ABSTRACT
The monocled cobra (Naja kaouthia) is among the most feared snakes in Southeast Asia due to its toxicity, which is predominantly derived from long-chain α-neurotoxins. The only specific treatment for snakebite envenoming is antivenom based on animal-derived polyclonal antibodies. Despite the lifesaving importance of these medicines, major limitations in safety, supply consistency, and efficacy create a need for improved treatments. Here, we describe the discovery and subsequent optimization of a recombinant human monoclonal immunoglobulin G antibody against α-cobratoxin using phage display technology. Affinity maturation by light chain-shuffling resulted in a significant increase in in vitro neutralization potency and in vivo efficacy. The optimized antibody prevented lethality when incubated with N. kaouthia whole venom prior to intravenous injection. This study is the first to demonstrate neutralization of whole snake venom by a single recombinant monoclonal antibody, thus providing a tantalizing prospect of bringing recombinant antivenoms based on human monoclonal or oligoclonal antibodies to the clinic.
Introduction
Snakebite envenoming is a neglected tropical disease, which each year claims hundreds of thousands of victims, who are either left permanently disfigured or meet an untimely death.Citation1 It is estimated that every year 1.8–2.7 million envenomings occur, which result in 81,000–138,000 deaths and more than 400,000 people left with permanent physical and psychological sequelae.Citation1 Asia is the continent where most envenomings and deaths occur, estimated to 1.2–2 million cases and 57,000–100,000 fatalities.Citation1 In Southern and Southeast Asia, the monocled cobra (Naja kaouthia) is responsible for a large number of the recorded severe snakebite cases,Citation2,Citation3 which is exemplified by the fact that 34% of snakebite-related deaths in Bangladesh from 1988 to 1989 were attributed to bites from either N. kaouthia or the closely related species, N. naja.Citation4 Life-threatening clinical manifestations of N. kaouthia envenomation include flaccid paralysis due to the actions of abundant long-chain α-neurotoxins, which block neuromuscular transmission by binding to the nicotinic acetylcholine receptor (nAChR) with high affinity, causing a curare-mimetic effect.Citation5,Citation6 These long-chain α-neurotoxins belong to the three-finger toxin superfamily, which dominate the venom in terms of abundance and toxicity, as judged by their high toxicity scores,Citation6–8 and are thus the main toxin targets to be neutralized for successful intervention in human snakebite cases.
Each year, the lack of access to affordable and effective treatment against snakebite envenoming leaves thousands of victims in despair, as revealed by the estimated number of 81,000 to 138,000 fatalities and the consequent social and economic impact in families and communities.Citation1 Animal-derived antivenoms remain the cornerstone of snakebite envenoming therapyCitation1 and are still produced by immunizing large mammals, usually horses, with snake venom, followed by the purification of antibodies from the blood plasma, resulting in polyclonal antibody preparations.Citation9 Being heterologous products, animal-derived antivenoms often lead to a range of adverse reactions whose incidence varies depending on the product.Citation10 Furthermore, it is estimated that only a fraction of the antibodies in current antivenoms contribute to neutralization of relevant toxins. Large amounts of antivenom are therefore required to treat a snakebite case, resulting in heterologous protein loads as high as 15 g per treatment in severe envenoming cases.Citation11,Citation12 Moreover, a discrepancy exists between the toxicity (high) and the immunogenicity (low) of these toxins, where antivenoms raised in animals have a sub-optimal concentration of therapeutic antibodies against low molecular weight elapid neurotoxins with high toxicity scores.Citation13–16 Despite many advances within antibody technology and biotechnology, a need remains for antivenoms with improved safety and efficacy.Citation17,Citation18
Recently, recombinant antivenoms based on oligoclonal mixtures of human antibodies have been proposed as a cost-competitive alternative to current antivenoms.Citation19–22 Such recombinant antivenoms may offer safer and more efficacious snakebite envenoming therapy due to their compatibility with the human immune system and the possibility of only including efficacious antibodies, targeting medically relevant snake toxins, in the antivenom mixture.Citation18 Moreover, it has been demonstrated that such oligoclonal recombinant antivenoms, consisting of carefully selected immunoglobulin Gs (IgGs), can be developed using phage display technology.Citation19 This technique has also been used to discover nanobodiesCitation23 and antibody fragmentsCitation24 against long-chain α-neurotoxins in the past. However, to date, no neutralizing human monoclonal IgG has been reported for whole venom from any animal following intravenous (i.v.) injection.
Here, we report the discovery and affinity maturation of a recombinant human monoclonal IgG targeting α-cobratoxin, the most medically relevant toxin from N. kaouthia venom. Using chain-shuffling, the affinity of the antibody was improved eightfold, resulting in enhanced neutralization both in vitro and in vivo. While the parent antibody could prolong survival of mice, when a preincubated mixture of purified α-cobratoxin and the antibody was administered i.v., the affinity-matured antibody was improved to a level where it could neutralize the lethal effect of whole N. kaouthia venom in mice.
Results
A phage display-derived fully human antibody that prolongs survival in vivo
A naïve single-chain variable fragment (scFv) phage display library containing 4 × 1010 clones was used for selections. This library was created using the variable heavy (VH) and variable light (VL) antibody genes from the naïve IgM repertoire of 43 healthy human donors.Citation25 Three rounds of panning were performed against biotinylated α-cobratoxin from N. kaouthia immobilized on a streptavidin-coated surface. Antibody-encoding genes (scFv format) were isolated from both the second and third panning rounds and subcloned into a bacterial expression plasmid.Citation26 In total, 282 clones harboring this expression plasmid were picked for antibody expression and subjected to binding analysis by dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA)-based assays as previously described.Citation19 Of these, 36 clones displaying a specific binding signal against α-cobratoxin (using an arbitrary cutoff of 25 times above the background binding signal) were picked for DNA sequencing and further characterization ().
Figure 1. Affinity ranking of scFvs and Kaplan–Meier survival curves for mice co-administered with IgG antibodies and α-cobratoxin. (a) direct and ENC DELFIA of 36 monoclonal scFv-containing supernatants. (b) direct and ENC DELFIA of the top six monoclonal scFv-containing supernatants. (c) Kaplan–Meier survival curves for mice co-administered with IgG antibodies and α-cobratoxin. α-cobratoxin refers to mice injected with α-cobratoxin alone.
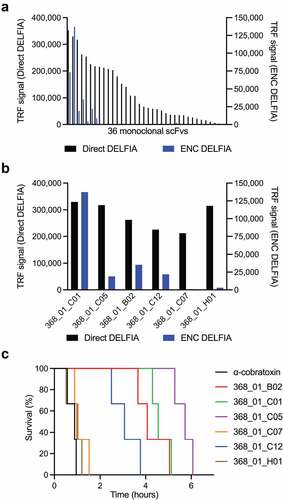
Of these 36 scFv clones, a total of 29 (80.5%) scFvs had unique VH and VL CDR3 sequences with typical VH CDR3 lengths of 10–25 amino acid residues. Interestingly, a high proportion of these clones (>50%) showed a cysteine pair in their VH CDR3 sequences (Supplementary Table S1). These scFvs were evaluated in an expression-normalized capture (ENC) DELFIA assay, which eliminates the signal dependence on the expression level of the clones, and allows ranking based on affinity. The six α-cobratoxin-binding scFvs that yielded the highest binding signals () were selected for expression in IgG format and transiently expressed using Expi293F™ cells. All six α-cobratoxin-targeting antibodies retained binding to α-cobratoxin upon conversion (data not shown). However, when the antibodies and α-cobratoxin were preincubated and administered i.v. to mice, the antibodies failed to prevent lethality, although they did succeed in prolonging survival significantly (). The limited efficacy of these antibodies could be due to sub-optimal binding affinity to α-cobratoxin.
Affinity maturation using chain-shuffling
In vitro affinity maturation strategies involve two key steps, diversification of the primary antibody sequence and enrichment of affinity-improved antibody variants using a selection platform such as phage display technology. Diversification of primary antibody sequence can be achieved by introducing mutations to the variable regions using random or targeted mutagenesis. Alternatively, new combinations of heavy and light variable regions can be made by recombining selected heavy or light chains with a repertoire of partner chains by a process known as chain shuffling.Citation27 Here, the 368_01_C01 antibody was selected for affinity maturation due to its combined performance in the ENC DEFLIA and in vivo experiments. This antibody was subjected to light-chain shuffling to diversify its sequence by pairing the VH chain with a library of naïve kappa and lambda light chains. Following library generation, three rounds of stringent panning against α-cobratoxin were completed. For precise control of antigen concentration, phage display selections were carried out in solution phase. The phage antibodies were allowed to bind to biotinylated α-cobratoxin in solution, and the bound phage was subsequently captured using streptavidin-coated beads for washing and elution. The antigen concentration was lowered 10- and 50-fold in each round to selectively enrich antibodies with high affinity. The polyclonal phage outputs for the selections were tested for α-cobratoxin binding through polyclonal DELFIA, revealing that α-cobratoxin-binding scFvs were present in all three rounds, while negligible binding to streptavidin was detected (Figure S1A). Then, scFv genes from the third panning rounds were subcloned into the pSANG10-3F expression vector. A total of 184 monoclonal colonies from two third-round selections were picked, and the soluble scFvs were assessed for binding to α-cobratoxin, revealing 290 of the 368 (79%) displaying a binding signal against the antigen (Figure S1B). Among the 290 hits, 60 clones were randomly picked and further characterized by ENC DELFIA () and DNA sequenced, revealing that 13 of the 60 scFvs had unique CDRL3 sequences (Supplementary Table 2). Based on their ranking in the ENC DELFIA, the 13 most promising scFvs were converted to the fully human IgG1 format and expressed in Expi293F™ cells. Purity and monomeric content of these antibodies were confirmed to be above 90% using HPLC-SEC analysis (Supplementary Table 3). A direct DELFIA was used to confirm that all 13 antibodies retained binding in the IgG format. An ENC DELFIA was used to rank the binding of the IgGs (). Due to a more than twofold higher expression yield than 2552_01_F12, combined with favorable ranking in the ENC DELFIA, 2552_02_B02 was picked as the top candidate for further study.
Figure 2. Binding, expression, and binding kinetics characterization of discovered antibodies. (a) direct monoclonal DELFIA signals (in black on the left Y-axis) and ENC DELFIA signals (in blue on the right Y-axis) of the 60 monoclonal scFvs selected for sequencing. (b) ENC DELFIA signals of IgG-containing supernatants of the 13 clones that were converted to the IgG format. 2552_02_B02 was selected for further characterization. for (A) and (B), data from the parent scFv (368_01_C01) is shown furthest to the right. (c) 1:1 binding model (black lines) fitted to the SPR data kinetics of parental antibody (368_01_C01) in the fab format. (d) 1:1 binding model (black lines) fitted to the SPR data of the affinity matured clone (2552_02_B02) in the fab format.
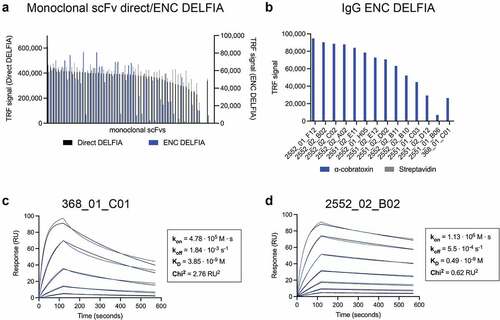
Surface plasmon resonance (SPR) was used to assess affinity improvement derived from the maturation of the parental antibody light chain. Here, the parent (368_01_C01) and 2552_02_B02 were reformatted as monovalent antigen-binding fragments (Fabs) and used for affinity measurements to avoid avidity effects. For the parent clone, affinity (equilibrium dissociation constant, KD) was determined to be 3.9 nM, whereas the matured clone displayed an affinity of 490 pM, an approximate eightfold increase. This increase in affinity resulted from the improvement of both the on and off rate of the original antibody by 2.4-fold and 3.3-fold, respectively ( c and d).
Affinity matured IgGs show potent blocking of toxin:receptor binding interaction
To assess if an increase in affinity translated to improved blocking of the binding interaction between the AChR and α-cobratoxin, a receptor blocking assay was performed ().
Figure 3. Inhibition of the binding interaction between the α7-AChR and α-cobratoxin using IgGs. (a) schematic representation of a receptor blocking assay, wherein the ability of IgG antibodies to inhibit the interaction between the α7 subunit of AChR (α7-AChR) and α-cobratoxin can be quantified. (b) antibodies block α-cobratoxin binding to its receptor (α7-AChR) in a concentration-dependent manner. as a negative control an IgG specific to dendrotoxins was used and showed no blocking (data not shown). Y-axis is signal relative to that of the positive control.
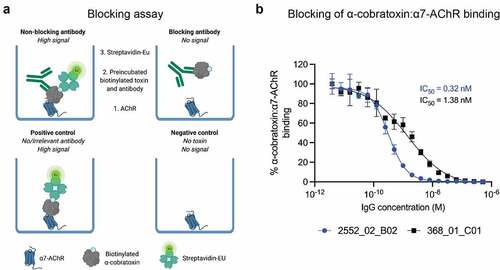
Results revealed that both IgGs were able to fully abrogate the binding between the receptor subunit and α-cobratoxin, though the affinity-matured clone was able to prevent the binding at lower concentrations than the parent antibody. The IC50 values were 0.32 nM (CI95 0.28–0.36 nM) and 1.38 nM (CI95 1.02–1.86 nM) for 2552_02_B02 and 368_01_C01, respectively (). The 8.1-fold increase in affinity to α-cobratoxin, therefore, resulted in a 4.3-fold improvement of IC50 in this blocking assay.
Affinity matured IgG shows increased neutralization potency in vitro
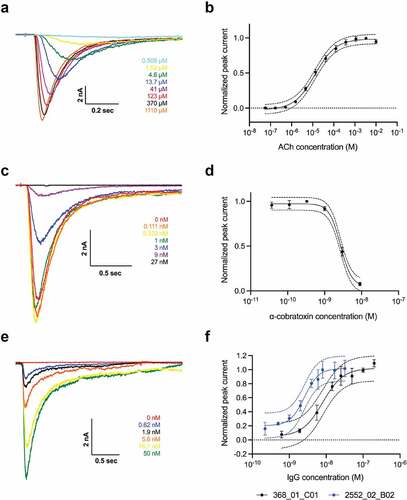
To assess if the increase in affinity and ability to block the α7-AChR:α-cobratoxin interaction for 2552_02_B02 also resulted in an improved ability to protect nAChR function, functional neutralization assays were conducted using automated patch-clamp electrophysiology. First, the EC80 value for ACh was established (70 µM, a and b), and the IC80 for α-cobratoxin was determined (4 nM, c and d). Then, titrated 368_01_C01 and 2552_02_B02 were preincubated with α-cobratoxin and tested for the ability to neutralize the current-inhibiting activity of α-cobratoxin. As a negative control, a dendrotoxin-binding IgG was included. This irrelevant IgG showed no effects, while the α-cobratoxin-recognizing antibodies were able to fully abrogate α-cobratoxin activity (data not shown). Furthermore, the affinity-matured clone, 2552_02_B02, was a more potent neutralizer with an IC50 of 2.6 nM (CI95 2.3–2.9 nM), while the parental clone (368_01_C01) exhibited an IC50 of 8.1 nM (CI95 6.6–10.0 nM). Relative to the concentration of α-cobratoxin used, these data indicate that 0.65 IgG molecules were needed per toxin molecule for 50% neutralization for 2552_02_B02, whereas 2.03 IgG molecules were needed per toxin to achieve the same effect with 368_01_C01. Hence, the increased affinity between 2552_02_B02 and α-cobratoxin resulted in increased functional neutralization in vitro.
Figure 4. In vitro neutralization of inhibition of nAChR by α-cobratoxin. automated patch-clamp experiments conducted using a QPatch (Sophion Bioscience). (a) example of sweep plot and (b) concentration–response curve showing the relationship between increased ACh concentration and the measured current running across the cell membrane. 70 µM ACh was used throughout the rest of the experiments. (c) Example of sweep plot and (d) concentration–response curve showing how increasing concentrations of α-cobratoxin result in a decrease in the current measured. 4 nM α-cobratoxin was used, resulting in approximately 80% inhibition of the current. (e) example of sweep plot (368_01_C01) and (f) concentration–response curves showing how increasing concentrations of the two IgGs preincubated with α-cobratoxin result in better protection of the nAChR, as the loss of current mediated by α-cobratoxin is prevented in a dose-dependent manner. An irrelevant IgG was used as a control, which did not prevent the inhibitory effects of α-cobratoxin (data not shown).
Affinity matured antibody neutralizes Naja kaouthia whole venom in vivo
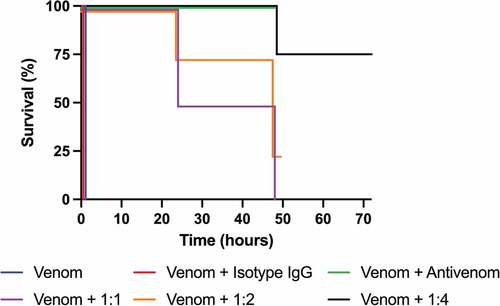
To determine the ability of 2552_02_B02 to neutralize the effects of N. kaouthia whole venom, a mouse lethality assay was performed, as this is the gold standard of the World Health Organization (WHO) for the assessment of the preclinical efficacy of antivenoms. Two LD50s of N. kaouthia whole venom (containing approximately 1.2 nmol of α-cobratoxin) were preincubated with the IgG in different molar ratios for 30 minutes before injecting the mixture i.v. in mice (four mice per group). Control groups consisted of 1) venom only, 2) venom incubated with an irrelevant IgG isotype control, or 3) venom incubated with a commercially available polyclonal antivenom. Survival was recorded and is illustrated in . All mice injected with either venom alone or in combination with the isotype control antibody died within 30 minutes after injection with signs of neuromuscular paralysis (). At the end of the experiment (48 hours post injection), the four mice receiving venom and polyclonal antivenom were alive and showed no signs of neurotoxicity, at which point the study on this group was concluded. For the groups of 1:1 and 1:2 toxin:IgG dosages, the surviving mice at 24 hours showed clear signs of toxicity, and it was decided to extend the observation period until 48 hours owing to the novel nature of these antibodies and the need to know whether the neutralization observed in the first hours could be reverted. At the 48-hour time point, all four mice in the 1:1 ratio were dead, and only one of four mice in the 1:2 toxin:IgG dosage group remained alive. Therefore, a new experiment increasing the dosage to 1:4 toxin:IgG was set up. Control mice injected with venom alone died within 30 min, as expected, whereas all four mice receiving venom and antibody survived the 24-hour period, and one out of four mice was dead at 48 hours. To ensure that protection was maintained beyond the 48-hour observation time, this group of mice was kept until 72 hours, and no further deaths had occurred (). These results clearly demonstrate that the therapeutic effect of 2552_02_B02 on mice injected with a lethal amount of N. kaouthia whole venom was dose-dependent in vivo, as expected for specific antibody therapeutics. Even further, at an α-neurotoxin to IgG molar ratio of 1:4, all four mice survived the observation period recommended by the WHO for this type of study (24 hours), and even at the 72-hour mark, three out of four mice had survived.
Figure 5. Kaplan–Meier curves showing survival of mice co-administered with N. kaouthia whole venom and IgG 2552_02_B02. 1:1, 1:2, and 1:4 designates the toxin:IgG molar ratio that 2552_02_B02 was injected in. The death of the venom only and antibody isotype control groups occurred within 30 minutes after injection, whereas those receiving venom and antivenom survived the 48-hour observation period. All groups were observed for 48 hours, except mice receiving 2552_02_B02 in a 1:4 molar ratio, which were observed for 72 hours.
Discussion
Previously, we described the discovery of an oligoclonal mixture of human antibodies capable of neutralizing dendrotoxin-mediated neurotoxicity of black mamba venom in a rodent model.Citation19 Although the cocktail of antibodies tested in that study did neutralize whole venom, the model, using intracerebroventricular injection (i.c.v.), did not account for the effects elicited by α-neurotoxins, since their main target is the nAChR in the neuromuscular junctions. Thus, the i.c.v. model is not as clinically relevant as i.v. injection, which is recommended by WHO as the standard for assessing antivenoms. Another study has reported in vivo neutralization of α-cobratoxin-induced lethality by a VHH and a VHH2-Fc following intraperitoneal injection in mice.Citation23 However, to date, no study has successfully demonstrated the neutralization of lethality caused by a whole venom (or a purified toxin) preincubated with a recombinant human monoclonal IgG antibody following i.v. injection.
In this study, we demonstrate that a recombinant human monoclonal IgG antibody, discovered and optimized entirely in vitro by phage display technology, was able to neutralize lethality in mice challenged i.v. with whole venom from N. kaouthia when venom and antibody were preincubated before administration. This clearly showcases the utility of in vitro selection methods for the discovery of efficacious antivenom antibodies against animal toxins with reduced immunogenicity, which may be challenging for traditional in vivo-based discovery approaches. Moreover, this study has also elucidated the mechanism of action of the neutralizing antibody using receptor blocking and automated patch clamp electrophysiology assays, which revealed that the antibody could abrogate neurotoxicity by preventing the medically most important toxin present in the venom of N. kaouthia, α-cobratoxin from interacting with the nAChR. This first report of a recombinant monoclonal antibody neutralizing whole venom from a snake thus presents an integrated approach for future discovery and evaluation of recombinant antibodies against toxins from snake and other animal venoms. Our study was designed to assess whether a neutralizing antibody could be generated using the experimental platform described, which was clearly demonstrated. Assessing the therapeutic efficacy of this antibody in a setting that would more closely resemble the actual circumstances of envenoming, i.e., by using a rescue-type neutralization protocol, was beyond the goals of this work and should be approached in future investigations.
It is important to note that the neutralization of this particular venom by a single IgG cannot be extrapolated to all other snake venoms due to their complex composition of different medically relevant toxin families that each may require one or more antibodies for neutralization.Citation28 In many cases, co-administration with other antibodies or small-molecule inhibitors, such as varespladib, batimastat, or marimastat,Citation29,Citation30 might be necessary to achieve full protection.Citation18 Furthermore, the targeted epitope and pharmacokinetic properties of the IgG reported in this study have not been investigated and therefore these properties remain unknown. The toxin epitope could have an influence on how effectively the toxin is neutralized.Citation31 Moreover, it is also known that antibody pharmacokinetics plays a significant role in drug efficacy,Citation32 while antibody biophysics can have a major influence on antibody manufacturability and stability.Citation33,Citation34,Citation35 Additional investigation and potential optimization of these properties and their effects on in vivo neutralization efficacy and downstream development are therefore warranted prior to further preclinical and clinical assessments.
Finally, it is currently not clear whether recombinant antivenoms based on monoclonal or oligoclonal antibodies will be regulated as blood products, similar to existing plasma-derived antivenoms, or whether recombinant antivenoms will be viewed as biotherapeutic products to be regulated as biopharmaceuticals by relevant authorities. Establishment of a regulatory framework for recombinant antivenom products is thus a necessity for bringing such new snakebite envenoming therapies swiftly to the clinic.
Nonetheless, the advances in the discovery, optimization, and assessment of monoclonal antibodies against snake toxins described in this study represent an important technical milestone toward the application of in vitro developed recombinant antivenoms as a therapeutic intervention in snakebite envenoming in the future.
Materials and methods
Toxin preparation
α-cobratoxin was obtained in lyophilized form from Latoxan SAS, France, Product ID L8114. The toxin was reconstituted in phosphate-buffered saline (PBS) and biotinylated using a 1:1 (toxin:biotinylation reagent) molar ratio as previously described.Citation19 Following biotinylation, Amicon® Ultra-4 Centrifugal Filter Units with a 3 kDa membrane were used for purification of the biotinylated toxin. Purification was performed at 8 °C and consisted of three washes of 4 mL PBS. The protein concentration was measured by the absorbance at 280 nm using a NanoDrop and adjusted using the extinction coefficient. The degree of biotinylation was analyzed using MALDI-TOF in a Proteomics Analyzer 4800 Plus mass spectrometer (Applied Biosystems).
Initial discovery and assessment of parent clone
The initial discovery of the parent antibody clone 368_01_C01 was performed by panning the IONTAS phage display library (diversity of 4 × 1010 human scFv clones) against biotinylated α-cobratoxin captured by streptavidin in a MaxiSorp vial, followed by subcloning and expression of scFv genes in BL21 (DE3) E. coli and DELFIA-based screening of the scFv-containing supernatants as previously described.Citation19 Thirty-six binding clones were cherry-picked and sequenced (Eurofins Genomics sequencing service) using the S10b primer (GGCTTTGTTAGCAGCCGGATCTCA). The binding strengths of the clones were ranked using an expression-normalized capture (ENC) assay, and the top six scFvs displaying the highest binding signals were reformatted into the IgG format and expressed in Expi293™ cells (Thermo Fisher) and subsequently purified using an Äkta Pure system (GE Healthcare) as previously described.Citation19 The binding of purified IgGs was confirmed using a DELFIA-based binding assay.Citation19
Library generation using chain-shuffling
The VH region of antibody 368_01_C01 was PCR amplified from pINT3 plasmid DNA using pINT3 Nco FWD (TCTCTCCACAGGCGCCATGG) and IgG1 CH1 Xho Rev (CCCTTGGTGGAGGCACTCGAG) primers using Platinum™ SuperFi II Green PCR Master Mix (Invitrogen, 12369010). The PCR product was cloned into pIONTAS1Citation25 vector harboring the naïve VL lambda and kappa chain libraries using NcoI and XhoI restriction endonucleases. Ligation reactions were carried out for 16 hours at 16 °C and contained 160 ng of insert and 400 ng of vector DNA in a total volume of 40 µL. Ligations were purified using the MinElute PCR Purification Kit (Qiagen, 28004) and eluted in 10 µL nuclease-free water. The purified ligation product was transformed into 200 µL of electrocompetent TG1 cells (Lucigen, 60000-PQ763-F) followed by addition of 6 mL of recovery medium (Lucigen, F98226-1) and incubation at 37 °C for 1 hour at 280 rpm rotation. Cells were plated on 2xTY agar plates supplemented with 100 µg/mL ampicillin and 2% glucose. Dilutions of the transformations were also plated to determine library size, which was 1.01 × 108 for the lambda library and 1.67 × 108 for the kappa library with more than 96% of the transformants being positive for insertion of heavy-chain insert, as determined by colony PCR.
Library rescue and solution-based phage display selection
Rescue of phages from the chain-shuffled libraries and the three rounds of selections were performed as described elsewhere,Citation25 except that the phages were not concentrated using PEG precipitation, but phage-containing supernatants were used directly for selections. Deselection of streptavidin-specific phages was performed before each round of selection using 80 µL of streptavidin-coated Dynabeads (Invitrogen, M-280). Additionally, the selections were conducted using biotinylated α-cobratoxin that was captured using 80 µL of streptavidin-coated Dynabeads (Invitrogen, M-280). The concentration of α-cobratoxin was decreased through the three rounds of selections starting at 10 nM in the first round and ending at 20 pM in the third round. The kappa and lambda libraries were mixed before the first round of selections.
Subcloning, primary screening, and sequencing of scFvs
Subcloning of the α-cobratoxin-binding selection output into pSANG10-3 F and primary screening of candidates was performed as described elsewhere.Citation19 In brief, scFv genes from the selection outputs were subcloned from the pIONTAS1 phagemid vector to the pSANG10-3 F expression vector using NcoI and NotI restriction endonuclease sites and transformed into E. coli strain BL21 (DE3) (New England Biolabs). From each of the two subcloned selection outputs, 184 colonies were picked and expressed in 96-well plates. The scFvs were assessed for their binding to biotinylated α-cobratoxin (5 µg/mL) indirectly immobilized on black MaxiSorp plates (Nunc) with streptavidin (10 µg/mL) using a DELFIA-based assay. After immobilization of the toxin, the wells were blocked with PBS supplemented with milk protein (MPBS). The scFvs were expressed overnight using autoinduction media. The scFv-containing supernatants were blocked with MPBS and added to the toxin-coated wells. Binding was detected using an anti-FLAG IgG (Sigma, F3165) conjugated with europium and DELFIA Enhancement solution (Perkin Elmer, 4001–0010). In total, 60 clones binding to α-cobratoxin were cherry-picked and sequenced (Eurofins Genomics sequencing service) using S10b primer (GGCTTTGTTAGCAGCCGGATCTCA). The antibody framework and CDR regions were annotated, and light-chain CDR3 regions were used to identify 14 unique clones.
IgG expression, purification, and characterization
VH and VL genes of 13 unique α-cobratoxin-binding scFvs were converted to the fully human IgG1 format as previously described.Citation19 IgG1 antibodies were expressed in HEK293 cells (Expi293, Thermo Fisher) and purified using Protein A affinity chromatography (Generon, PC-100). Purity and monomeric content of the antibodies were analyzed using SDS-PAGE and analytical size exclusion chromatography (Agilent 1100 HPLC instrument, Superdex 200 Increase 5/150 column at a flow rate of 0.25 mL/min). The binding of the IgGs was confirmed and ranked using an expression-normalized capture (ENC) assay. Briefly, black MaxiSorp plates (Nunc) were coated overnight with an anti-human IgG (Jackson ImmunoResearch, 109–005-098). Plates were washed thrice with PBS and blocked with PBS supplemented with 3% milk protein. Plates were washed thrice with PBS and 0.25x unpurified IgG-containing culture supernatant in PBS supplemented with 3% milk protein was added before incubating for 1 hour at room temperature. Plates were washed thrice with PBS-T and thrice with PBS before adding either 1 nM or 100 pM biotinylated α-cobratoxin in PBS supplemented with 3% milk protein to each well. After 1 hour of incubation, the plates were washed thrice with PBS-T and thrice with PBS. Then, 1 µg/mL of Europium-labeled Streptavidin (Perkin Elmer, 1244–360) in DELFIA Assay Buffer (Perkin Elmer, 4002–0010) was added. Following 30 minutes of incubation, plates were washed thrice with PBS-T and thrice with PBS, and DELFIA Enhancement Solution (Perkin Elmer, 4001–0010) was added for detection of binding. Based on these results, the top clone, 2552_02_B02, was expressed and purified as described previously.Citation19
Fab expression and purification
VH and VL genes of 13 unique α-cobratoxin-binding scFvs were converted to the Fab format as performed for IgGs as described previously,Citation19 except the Fab-vector, pINT12, was used instead of the pINT3 IgG1 vector.
Surface plasmon resonance
The binding affinity of the discovered antibodies for α-cobratoxin was determined using SPR (BIAcore T100, GE Healthcare). All measurements were performed at 25 °C using 10 mM HEPES, 150 mM NaCl, and 3 mM EDTA at pH 7.4 as running buffer. Immobilization of α-cobratoxin on CM5 sensor chips (Cytiva, BR100530) was performed by amine coupling using 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)/N-hydroxysuccinimide surface activation followed by injection of 5 µg/mL of α-cobratoxin in 10 mM NaOAc pH 4 to obtain a final immobilization level of 23 response units (RU). The sensor chip was inactivated using ethanolamine. The Fabs in concentrations ranging from 81 nM to 390 pM were injected at 40 µL/minute for 120 seconds and dissociation was recorded for 450 seconds. Following dissociation, the sensor was regenerated using two injections (15–20 seconds) of 20 mM NaOH. Measurements were conducted using 5–7 analyte concentrations for each antibody. The blank subtracted data was analyzed using the BIAcore T100 Evaluation Software employing a 1:1 Langmuir binding model.
Receptor blocking DELFIA
The receptor blocking assay was adapted from Ratanabanangkoon et al.Citation36 Black MaxiSorp plates (Nunc) were coated overnight with 100 µL of 5 µg/mL human α7-acetylcholine receptor chimera (adapted from Ref.Citation37) in PBS. Plates were washed thrice with PBS and blocked with 1% bovine serum albumin (BSA) in PBS. About 4 nM of biotinylated α-cobratoxin with various concentrations of 368_01_C01, 2552_02_B02, or a negative control IgG specific to dendrotoxins in 0.1% BSA was prepared and preincubated for 30 minutes at room temperature. Plates were washed thrice, and 100 µL of the preincubated toxin and antibody mixture was added to the blocked wells. Following incubation for 1 hour, the plates were washed thrice with PBS-T (PBS, 0.1% Tween-20) and thrice with PBS, and 100 µL of 1 µg/mL of Europium-labeled Streptavidin (Perkin Elmer, 1244–360) in 0.1% BSA was added. Following 30 minutes of incubation, plates were washed thrice with PBS-T and thrice with PBS and 100 µL of DELFIA Enhancement Solution (Perkin Elmer, 4001–0010) was added to each well. Signals were measured using a VICTOR Nivo Multimode Microplate Reader using excitation at 320 nm and emission at 615 nm. Each antibody concentration was tested in quadruplicate.
Electrophysiology
Planar whole-cell patch-clamp experiments were carried out on a QPatch II automated electrophysiology platform (Sophion Bioscience), where 48-channel patch chips with 10 parallel patch holes per channel (patch hole diameter ∼1 μm, resistance 2.00 ± 0.02 MΩ) were used.
The cell line used was a human-derived Rhabdomyosarcoma RD cell line (CCL-136, from ATCC), endogenously expressing the muscle-type nAChR, composed of the α1, β1, δ, γ, and ε subunits. The cells were cultured according to the manufacturer’s guideline, and on the day of the experiment, enzymatically detached from the culture flask and brought into suspension.
For patching, the extracellular solution contained 145 mM NaCl, 10 mM HEPES, 4 mM KCl, 1 mM MgCl2, 2 mM CaCl2, and 10 mM glucose, pH adjusted to 7.4 and osmolality adjusted to 296 mOsm. The intracellular solution contained 140 mM CsF, 10 mM HEPES, 10 mM NaCl, 10 mM EGTA, pH adjusted to 7.3, and osmolality adjusted to 290 mOsm.
In the experiments, an nAChR-mediated current was elicited by 70 µM acetylcholine (ACh, Sigma-Aldrich), approximately the EC80 value, and after compound wash-out, 2 U acetylcholinesterase (Sigma-Aldrich) was added to ensure complete ACh removal. The ACh response was allowed to stabilize over three ACh additions, before the fourth addition was used to evaluate the effect of α-cobratoxin (4 nM α-cobratoxin, reducing the ACh response by 80%), preincubated with varying concentrations of IgGs. α-cobratoxin and IgGs were preincubated at room temperature for at least 30 minutes before application, and the patched cells were preincubated with α-cobratoxin and IgG for 5 minutes prior to the fourth ACh addition. As a negative control, an IgG specific to dendrotoxins was included.
The inhibitory effect of α-cobratoxin was normalized to the full ACh response (fourth response normalized to third response), plotted in a non-cumulative concentration-response plot, and a Hill fit was used to obtain IC50 values for each IgG. The data analysis was performed in Sophion Analyzer (Sophion Bioscience) and GraphPad Prism (GraphPad Software).
Animals
In vivo assays were conducted in CD-1 mice of both sexes of 18–20 g body weight, supplied by Instituto Clodomiro Picado, following protocols approved by the Institutional Committee for the Use and Care of Animals (CICUA), University of Costa Rica (approval number CICUA 82–08). Mice were housed in cages in groups of 3–4 and were provided food and water ad libitum.
In vivo preincubation experiments
The neutralization activity of the naïve IgGs against α-cobratoxin was tested by i.v. injection in groups of three mice. About 4 µg of α-cobratoxin (corresponding to 2 LD50s) and 150 µg of corresponding IgG (α-cobratoxin:IgG = 1:2 molar ratio) were dissolved in PBS, preincubated (30 min at 37 °C), and injected into the caudal vein, using an injection volume of 100 µL. Control mice were injected with either isotype control IgG and α-cobratoxin or α-cobratoxin alone. Deaths were recorded, and Kaplan–Meier curves were used to represent mouse survival along time.
For the affinity-matured clone, similar in vivo experiments were conducted, except the IgG was preincubated with 9.12 µg of N. kaouthia whole venom (Latoxan, L1323), corresponding to 2 LD50s and containing approximately 1.2 nmol of α-cobratoxin, at a 1:1, 1:2, and 1:4 α-neurotoxin:IgG molar ratio. For calculating molar ratios, it was estimated that 55% of N. kaouthia venom consists of α-neurotoxins, based on a toxicovenomic study of the venom.Citation7 All injections were performed as described above on groups of four mice. Control mice were injected with either an isotype control IgG incubated with N. kaouthia venom or N. kaouthia venom alone. As a positive control for N. kaouthia venom neutralization, Snake Venom Antiserum from VINS Bioproducts Limited (Batch number: 01AS13100) was used. According to the manufacturer, the potency of this antivenom against the venom of Naja naja is 0.6 mg venom neutralized per mL antivenom. Since no information is provided on the neutralization of N. kaouthia venom, we used a ratio of 0.2 mg venom per mL antivenom to ensure neutralization. Survival was monitored for 24–72 hours, and results are presented in Kaplan–Meier curves.
Abbreviations
Ach, Acetylcholine; BSA, Bovine serum albumin; CDR, Complementarity-determining region; CICUA, Institutional Committee for the Use and Care of Animals; CI95, 95% confidence interval; DELFIA, Dissociation-enhanced lanthanide fluorescence immunoassay; DNA, Deoxyribonucleic acid; EC80, 80% of maximal effective concentration; EDC, Ethyl-3-(3-dimethylaminopropyl)carbodiimide; ENC DELFIA, Expression-normalized capture DELFIA; ERC, European Research Council; Fab, Fragment antigen-binding; HEPES, (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; IC50, Half-maximal inhibitory concentration; IC80, 80% of maximal inhibitory concentration; IgG, Immunoglobulin G; i.c.v, Intracerebroventricular; i.v, intravenous; LD50, Median lethal dose; MALDI-TOF, Matrix-assisted laser desorption/ionization time-of-flight; MPBS, PBS supplemented with milk protein; nAChR, nicotinic acetylcholine receptor; PBS, Phosphate-buffered saline; PBS-T, PBS supplemented with 0.1% Tween-20; PCR, Polymerase chain reaction; PEG, Polyethylene glycol; RU, Response units; scFv, Single-chain variable fragment; SPR, Surface plasmon resonance; VH, Human immunoglobulin heavy-chain-variable; VL, Human immunoglobulin light-chain-variable; WHO, World Health Organization
Supplemental Material
Download Zip (31.4 KB)Acknowledgments
The authors are thankful to Georgia Bullen from IONTAS Ltd. for general guidance in the laboratory as well as to Yessica Wouters from the Technical University of Denmark for help with data analysis. Birte Svensson from the Technical University of Denmark is thanked for discussions on SPR analysis. is created using BioRender.com.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19420862.2022.2085536
Additional information
Funding
References
- Gutiérrez JM, Calvete JJ, Habib AG, Harrison RA, Williams DJ, Warrell DA. Snakebite envenoming. Nat Rev Dis Primer. 2017;3:1–11.
- Viravan C, Veeravat U, Warrell MJ, Theakston RDG, Warrell DA. ELISA confirmation of acute and past envenoming by the monocellate Thai Cobra (Naja kaouthia). Am J Trop Med Hyg. 1986;35:173–81. doi:10.4269/ajtmh.1986.35.173
- Warrell D. 1995. Clinical Toxicology of Snakebite in Asia. In: Meier J, White J, editors. . Handbook of clinical toxicology of animal venoms and poisons. Boca Raton, FL, USA: CRC Press. pp. 493–594.
- Asia RO for S-E ARO, Organization WH. 2016. Guidelines for the management of snakebites [Internet]. WHO Regional Office for South-East Asia; [accessed 2022 June 8]. https://apps.who.int/iris/handle/10665/249547
- Warrell D. 1995. Handbook of clinical toxicology of animal venoms and poisons. In: Meier J, White J, editors. Toxicon. Vol. 35. Boca Raton (FL): CRC Press; p. 752.
- Alkondon M, Albuquerque EX. alpha-Cobratoxin blocks the nicotinic acetylcholine receptor in rat hippocampal neurons. Eur J Pharmacol. 1990;191:505–06. doi:10.1016/0014-2999(90)94190-9
- Laustsen AH, Gutiérrez JM, Lohse B, Rasmussen AR, Fernández J, Milbo C, Lomonte B. Snake venomics of monocled cobra (Naja kaouthia) and investigation of human IgG response against venom toxins. Toxicon Off J Int Soc Toxinology. 2015a;99:23–35. doi:10.1016/j.toxicon.2015.03.001
- Laustsen AH, Lohse B, Lomonte B, Engmark M, Gutiérrez JM. Selecting key toxins for focused development of elapid snake antivenoms and inhibitors guided by a Toxicity Score. Toxicon Off J Int Soc Toxinology. 2015b;104:43–45. doi:10.1016/j.toxicon.2015.07.334
- León G, Vargas M, Segura Á, Herrera M, Villalta M, Sánchez A, Solano G, Gómez A, Sánchez M, Estrada R, et al. Current technology for the industrial manufacture of snake antivenoms. Toxicon Off J Int Soc Toxinology. 2018;151:63–73. doi:10.1016/j.toxicon.2018.06.084
- León G, Herrera M, Segura Á, Villalta M, Vargas M, Gutiérrez JM. Pathogenic mechanisms underlying adverse reactions induced by intravenous administration of snake antivenoms. Toxicon Off J Int Soc Toxinology. 2013;76:63–76. doi:10.1016/j.toxicon.2013.09.010
- Laustsen AH. How can monoclonal antibodies be harnessed against neglected tropical diseases and other infectious diseases? Expert Opin Drug Discov. 2019;14:1103–12. doi:10.1080/17460441.2019.1646723
- Harrison RA, Gutiérrez JM. Priority actions and progress to substantially and sustainably reduce the mortality, morbidity and socioeconomic burden of tropical snakebite. Toxins. 2016;8:351.
- Laustsen AH, Engmark M, Clouser C, Timberlake S, Vigneault F, Gutiérrez JM, Lomonte B. Exploration of immunoglobulin transcriptomes from mice immunized with three-finger toxins and phospholipases A2 from the Central American coral snake, Micrurus nigrocinctus. PeerJ. 2017;5:e2924. doi:10.7717/peerj.2924
- Tan NH, Wong KY, Tan CH. Venomics of Naja sputatrix, the Javan spitting cobra: a short neurotoxin-driven venom needing improved antivenom neutralization. J Proteomics. 2017;157:18–32. doi:10.1016/j.jprot.2017.01.018
- Laustsen AH, Gutiérrez JM, Rasmussen AR, Engmark M, Gravlund P, Sanders KL, Lohse B, Lomonte B. Danger in the reef: proteome, toxicity, and neutralization of the venom of the olive sea snake, Aipysurus laevis. Toxicon. 2015;107:187–96. doi:10.1016/j.toxicon.2015.07.008
- Tan CH, Tan KY, Lim SE, Tan NH. Venomics of the beaked sea snake, Hydrophis schistosus: a minimalist toxin arsenal and its cross-neutralization by heterologous antivenoms. J Proteomics. 2015;126:121–30. doi:10.1016/j.jprot.2015.05.035
- Laustsen AH, Engmark M, Milbo C, Johannesen J, Lomonte B, Gutiérrez JM, Lohse B. From fangs to pharmacology: the future of snakebite envenoming therapy. Curr Pharm Des. 2016;22:5270–93. doi:10.2174/1381612822666160623073438
- Kini RM, Sidhu SS, Laustsen AH. Biosynthetic Oligoclonal Antivenom (BOA) for snakebite and next-generation treatments for snakebite victims. Toxins. 2018;10:534. doi:10.3390/toxins10120534
- Laustsen AH, Karatt-Vellatt A, Masters EW, Arias AS, Pus U, Knudsen C, Oscoz S, Slavny P, Griffiths DT, Luther AM, et al. In vivo neutralization of dendrotoxin-mediated neurotoxicity of black mamba venom by oligoclonal human IgG antibodies. Nat Commun. 2018;9:3928. doi:10.1038/s41467-018-06086-4
- Laustsen AH. Snakebites: costing recombinant antivenoms. Nature. 2016;538:41–41. doi:10.1038/538041e
- Laustsen AH, Johansen KH, Engmark M, Andersen MR. Recombinant snakebite antivenoms: a cost-competitive solution to a neglected tropical disease? PLoS Negl Trop Dis. 2017;11:e0005361. doi:10.1371/journal.pntd.0005361
- Jenkins TP, Laustsen AH. 2020. Cost of manufacturing for recombinant snakebite antivenoms. Front Bioeng Biotechnol. 8: 703. https://www.frontiersin.org/articles/10.3389/fbioe.2020.00703/full doi:10.3389/fbioe.2020.00703. 32766215.
- Richard G, Meyers AJ, McLean MD, Arbabi-Ghahroudi M, MacKenzie R, Hall JC. In vivo neutralization of α-cobratoxin with high-affinity llama single-domain antibodies (VHHs) and a VHH-Fc Antibody. PLOS ONE. 2013;8:e69495. doi:10.1371/journal.pone.0069495
- Kulkeaw K, Sakolvaree Y, Srimanote P, Tongtawe P, Maneewatch S, Sookrung N, Tungtrongchitr A, Tapchaisri P, Kurazono H, Chaicumpa W. Human monoclonal ScFv neutralize lethal Thai cobra, Naja kaouthia, neurotoxin. J Proteomics. 2009;72:270–82. doi:10.1016/j.jprot.2008.12.007
- Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple SD, Clarke KF, Conquer JS, Crofts AM, Crowther SR, Dyson MR, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi:10.1186/gb-2007-8-11-r254
- Martin CD, Rojas G, Mitchell JN, Vincent KJ, Wu J, McCafferty J, Schofield DJ. A simple vector system to improve performance and utilisation of recombinant antibodies. BMC Biotechnol. 2006;6:46. doi:10.1186/1472-6750-6-46
- Marks JD, Griffiths AD, Malmqvist M, Clackson TP, Bye JM, Winter G. By–passing immunization: building high affinity human antibodies by Chain shuffling. Bio/Technology. 1992;10:779–83.
- Casewell NR, Jackson TNW, Laustsen AH, Sunagar K. Causes and consequences of snake venom variation. Trends Pharmacol Sci. 2020;41:570–81. doi:10.1016/j.tips.2020.05.006
- Lewin M, Samuel S, Merkel J, Bickler P. Varespladib (LY315920) appears to be a potent, broad-spectrum, inhibitor of snake venom phospholipase A2 and a possible pre-referral treatment for envenomation. Toxins. 2016;8:248. doi:10.3390/toxins8090248
- Rasmussen HS, McCann PP. Matrix metalloproteinase inhibition as a novel anticancer strategy: a review with special focus on batimastat and marimastat. Pharmacol Ther. 1997;75:69–75. doi:10.1016/S0163-7258(97)00023-5
- Laustsen AH, Gutiérrez JM, Knudsen C, Johansen KH, Bermúdez-Méndez E, Cerni FA, Jürgensen JA, Ledsgaard L, Martos-Esteban A, Øhlenschlæger M, et al. Pros and cons of different therapeutic antibody formats for recombinant antivenom development. Toxicon. 2018;146:151–75. doi:10.1016/j.toxicon.2018.03.004
- Dobson CL, Devine PWA, Phillips JJ, Higazi DR, Lloyd C, Popovic B, Arnold J, Buchanan A, Lewis A, Goodman J, et al. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci Rep. 2016;6:38644. doi:10.1038/srep38644
- Dyson MR, Masters E, Pazeraitis D, Perera RL, Syrjanen JL, Surade S, Thorsteinson N, Parthiban K, Jones PC, Sattar M, et al. Beyond affinity: selection of antibody variants with optimal biophysical properties and reduced immunogenicity from mammalian display libraries. mAbs. 2020;12:1829335. doi:10.1080/19420862.2020.1829335
- Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci. 2017;114:944–49. doi:10.1073/pnas.1616408114
- Bailly M, Mieczkowski C, Juan V, Metwally E, Tomazela D, Baker J, Uchida M, Kofman E, Raoufi F, Motlagh S, et al. Predicting antibody developability profiles through early stage discovery screening. mAbs. 2020;12:1743053. doi:10.1080/19420862.2020.1743053
- Ratanabanangkoon K, Simsiriwong P, Pruksaphon K, Tan KY, Chantrathonkul B, Eursakun S, Tan CH. An in vitro potency assay using nicotinic acetylcholine receptor binding works well with antivenoms against Bungarus candidus and Naja naja. Sci Rep. 2018;8:9716. doi:10.1038/s41598-018-27794-3
- S-X L, Huang S, Bren N, Noridomi K, Dellisanti CD, Sine SM, Chen L. Ligand-binding domain of an α7-nicotinic receptor chimera and its complex with agonist. Nat Neurosci. 2011;14:1253–59. doi:10.1038/nn.2908
