products selected were the NIST reference material RM8671 (NISTmAb),Citation28 and three commercially available mAb products from the same manufacturer (Genentech, San Francisco, CA): bevacizumab (Lot# 3049679), trastuzumab (Lot# 3049782), and two separate lots of rituximab (Lot# 461034 and 3031077). Excluding trastuzumab, all samples were supplied as liquid formulations for storage at 2–8°C. Trastuzumab was supplied as a lyophilized product, for storage at 2–8°C, which was reconstituted per manufacturer guidelines at the start of the studies. Additional details regarding product formulations, including concentrations, are provided in . With the exception of one lot of rituximab (Lot#:461034, expiration date: July 2014) chosen as a negative control, all therapeutic mAbs were used within one month of their expiration date by the end of the study (August 2017).
According to ICH guidelines Q1ACitation76 and Q5C,Citation23 samples were aliquoted undiluted into 2 ml amber borosilicate HPLC vials (Agilent, Santa Clara, CA) to mimic the final product packing material used by the manufacturer. Vials were capped using lids having a PTFE insert, and purged with nitrogen. Four separate vials of each mAb were incubated at the ICH-specified accelerated storage conditions for refrigerated products, 25 ± 2°C and 60 ± 5% RH, for the specified minimum of 6 months, in the absence of light using a TPS (New Columbia, PA) C-EVO temperature/humidity test chamber.Citation23,Citation76 Additional triplicate tests were performed on separate samples, incubated under the same ICH-specified conditions, at 1 and 3 months. Control samples, stored in the absence of light at the manufacturer-recommended storage conditions (2–8°C, ambient humidity), were tested in triplicate at 0 and 6 months. All samples were stored vertically to prevent interactions between the sample and the lid.
All IM-MS experiments, including CIU, were performed using a Waters (Milford, MA) Synapt G2-Si. IM-MS instrumental parameters were optimized to prevent unnecessary gas phase activation, consistent with previously published methods.Citation33 Immediately prior to analysis, samples were buffer exchanged into 150 mM ammonium acetate (Fisher Chemical, A114) (pH 6.9) using Amicon Ultra centrifugal filters (EMD Millipore, Burlington, MA) with a 10 kDa cut-off and diluted to 5 µM using the same buffer for analysis. Samples were ionized using a nano-electrospray source operated in positive ion mode, using coated borosilicate capillaries (New Objective, Woburn, MA), with the source cone set at 40 V operating at a temperature of 30°C. Nitrogen traveling wave ion mobility collision cross section (TWCCSN2) values were calculated as the average of each technical replicate acquired under 3 different ion-mobility parameter sets, using previously published methods.Citation33,Citation77 CIU experiments were performed on the quadrupole selected +25 precursor ion, with the trap activation energy ramped from 20V to 120V in 2.5V increments. CIU fingerprint, difference plot and SDS scoring analysis were performed using an in-house MATLAB (MathWorks, Natick, MA) script adapted from the CIUSuite analysis package originally written in the Python coding language.Citation43 Data were imported into MATLAB using datasets normalized in Microsoft Excel (Redmond, WA), with minimal smoothing applied using MassLynx (Waters, Milford, MA). Data presented are the averaged technical replicates for all samples studied.
All HDX-MS experiments were performed using a Waters (Milford, MA) Synapt G2-Si coupled to a Waters ACQUITY UPLC M-Class with hydrogen-deuterium exchange technology. Deuterated samples (3 µg) were quenched at 0, 10 seconds, 1, 10, 60 or 240 minutes, before injection onto the UPLC and digested online using a Waters Enzymate Pepsin column (20°C) prior to trapping on a Waters Vanguard C18 trap column (0.5°C). The digested samples were then chromatographically separated using a Waters 1 × 100 mm ACQUITY UPLC BEH C18 Column (130 Ȧ, 1.7 µM) with a 9.5-minute water/acetonitrile (0.1% formic acid) gradient (5–35% acetonitrile) (0.5°C). The column was re-equilibrated by first holding at 75% acetonitrile for 1.5 minutes, then dropping to 5% acetonitrile for another 2 minutes (0.5°C). Eluted samples were ionized using an electrospray ionization source operating at positive ion mode, with the source cone set at 40 V and 80°C. All remaining instrumental, sample handling parameters and procedures were optimized based on previously published methods.Citation78 Studies were performed using a data-independent LC-IM-MS approach (HDMSE) shown to improve proteome coverage, which was consistent with the additional layer of separation provided by the ion mobility drift time measurement.Citation79 Output data and deuterium uptake levels were processed, calculated and manually verified using a combination of Protein Lynx Global Server 3.0 (Waters, Milford, MA) and DynamX 3.0 (Waters, Milford, MA).
RP-LC-MS was performed using a Thermo Fisher Scientific (Waltham, MA) Ultimate 3000 UPLC coupled to an Orbitrap Fusion. Samples were prepared for analysis by buffer exchanging 20 µg of sample into 5% acetonitrile with 0.1% formic acid using Amicon Ultra centrifugal filters with a 10 kDa cut-off. Samples were diluted to 200 µg/ml using the same solvent prior to analysis. Samples (1 µg, 5 µL) were injected and chromatographically separated using a temperature controlled (65 ± 5°C) Waters (Milford, MA) 1 × 150 mm ACQUITY UPLC Protein BEH C4 (300Ȧ 1.7 µM) column, eluting with a 13-minute water/acetonitrile (0.1% formic acid) gradient (20–60% acetonitrile) at a flow rate of 0.15 ml/minute. The column was re-equilibrated by first holding at 95% acetonitrile for 2 minutes, then dropping to 5% acetonitrile for another minute. Eluted samples were subsequently ionized using an electrospray ionization source operating in positive ion mode, with the in-source fragmentation set to 20V, operating at ion transfer tube and vaporizer temperatures of 400 and 200°C, respectively. All mass spectra were acquired using the Orbitrap at a resolution of 15000. Output data were deconvoluted using the Auto ReSpect algorithm within Protein Deconvolution 3.0 (Thermo Fisher Scientific), prior to additional processing and averaging in Microsoft Excel. Protein sequences for all samples, except trastuzumab, were taken from drugbank.ca.Citation80 Trastuzumab sequence data were taken from work published by Beck and co-workers,Citation81 consistent with heavy chain sequence differences. Common sequence modifications assumed for all mAbs studied included C-terminal lysine cleavages and N-terminal glutamines to pyroglutamine conversion, where relevant.Citation82,Citation83
Sample selection
This study was designed with several considerations regarding the samples selected. First, all samples selected were IgG1 type antibodies.49 Second, consistent with ICH recommendations regarding the use of suitable reference materialsCitation84 to support method evaluation, these studies were expanded to include the NISTmAb.Citation28 Third, as we were interested in comparing the effect of thermal stability as a function of accelerated aging on in-date samples, an unopened negative control was included for study against an unexpired sample from the same manufacturer: rituximab lot 1 (Lot# 3031077) expiration August 2017, rituximab lot 2 (Lot# 461034) expiration July 2014 (study ended August 2017). In addition, to evaluate the effect of concentration on sample ageing, we chose the biotherapeutic bevacizumab, formulated as a 25 mg/ml solution compared to the 10 mg/ml for both rituximab and the NISTmAb. Last, to compare the effect of lyophilization on long-term product stability, we additionally investigated trastuzumab, which was supplied as a commercial lyophilized product, with a shelf life of 1 month past the reconstitution date at a concentration of 21 mg/ml. All other formulation differences, and brand names, are given in .
Abbreviations
Supplemental Material
Download MS Word (33.4 MB)Acknowledgments
This project was supported in part by an appointment to the Research Participation Program at the Center for Drug Evaluation and Research administered by the Oak Ridge Institute for Science and Engineering and the U.S. Food and Drug Administration for R.A.K. The authors would further like to thank Brandon Ruotolo and Daniel Polasky from The University of Michigan for useful discussions regarding implementation of several CIUSuite features into MATLAB.
This publication reflects the views of the authors and should not be construed to represent FDA’s views or policies.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Tsong Y. Recent issues in stability study. J Biopharm Stat. 2003;13(3):vii–ix. doi:10.1081/BIP-120022758.
- ICH Harmonised Tripartate Guideline: Pharmaceutical Development Q8(R2). International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use; 2009.
- Rathore AS, Winkle H. Quality by design for biopharmaceuticals. Nat Biotechnol. 2009;27:26. doi:10.1038/nbt0109-26.
- Xu Y, Wang D, Mason B, Rossomando T, Li N, Liu D, Cheung JK, Xu W, Raghava S, Katiyar A, e al. Structure, heterogeneity and developability assessment of therapeutic antibodies. mAbs. 2019;11(2):239–64.
- Ambrogelly A, Gozo S, Katiyar A, Dellatore S, Kune Y, Bhat R, Sun J, Li N, Wang D, Nowak C, et al. Analytical comparability study of recombinant monoclonal antibody therapeutics. mAbs. 2018;10(4):513–38. doi:10.1080/19420862.2018.1438797.
- Leblanc Y, Bihoreau N, Jube M, Andre MH, Tellier Z, Chevreux G. Glycation of polyclonal IgGs: effect of sugar excipients during stability studies. Eur J Pharm Biopharm. 2016;102:185–90. doi:10.1016/j.ejpb.2016.03.016.
- Agarkhed M, O’Dell C, Hsieh M-C, Zhang J, Goldstein J, Srivastava A. Effect of polysorbate 80 concentration on thermal and photostability of a monoclonal antibody. AAPS PharmSciTech. 2013;14(1):1–9. doi:10.1208/s12249-012-9878-0.
- Vlasak J, Ionescu R. Fragmentation of monoclonal antibodies. mAbs. 2011;3:253–63.
- Reusch D, Tejada ML. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology. 2015;25(12):1325–34. doi:10.1093/glycob/cwv065.
- Fang J, Richardson J, Du Z, Zhang Z. Effect of Fc-Glycan structure on the conformational stability of IgG revealed by hydrogen/deuterium exchange and limited proteolysis. Biochemistry. 2016;55(6):860–68. doi:10.1021/acs.biochem.5b01323.
- Lam XM, Yang JY, Cleland JL. Antioxidants for prevention of methionine oxidation in recombinant monoclonal antibody HER2. J Pharm Sci. 1997;86(11):1250–55. doi:10.1021/js970143s.
- Haywood J, Mozziconacci O, Allegre KM, Kerwin BA, Schöneich C. Light-induced conversion of Trp to gly and gly hydroperoxide in IgG1. Mol Pharm. 2013;10(3):1146–50. doi:10.1021/mp300680c.
- Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22:1393. doi:10.1038/nbt1026.
- Goetze AM, Schenauer MR, Flynn GC. Assessing monoclonal antibody product quality attribute criticality through clinical studies. mAbs. 2010;2:500–07.
- Koren E, Zuckerman LA, Mire-Sluis AR. Immune responses to therapeutic proteins in humans - clinical significance, assessment and prediction. Curr Pharm Biotechnol. 2002;3:349–60.
- Moussa EM, Panchal JP, Moorthy BS, Blum JS, Joubert MK, Narhi LO, Topp EM. Immunogenicity of therapeutic protein aggregates. J Pharm Sci. 2016;105(2):417–30. doi:10.1016/j.xphs.2015.11.002.
- Maa Y-F, Hsu CC. Investigation on fouling mechanisms for recombinant human growth hormone sterile filtration. J Pharm Sci. 1998;87(7):808–12. doi:10.1021/js980114x.
- Maa Y-F, Hsu CC. Protein denaturation by combined effect of shear and air-liquid interface. Biotechnol Bioeng. 1997;54(6):503–12. doi:10.1002/(SICI)1097-0290(19970620)54:6<503::AID-BIT1>3.0.CO;2-N.
- Passot S, Fonseca F, Barbouche N, Marin M, Alarcon-Lorca M, Rolland D, Rapaud M. Effect of product temperature during primary drying on the long-term stability of lyophilized proteins. Pharm Dev Technol. 2007;12(6):543–53. doi:10.1080/10837450701563459.
- Eckhardt BM, Oeswein JQ, Bewley TA. Effect of freezing on aggregation of human growth hormone. Pharm Res. 1991;8:1360–64.
- Cao E, Chen Y, Cui Z, Foster PR. Effect of freezing and thawing rates on denaturation of proteins in aqueous solutions. Biotechnol Bioeng. 2003;82(6):684–90. doi:10.1002/bit.10612.
- Porter D, Vollrath F. The role of kinetics of water and amide bonding in protein stability. Soft Matter. 2008;4(2):328–36. doi:10.1039/B713972A.
- ICH Harmonised Tripartite Guideline: Quality of Biotechnological Products; Stability Testing of Biotechnological/Biological Products Q5C. International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use; 1995.
- Nowak C,K, Cheung J,M, Dellatore S, Katiyar A, Bhat R, Sun J, Ponniah G, Neill A, Mason B, Beck A, et al. Forced degradation of recombinant monoclonal antibodies: A practical guide. mAbs. 2017;9(8):1217–30. doi:10.1080/19420862.2017.1368602.
- Krishnan S, Pallito MM, Ricci MS. Development of formulations for therapeutic monoclonal antibodies and Fc fusion proteins. In: Jameel F, Hershenson S, editors. Formulation and process development strategies for manufacturing biopharmaceuticals. Haboken (NJ): John Wiley & Sons, Inc.; 2010. p. 383–427.
- Singh SM, Bandi S, Jones DNM, Mallela KMG. Effect of polysorbate 20 and polysorbate 80 on the higher-order structure of a monoclonal antibody and its fab and Fc fragments probed using 2d nuclear magnetic resonance spectroscopy. J Pharm Sci. 2017;106(12):3486–98. doi:10.1016/j.xphs.2017.08.011.
- Ionescu RM, Vlasak J, Price C, Kirchmeier M. Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. J Pharm Sci. 2008;97(4):1414–26. doi:10.1002/jps.21104.
- State-of-the-Art and Emerging Technologies for Therapeutic Monoclonal Antibody Characterization Volume 2. Biopharmaceutical Characterization: The NISTmAb Case Study. First ed: American Chemical Society; 2015. 427 p.
- Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci: A Publ Protein Soc. 2009;18(1):24–36. doi:10.1002/pro.3.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi:10.1016/S0022-2836(05)80360-2.
- Eschweiler JD, Kerr R, Rabuck-Gibbons J, Ruotolo BT. Sizing up protein–ligand Complexes: the rise of structural mass spectrometry approaches in the pharmaceutical sciences. Annu Rev Anal Chem. 2017;10(1):25–44. doi:10.1146/annurev-anchem-061516-045414.
- Campuzano IDG, Lippens JL. Ion mobility in the pharmaceutical industry: an established biophysical technique or still niche? Curr Opin Chem Biol. 2018;42:147–59. doi:10.1016/j.cbpa.2017.11.008.
- Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV. Ion mobility–mass spectrometry analysis of large protein complexes. Nat Protoc. 2008;3:1139. doi:10.1038/nprot.2008.78.
- Huang Y, Salinas ND, Chen E, Tolia NH, Gross ML. Native mass spectrometry, ion mobility, and collision-induced unfolding categorize malaria antigen/antibody binding. J Am Soc Mass Spectrom. 2017;28(11):2515–18. doi:10.1007/s13361-017-1782-0.
- Scarff CA, Thalassinos K, Hilton GR, Scrivens JH. Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements. Rapid Commun. Mass Spectrom. 2008;22(20):3297–304. doi:10.1002/rcm.3737.
- Campuzano IDG, Larriba C, Bagal D, Schnier PD. Ion mobility and Mass Spectrometry Measurements of the humanized IgGk NIST monoclonal antibody. In: Schiel J, Davis D, Borisov OV, editors. State-of-the-art and emerging technologies for therapeutic monoclonal antibody characterization volume 3 defining the next generation of analytical and biophysical techniques. ACS Symposium Series. 1202. Washington (DC): American Chemical Society; 2015. p. 75–112.
- Pacholarz KJ, Porrini M, Garlish RA, Burnley RJ, Taylor RJ, Henry AJ, Barran PE. Dynamics of intact immunoglobulin g explored by drift-tube ion-mobility mass spectrometry and molecular modeling. Angew Chem Int Ed. 2014;53(30):7765–69. doi:10.1002/anie.201402863.
- Dixit SM, Polasky DA, Ruotolo BT. Collision induced unfolding of isolated proteins in the gas phase: past, present, and future. Curr Opin Chem Biol. 2018;42:93–100. doi:10.1016/j.cbpa.2017.11.010.
- Tian Y, Lippens JL, Netirojjanakul C, Campuzano IDG, Ruotolo BT. Quantitative collision-induced unfolding differentiates model antibody–drug conjugates. Protein Sci. 2018.
- Hernandez-Alba O, Wagner-Rousset E, Beck A, Cianférani S. Native mass spectrometry, ion mobility, and collision-induced unfolding for conformational characterization of IgG4 monoclonal antibodies. Anal Chem. 2018;90(15):8865–72. doi:10.1021/acs.analchem.8b00912.
- Temel DB, Landsman P, Brader ML. Orthogonal methods for characterizing the unfolding of therapeutic monoclonal antibodies: differential scanning calorimetry, isothermal chemical denaturation, and intrinsic fluorescence with concomitant static light scattering. In: Feig AL, editor. Methods in enzymology. 567. Cambridge (MA): Academic Press; 2016. p. 359–89. doi:10.1016/bs.mie.2015.08.029
- Brader ML, Estey T, Bai S, Alston RW, Lucas KK, Lantz S, Landsman P, Maloney KM. Examination of thermal unfolding and aggregation profiles of a series of developable therapeutic monoclonal antibodies. Mol Pharm. 2015;12(4):1005–17. doi:10.1021/mp400666b.
- Eschweiler JD, Rabuck-Gibbons JN, Tian Y, Ruotolo BT. CIUSuite: A quantitative analysis package for collision induced unfolding measurements of gas-phase protein ions. Anal Chem. 2015;87(22):11516–22. doi:10.1021/acs.analchem.5b03292.
- Roberts CJ. Therapeutic protein aggregation: mechanisms, design, and control. Trends Biotechnol. 2014;32(7):372–80. doi:10.1016/j.tibtech.2014.05.005.
- Rosenberg AS. Effects of protein aggregates: an immunologic perspective. Aaps J. 2006;8(3):E501–E7. doi:10.1208/aapsj080359.
- Borotto NB, Zhou Y, Hollingsworth SR, Hale JE, Graban EM, Vaughan RC, Vachet RW. Investigating therapeutic protein structure with diethylpyrocarbonate labeling and mass spectrometry. Anal Chem. 2015;87(20):10627–34. doi:10.1021/acs.analchem.5b03180.
- Kaur P, Tomechko SE, Kiselar J, Shi W, Deperalta G, Wecksler AT, Gokulrangan G, Ling V, Chance MR. Characterizing monoclonal antibody structure by carboxyl group footprinting. mAbs. 2015;7(3):540–52. doi:10.1080/19420862.2015.1023683.
- Yan Y, Chen G, Wei H, Huang RYC, Mo J, Rempel DL, Tymiak AA, Gross ML. Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. J Am Soc Mass Spectrom. 2014;25(12):2084–92. doi:10.1007/s13361-014-0993-x.
- Deng B, Lento C, Wilson DJ. Hydrogen deuterium exchange mass spectrometry in biopharmaceutical discovery and development – A review. Anal Chim Acta. 2016;940:8–20. doi:10.1016/j.aca.2016.08.006.
- Iacob RE, Engen JR. Hydrogen exchange mass spectrometry: are we out of the quicksand? J Am Soc Mass Spectrom. 2012;23(6):1003–10. doi:10.1007/s13361-012-0377-z.
- Vidarsson G, Dekkers G, Rispens T. igg subclasses and allotypes: from structure to effector functions. Front Immunol. 2014;5:520. doi:10.3389/fimmu.2014.00520.
- Irani V, Guy AJ, Andrew D, Beeson JG, Ramsland PA, Richards JS. Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol Immunol. 2015;67(2,Part A):171–82. doi:10.1016/j.molimm.2015.03.255.
- Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianférani S. Characterization of therapeutic antibodies and related products. Anal Chem. 2013;85(2):715–36. doi:10.1021/ac3032355.
- Zhang A, Singh SK, Shirts MR, Kumar S, Fernandez EJ. Distinct aggregation mechanisms of monoclonal antibody under thermal and freeze-thaw stresses revealed by hydrogen exchange. Pharm Res. 2012;29(1):236–50. doi:10.1007/s11095-011-0538-y.
- Liu H, Gaza-Bulseco G, Lundell E. Assessment of antibody fragmentation by reversed-phase liquid chromatography and mass spectrometry. J Chromatogr B. 2008;876(1):13–23. doi:10.1016/j.jchromb.2008.10.015.
- Liu H, Gaza-Bulseco G, Sun J. Characterization of the stability of a fully human monoclonal IgG after prolonged incubation at elevated temperature. J Chromatogr B. 2006;837(1):35–43. doi:10.1016/j.jchromb.2006.03.053.
- Xiang T, Lundell E, Sun Z, Liu H. Structural effect of a recombinant monoclonal antibody on hinge region peptide bond hydrolysis. J Chromatogr B. 2007;858(1):254–62. doi:10.1016/j.jchromb.2007.08.043.
- Hehle Verena K, Lombardi R, Dolleweerd Craig J, Paul Mathew J, Di Micco P, Morea V, Benvenuto E, Donini M, Ma JK-C. Site‐specific proteolytic degradation of IgG monoclonal antibodies expressed in tobacco plants. Plant Biotechnol J. 2014;13(2):235–45. doi:10.1111/pbi.12266.
- Bailey AO, Han G, Phung W, Gazis P, Sutton J, Josephs JL, Sandoval W. Charge variant native mass spectrometry benefits mass precision and dynamic range of monoclonal antibody intact mass analysis. mAbs. 2018;10(8):1214–25. doi:10.1080/19420862.2018.1521131.
- Redman EA, Batz NG, Mellors JS, Ramsey JM. Integrated microfluidic capillary electrophoresis-electrospray ionization devices with online MS detection for the separation and characterization of intact monoclonal antibody variants. Anal Chem. 2015;87(4):2264–72. doi:10.1021/ac503964j.
- Sandin S, Öfverstedt L-G, Wikström A-C, Wrange Ö, Skoglund U. Structure and flexibility of individual immunoglobulin G molecules in solution. Structure. 2004;12(3):409–15. doi:10.1016/j.str.2004.02.011.
- Zhang X, Zhang L, Tong H, Peng B, Rames MJ, Zhang S, Ren G. 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography. Sci Rep. 2015;5:9803. doi:10.1038/srep09803.
- Tian Y, Han L, Buckner AC, Ruotolo BT. Collision induced unfolding of intact antibodies: rapid characterization of disulfide bonding patterns, glycosylation, and structures. Anal Chem. 2015;87(22):11509–15. doi:10.1021/acs.analchem.5b03291.
- Paborji M, Pochopin NL, Coppola WP, Bogardus JB. Chemical and physical stability of chimeric L6, a mouse−human monoclonal antibody. Pharm Res. 1994;11:764–71.
- Lacy ER, Baker M, Brigham-Burke M. Free sulfhydryl measurement as an indicator of antibody stability. Anal Biochem. 2008;382(1):66–68. doi:10.1016/j.ab.2008.07.016.
- Moritz B, Stracke JO. Assessment of disulfide and hinge modifications in monoclonal antibodies. Electrophoresis. 2017;38(6):769–85. doi:10.1002/elps.201600425.
- Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides in protein chemical and physical stability. Curr Protein Pept Sci. 2009;10(6):614–25. doi:10.2174/138920309789630534.
- Lakbub JC, Shipman JT, Desaire H. 2017. Recent mass spectrometry-based techniques and considerations for disulfide bond characterization in proteins. Analytical and bioanalytical chemistry.
- Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics. 2010;9(8):1716–28. doi:10.1074/mcp.M900540-MCP200.
- Mo J, Yan Q, So CK, Soden T, Lewis MJ, Hu P. Understanding the impact of methionine oxidation on the biological functions of IgG1 antibodies using hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2016;88(19):9495–502. doi:10.1021/acs.analchem.6b01958.
- Belov AM, Zang L, Sebastiano R, Santos MR, Bush DR, Karger BL, Ivanov AR. Complementary middle-down and intact monoclonal antibody proteoform characterization by capillary zone electrophoresis – mass spectrometry. Electrophoresis. 2018;39(16):2069–82. doi:10.1002/elps.201800067.
- Said N, Gahoual R, Kuhn L, Beck A, François Y-N, Leize-Wagner E. Structural characterization of antibody drug conjugate by a combination of intact, middle-up and bottom-up techniques using sheathless capillary electrophoresis – tandem mass spectrometry as nanoESI infusion platform and separation method. Anal Chim Acta. 2016;918:50–59. doi:10.1016/j.aca.2016.03.006.
- Tang X, Pikal MJ. Design of freeze-drying processes for pharmaceuticals: practical advice. Pharm Res. 2004;21:191–200.
- Goyon A, Fekete S, Beck A, Veuthey J-L, Guillarme D. Unraveling the mysteries of modern size exclusion chromatography - the way to achieve confident characterization of therapeutic proteins. J Chromatogr B. 2018;1092:368–78. doi:10.1016/j.jchromb.2018.06.029.
- Narhi LO, Corvari V, Ripple DC, Afonina N, Cecchini I, Defelippis MR, Garidel P, Herre A, Koulov AV, Lubiniecki T, et al. Subvisible (2–100 μm) particle analysis during biotherapeutic drug product development: part 1, considerations and strategy. J Pharm Sci. 2015;104(6):1899–908. doi:10.1002/jps.24437.
- ICH Hamonised Tripartate Guideline: Stability Testing of New Drug Subtances and Products Q1A(R2). International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use; 2003.
- Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82(22):9557–65. doi:10.1021/ac1022953.
- Houde D, Engen JR. Conformational analysis of recombinant monoclonal antibodies with hydrogen/deuterium exchange mass spectrometry. In: Beck A, editor. Glycosylation engineering of biopharmaceuticals: methods and protocols. Totowa (NJ): Humana Press; 2013. p. 269–89.
- Shliaha PV, Bond NJ, Gatto L, Lilley KS. Effects of traveling wave ion mobility separation on data independent acquisition in proteomics studies. J Proteome Res. 2013;12(6):2323–39. doi:10.1021/pr300775k.
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(suppl_1):D668–D72. doi:10.1093/nar/gkj067.
- Beck A, Debaene F, Diemer H, Wagner-Rousset E, Colas O, Dorsselaer AV, Cianférani S. Cutting-edge mass spectrometry characterization of originator, biosimilar and biobetter antibodies. J Mass Spectrom. 2015;50(2):285–97. doi:10.1002/jms.3554.
- Lewis DA, Guzzetta AW, Hancock WS, Costello M. Characterization of humanized anti-TAC, an antibody directed against the interleukin 2 receptor, using electrospray ionization mass spectrometry by direct infusion, LC/ MS,and MS/MS. Anal Chem. 1994;66:585–95.
- Werner WE, Wu S, Mulkerrin M. The removal of pyroglutamic acid from monoclonal antibodies without denaturation of the protein chains. Anal Biochem. 2005;342(1):120–25. doi:10.1016/j.ab.2005.04.012.
- ICH Harmonised Tripartite Guideline: Specifications; Test Procedures and Acceptance Criteria for Biotechnological/Biological Products Q6B. International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use; 1999.
