ABSTRACT
Two recent publications out of the same research laboratory report on structure-based in silico design of antibodies against viral targets without sequence disclosure. Cross-referencing the published data to patent databases, we established the sequence identity of said computationally designed antibodies. In both cases, the antibodies align with high sequence identity to previously reported antibodies of the same specificity. This clear underlying sequence relationship, which is far closer than the antibody templates reported to seed the computational design, suggests an alternative origin of the computationally designed antibodies. The lack of both reproducible computational algorithms and of output sequences in the initial publications obscures the relationship to previously reported antibodies, and sows doubt as to the genesis narrative described therein.
Introduction
Human antibodies are a major modality to treat human disease, and therefore the focus of significant technology development.Citation1 Historically, two main approaches have been used to discover antibodies against targets of therapeutic interest: 1) in vivo technologies based on isolating B cell diversity from animalsCitation2-Citation5 or humans,Citation6–Citation8 following immunization or exposure to infectious agents, respectively, and 2) in vitro technologies based on the display of synthetic or semi-synthetic human IgG diversity on the surface of phage or yeast.Citation9–Citation12
More recently, a third approach of designing human antibodies in silico against specific epitopes has been fueled by two major trends: 1) ever-increasing structural information of antibodies and their potential targets, as well as 2) access to more powerful computational tools. Notwithstanding the potential impact, the general lack of published successes in this area has highlighted the extreme challenge of designing antibodies in silico,Citation13,Citation14 although progress in the area of affinity maturation has been made.Citation15,Citation16
As such, two publicationsCitation17,Citation18 originating from the same research lab (based on the first and corresponding author), reporting the in silico design of epitope-specific, broadly neutralizing, human antibodies against two infectious disease targets, garnered our attention. Strikingly, in both cases the extraordinary accomplishments were not supported by a detailed description of methods or intermediate results, nor were the end-products of these efforts, namely the amino acid sequences of the designed antibodies, disclosed, making it impossible to independently reproduce the reported functional characterization. To understand how these results could have been achieved, we endeavored to better understand the identity and, potentially, the genesis of these antibodies. In this communication, we present evidence that in both cases, previously published antibody sequences and structures are the basis for the in silico designed antibodies.
Results
Influenza antibody
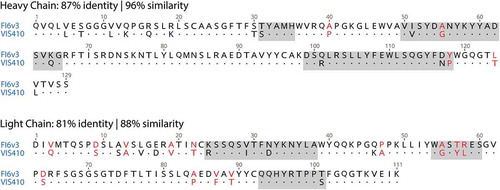
VIS410 is describedCitation17 as a broadly neutralizing antibody generated by a process that used “a database of nonredundant combinations of complementary determining region (CDR) canonical structures (antibody scaffolds), (to select) multiple antibody templates satisfying shape complementarity criteria and systematically engineered energetically favorable, hotspot-like interactions between CDR residues and these anchor residues on hemagglutinin (HA).” The authors then present experimental data on binding, neutralization, and protective efficacy in influenza animal models. However, the sequence of VIS410, the output of the design and engineering process, was neither provided in the publication, nor deposited into a public database. Using VIS410 as a search term in the USPTO database readily leads to a US patent applicationCitation19 that also designates VIS410 as Ab044. This application further establishes that the variable heavy- and light-chain (VH and VL) sequences correspond to sequence ID numbers 25 and 52, which are shown in . Searching the patent database with the VIS410 sequences produces exact matches to an earlier US patent publication from 2013 describing Ab044 with a similar inventorship group.Citation20 As shown in Figure S1, a comparison of tables from the two sources,Citation17,Citation20 confirms that VIS410 and Ab044 are in fact the same antibody.
Figure 1. VH and VL alignment of FI6v3 (PDB file 3ZTJ) and VIS410 (Ab 044) FI6. Non-conservative substitutions depicted in red font. CDRs are highlighted in gray according to Kabat’sCitation23 definition.
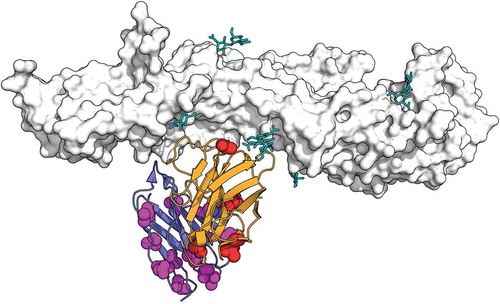
It is interesting that searching Genbank with the VH sequence, even today (April 2019, nearly four years after the original publication) does not yield a 100% match. However, the search does reveal FI6v3, a broadly neutralizing anti-influenza antibody first described in 2011 by Corti and et al.Citation21 An alignment of VIS410 and FI6v3 is shown in . The overall percent identity values are 87% and 81%, respectively, for the VH and VL domains. This is achieved with no gaps in the alignment, indicating that all the corresponding CDR lengths are identical; this result is particularly significant for the CDRH3 and for the CDRL1, with the latter showing a relatively rare two amino acid deletion relative to the human germline of origin, as presented in the original publication by Corti et al.Citation21 Including conservative substitutions (“positives” as in the default settings for BLASTCitation22) reveals an even closer relationship with a similarity of 96% and 88%, respectively, for VH and VL. The VIS410 publication includes in supplementary material a list of accession numbers to antibody variable regions as “top ranking templates” used for the design of anti-influenza antibodies.Citation17 In we present the corresponding VH sequences, retrieved from the NCBI database using those accession numbers, and aligned with both VIS410 and FI6v3. Focusing on the CDRs, the portion of the antibody sequence expected to be most important to determine specificity and govern binding to antigen,Citation23–Citation25 the closest template CDRH3 to the output VIS410 CDRH3 is 10% identical, while the FI6v3 CDRH3 to VIS410 CDRH3 is 85% identical. Given the remarkable diversity of CDRH3 sequences, it is hard to conceive how a computational method could have credibly and independently converged near the FI6v3 sequence. Further comparison of the other CDRs between VIS410 and FI6v3, shows just two non-conservative substitutions in VH CDRs, and three in CDRL2. According to the crystal structure, CDRL2 was deemed non-essential for antigen binding by FI6v3 and therefore an attractive location to introduce sequence alterations that are unlikely to compromise binding.Citation21 Based on the structure of FI6v3 in complex with influenza HA, we mapped those differences, as shown in . Only one of the non-conservatively substituted amino acid positions appears to be directly involved in antigen contact; this would be position 54 of the VH, which is Ala in FI6v3, but Gly in VIS410. As detailed in the original publication,Citation21 the precursor of FI6v3, called FI6, does have Gly at the same position, and this change is among a few others demonstrated to have no impact on functionality. Given the remarkable degree of similarity, it is important to remember that FI6v3, including its sequence and structure, was described almost a year before the submission date of the first patent application describing the discovery of VIS410 (Figure S2)
Figure 2. Design process for VH of VIS410: alignment of VH template sequences listed in table S1 of original publicationCitation17 with VIS410 and FI6v3. CDRs as defined in Kabat are highlighted in gray.

Zika antibody
More recently, the same research group with new collaborators reported to have “applied computational methods to engineer an antibody, ZAb_FLEP,” with broadly neutralizing activity against Zika virus.Citation18 As in the earlierCitation17 publication, no sequence information was provided for ZAb_FLEP. Following the same approach as in the previous section, we initially searched the patent literature with the term “ZAb_FLEP,” which proved unproductive. However, searching Zika together with some of the author’s names in a patent database led us to a published patent applicationCitation26 titled “Antibodies that bind Zika virus envelope protein and uses thereof”. All six named inventors are also authors of the publication describing ZAb_FLEP.Citation18
Comparison of figures in the publication and patent application leaves little doubt that ZAb_FLEP corresponds to mAb 8; see Figure S3.Citation18,Citation26 The sequences of mAb 8 are presented in the patent applicationCitation26 (sequence IDs 6 and 15), and are shown in .
Figure 4. VH and VL alignment of VH alignment of EDE1 C8 (PDB files 4UTA or 5LBS) and mAb 8 (likely ZAb_FLEP). Non-conservative substitutions depicted in red font. CDRs highlighted in gray according to Kabat’s definition.
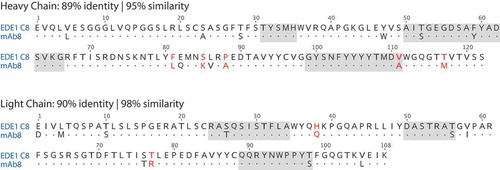
As in the influenza case, a Genbank search with mAb 8 sequences fails to retrieve an exact match, but results in sequence hits to EDE1 C8,Citation27–Citation30 a previously reported dengue/Zika cross-reactive antibody first described in 2015 by Dejnirattisai et al.Citation29 An alignment of EDE1 C8 and mAb 8 is shown in .
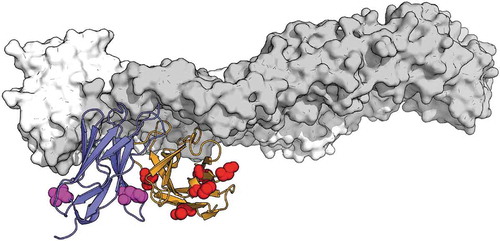
As in the influenza example, the similarity is remarkable, with 89% and 90% identity, for VH and VL, respectively. This is obtained without any gaps in the alignment, and thus all CDR lengths are identical between ZAb_FLEP and EDE1 C8. Considering conservative substitutions, as before, yields similarities of 95% and 98%, respectively, for VH and VL. Within the CDRs of the VH, there is a single V to A non-conservative substitution (as defined by BLAST default settings). There are only three conservative substitutions in VL CDRs; in fact, one in each CDR, all involving S/T exchanges. The non-conservative substitutions in the context of the known EDE1 C8 complex with Zika proteinCitation27 are depicted in . Once again, given the remarkable sequence similarity, it is important to emphasize the prior publication of the EDE1 C8 sequences, in the context of both dengue and Zika (Figure S2).Citation27,Citation30
Figure 5. Depiction of non-conservative substitutions (red for the VH and magenta for the VL) in the context of the structure of EDE1 C8 complexed with ZIKV E (gray). VH and VL are shown in orange and purple, respectively. PDB 5LBS, chains AHL.
It is interesting that EDE1 C8 is mentioned, among other antibodies, as a potential template for the design of ZAb_FLEP, as indicated in the supplementary material:Citation18 “Multiple antibody scaffolds (including mouse-derived pan-flavivirus 4G2, anti-EDE1 Dengue mAbs C8, C10 and anti-EDE2 Dengue mAb A11, anti-TDRD3 antibody and anti-HIV antibody PGT124) were used as starting templates for antibody engineering.”Citation18 The lack of sequence disclosure for ZAb_FLEP and any direct data comparisons to EDE1 C8, however, obscures from readers, as well as peer reviewers, the remarkable similarity of ZAb_FLEP and EDE1 C8. Given the apparent origin of ZAb_FLEP from EDE1 C8, we wonder why a direct comparison between the two was not reported, especially in light of the authors’ statementCitation:18 “The in vitro neutralization potential of ZAb_FLEP approaches the potency of select Zika antibodies” (emphasis added).
The narrative in the patent application,Citation26 which is intended to teach the skilled artisan how to practice the invention, only provides sequences from an anti-TDRD3 antibody as input template, and it makes no explicit mention of EDE1 C8 as input. Moreover, sequences identical to EDE1 C8, represented as mAb 3 in the patent document, are said to have been “designed by computing the epitope-paratope connectivity network,” whereby “variable regions and CDRs (are) generated (and) shown in (…) )” (see Figure S4Citation26). However, the designed mAb 3 has a non-traditional two amino acid addition (Arg-Ser) at the VL N-terminus. This sequence matches a non-coding cloning site present in the original EDE1 C8 VL expression vector.Citation29 It is inconceivable that an unsupervised algorithm would produce vestigial vector sequence unrelated to antigen recognition.
Discussion
In this report, we examine two instances in which the same research group has made representations of structure-based computational design of anti-viral antibodies with exceptional neutralization breadth and potency.Citation17,Citation18 In neither case were the sequences of the designed antibodies disclosed, leading us to question the enforcement of editorial policies regarding reproducibility. Perhaps more concerning is the potential for contamination of the scientific literature with claims by innocent third parties. For example, in a commentary articleCitation31 about the clinical evaluation of VIS410,Citation32 it is said that “VIS410, however, is not just another HA-stem specific human monoclonal antibody. This human IgG1 monoclonal antibody is the result of man-made design and protein engineering and so is not derived from a natural source.” Clearly, the author of this comment was not in possession of the comparison presented in .
We present with a high degree of confidence the actual sequence identity of the designed antibodies, and a more plausible genesis narrative. Comparisons of these sequences to those of previously described human B cell-derived antibodies to the same targets show striking similarities. By contrast, those designed sequences appear very dissimilar from the templates said to have been used to start the design process ( and S4); we leave it to the reader to judge the likelihood of these highly homologous sequences being re-discovered coincidentally, or simply derived from existing antibodies targeting the same epitopes as those of the computationally designed antibodies.
In conclusion, the presented fact pattern calls into question the publications’ claimed genesis of VIS410 and ZAb_FLEP. Furthermore, the lack of sequence disclosure exposes a serious weakness in the peer review process in the emerging field of computational antibody design.Citation13–Citation16 (It is instructive to compare the level of transparency provided by someCitation16 to the opaque disclosures in the publications examined here.Citation17,Citation18) Such obfuscation prevents independent confirmation, and is contrary to basic scientific norms. We find it difficult to view these authors’ approachCitation17,Citation18 in any light other than an intent to mislead as to the level of originality and significance of the published work.
Disclosure of potential conflicts of interest
All authors are equity stakeholders in, and or employed by, Adimab LLC. Adimab provides commercial antibody discovery and optimization services for the biotechnology industry, which includes infectious disease programs. Adimab also has research interests in aspects of computational antibody design. All authors are named inventors or co-inventors in multiple patent filing concerning antibody discovery and engineering. TG was a co-founder of Arsanis, a company with an infectious disease focus that recently merged with X4 Pharma.
Supplemental Material
Download PDF (2.3 MB)Supplemental material
Supplemental data for this article can be accessed on the publisher’s website
References
- Kaplon H, Reichert JM. Antibodies to watch in 2019. MAbs. 2019;11:219–38. doi:10.1080/19420862.2018.1556465.
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–97.
- Lonberg N, Huszar D. Human antibodies from transgenic mice. Int Rev Immunol. 1995;13:65–93.
- Fishwild DM, O‘Donnell SL, Bengoechea T, Hudson DV, Harding F, Bernhard SL, Jones D, Kay RM, Higgins KM, Schramm SR, et al. High-avidity human IgG kappa monoclonal antibodies from a novel strain of minilocus transgenic mice. Nature Biotechnology. 1996;14:845–51. doi:10.1038/nbt0796-845.
- Mendez MJ, Green LL, Corvalan JR, Jia XC, Maynard-Currie CE, Yang XD, Gallo ML, Louie DM, Lee DV, Erickson KL, et al. Functional transplant of megabase human immunoglobulin loci recapitulates human antibody response in mice. Nat Genet. 1997;15:146–56. doi:10.1038/ng0297-146.
- Lanzavecchia A, Corti D, Sallusto F. Human monoclonal antibodies by immortalization of memory B cells. Curr Opin Biotechnol. 2007;18:523–28. doi:10.1016/j.copbio.2007.10.011.
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–40. doi:10.1038/nature07930.
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–71. doi:10.1038/nature06890.
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–54. doi:10.1038/348552a0.
- Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–28. doi:10.1038/352624a0.
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nature Biotechnology. 1997;15:553–57. doi:10.1038/nbt0697-553.
- Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nature Biotechnology. 2003;21:163–70. doi:10.1038/nbt785.
- Fischman S, Ofran Y. Computational design of antibodies. Curr Opin Struct Biol. 2018;51:156–62. doi:10.1016/j.sbi.2018.04.007.
- Sormanni P, Aprile FA, Vendruscolo M. Third generation antibody discovery methods: in silico rational design. Chem Soc Rev. 2018;47:9137–57. doi:10.1039/c8cs00523k.
- Lippow SM, Wittrup KD, Tidor B. Computational design of antibody-affinity improvement beyond in vivo maturation. Nature Biotechnology. 2007;25:1171–76. doi:10.1038/nbt1336.
- Sevy AM, Wu NC, Gilchuk IM, Parrish EH, Burger S, Yousif D, Nagel MBM, Schey KL, Wilson IA, Crowe JE, et al. Multistate design of influenza antibodies improves affinity and breadth against seasonal viruses. Proc Natl Acad Sci U S A. 2019;116:1597–602. doi:10.1073/pnas.1806004116.
- Tharakaraman K, Subramanian V, Viswanathan K, Sloan S, Yen H-L, Barnard DL, Leung YHC, Szretter KJ, Koch TJ, Delaney JC, et al. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc Natl Acad Sci U S A. 2015;112:10890–95. doi:10.1073/pnas.1502374112.
- Tharakaraman K, Watanabe S, Chan KR, Huan J, Subramanian V, Chionh YH, Raguram A, Quinlan D, McBee M, Ong EZ, et al. Rational engineering and characterization of an mAb that neutralizes zika virus by targeting a mutationally constrained quaternary epitope. Cell Host Microbe. 2018;23:618–27 e6. doi:10.1016/j.chom.2018.04.004.
- Wollacott AM, Viswanathan K, Trevejo J, Sloan S, Shriver Z, Boni M Compositions and methods for treating and preventing influenza. Visterra. United States: US20170137498. 2017.
- Shriver Z, Viswanathan K, Subramanian V, Raguram S Novel HA binding agents. Visterra. United States: US20130302349. 2013.
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–56. doi:10.1126/science.1205669.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi:10.1016/S0022-2836(05)80360-2.
- Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. The Journal of Experimental Medicine. 1970;132:211–50.
- Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321:522–25. doi:10.1038/321522a0.
- Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature. 1988;332:323–27. doi:10.1038/332323a0.
- Sasisekharan R, Tharakaraman K, Chan KR, Watanabe S, Vasudevan SG, Ooi EE Antibodies that bind Zika virus envelope protein and uses thereof. Massachusetts Institute of Technology; National University of Singapore. United States: US20180105583. 2018.
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Lorière E, Sakuntabhai A, Cao-Lormeau V-M, Haouz A, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi:10.1038/nature18938.
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau V-M, Malasit P, Rey FA, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nature Immunology. 2016;17:1102–08. doi:10.1038/ni.3515.
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nature Immunology. 2015;16:170–77. doi:10.1038/ni.3058.
- Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520:109–13. doi:10.1038/nature14130.
- Saelens X. One against all: a broadly influenza neutralizing man-made monoclonal antibody passes phase I. EBioMedicine. 2016;5:16–17. doi:10.1016/j.ebiom.2016.02.036.
- Wollacott AM, Boni MF, Szretter KJ, Sloan SE, Yousofshahi M, Viswanathan K, Bedard S, Hay CA, Smith PF, Shriver Z, et al. Safety and upper respiratory pharmacokinetics of the hemagglutinin stalk-binding antibody VIS410 support treatment and prophylaxis based on population modeling of seasonal influenza A outbreaks. EBioMedicine. 2016;5:147–55. doi:10.1016/j.ebiom.2016.02.021.