ABSTRACT
The chromalveolate hypothesis presents an attractively simple explanation for the presence of red algal-derived secondary plastids in 5 major eukaryotic lineages: “chromista” phyla, cryptophytes, haptophytes and ochrophytes; and alveolate phyla, dinoflagellates and apicomplexans. It posits that a single secondary endosymbiotic event occurred in a common ancestor of these diverse groups, and that this ancient plastid has since been maintained by vertical inheritance only. Substantial testing of this hypothesis by molecular phylogenies has, however, consistently failed to provide support for the predicted monophyly of the host organisms that harbour these plastids—the “chromalveolates.” This lack of support does not disprove the chromalveolate hypothesis per se, but rather drives the proposed endosymbiosis deeper into the eukaryotic tree, and requires multiple plastid losses to have occurred within intervening aplastidic lineages. An alternative perspective on plastid evolution is offered by considering the metabolic partnership between the endosymbiont and its host cell. A recent analysis of metabolic pathways in a deep-branching dinoflagellate indicates a high level of pathway redundancy in the common ancestor of apicomplexans and dinoflagellates, and differential losses of these pathways soon after radiation of the major extant lineages. This suggests that vertical inheritance of an ancient plastid in alveolates is highly unlikely as it would necessitate maintenance of redundant pathways over very long evolutionary timescales.
Establishment of any endosymbiotic partnership between 2 free-living cells will inevitably result in initial duplication of many common basic cell functions. These include anabolic pathways for essential biomolecules such as lipids, nucleotides and amino acids. Duplicated anabolic pathways create initial redundancy, but the evolution of stable endosymbiotic organelles typically results in sharing of common metabolic resources and elimination of these duplicated functions. There appears to be no absolute rule, however, for whether the host cell's pathways are retained and the endosymbiont's versions lost, or vice versa. This has been clearly demonstrated by studies of the non-photosynthetic plastids in apicomplexan parasites such as Plasmodium, the causative agent of malaria. The Plasmodium plastid, dubbed the apicoplast, is no longer photosynthetic, but is now an essential organelle because the plastid pathways for both de novo fatty acid synthesis (type II FAS pathway), and isopentenyl pyrophosphate (IPP) synthesis for isoprenoids (1-deoxy-d-xylulose-5-phosphate [DOXP] pathway) were retained instead of the host cell cytosolic pathways (type I FAS and mevalonate pathway, respectively).Citation1-4 Furthermore, a partial tetrapyrrole biosynthetic pathway in the plastid complements missing elements of the canonical host cytosol/mitochondrion pathway.Citation5,6 Thus, in the case of tetrapyrrole synthesis, elimination of enzyme redundancy resulted in a chimeric pathway dependent on both the symbiont and host compartments. The process of rationalising host/endosymbiont metabolic redundancy would have initially had no cost, perhaps even benefits, and probably occurred haphazardly in many instances. But if one or more elements of the endosymbiont's metabolism are kept in place of the cytosolic equivalents, these functions can commit cells to enduring alliances with their endosymbiont. Many of these pathways are complex, consisting of several enzymatic steps, and thus cannot be easily regained in their entirety by horizontal gene transfers. Plastid loss after stable endosymbiosis, therefore, is seemingly very difficult to achieve, even when the function that drove the initial endosymbiosis, such as photosynthesis, is lost.
We have recently described a rare example of plastid loss in Hematodinium sp., a parasitic deep-branching dinoflagellate within the apicomplexan-dinoflagellate radiation ().Citation7 This is the second only clear case of plastid loss to date, the other being from the apicomplexan Cryptosporidium.Citation8 Key to the successful loss of a plastid in Hematodinium was the retention of some of the host cell-based metabolic pathways in place of plastid ones, thus providing host cell independence. In turn, presence of these host pathways, as alternatives to the plastid pathways found in Plasmodium, demonstrated that the common ancestor of apicomplexans and dinoflagellates must have still contained redundant metabolic pathways. Differential rationalisation of this redundancy evidently then occurred in the radiating lineages. By surveying molecular data for pathway presence from across extant apicomplexan and dinoflagellate taxa, a compilation of pathways present in the common ancestor can be made ().Citation6,7,9-11 This shows that redundant host and plastid pathways were present for fatty acid synthesis (type I and type II FAS) and tetrapyrrole synthesis (plastid-based C5 [glutamate] pathway and cytosol/mitochondrion-based C4 [Shemin] pathway). Furthermore, redundancy in IPP synthesis for isoprenoids must have been present before apicomplexans and dinoflagellates diverged because, while both lineages now exclusively use the plastid DOXP pathway, the sister Alveolata lineage, ciliates, retains the host mevalonate pathway ().Citation12 Our studies of Hematodinium also reveal the presence of a distinctive plastid-type diaminopimelate lysine biosynthetic pathway.Citation7,13,14 This pathway occurs in plastids of red algae and other lineages with red algal-derived plastids (e.g. ochrophytes, haptophytes),Citation7 suggesting that it was most likely present also in the original red plastid of the common ancestor of apicomplexans and dinoflagellates. This pathway is now only present in deep-branching dinoflagellates (Perkinsus, Oxyrrhis and Hematodinium),Citation7 and in all cases it is predicted to be relocated to the cytosol. This implies further differential evolution of a plastid function after apicomplexan-dinoflagellate divergence ().
Figure 1. Schematic phylogeny of alveolates (black) with inferred metabolic pathway presence, loss and redundancy indicated (colored lines). Plastid-derived pathways are shown right of phylogeny branches, host-derived pathways (located in the cytosol or mitochondrion) are shown to the left, and pathway loss is indicated by lines ending in dashes. The point of greatest inferred metabolic pathway redundancy is indicated by circles. Formation of a single chimeric tetrapyrrole pathway in apicomplexans is indicated by the merger of the plastid and cytosol/mitochondrial pathways to an undulating line. Inferred relocation of the diaminopimelate lysine pathway from the plastid to cytosol is shown by right to left switching (the number of relocations is unknown). Question marks indicate unconfirmed presence of type I fatty acid synthase in dinoflagellates and colpodellids where distinction from polyketide synthases is difficult from current incomplete gene sequence data. M, mevalonate; D, DOXP; I, type I; II, type II.
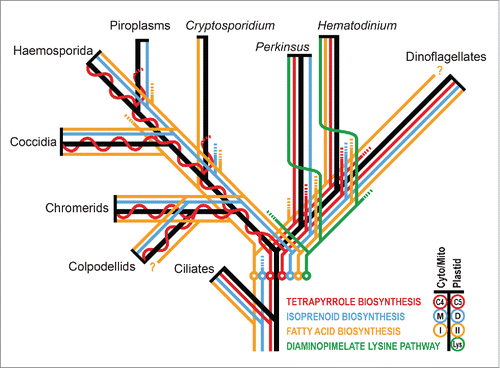
Metabolic reconstruction of the apicomplexan-dinoflagellate common ancestor thus provides a view of a cell that maintained a surprising level of host-plastid metabolic redundancy. Surprising because we do not find any extant taxa in either apicomplexan or dinoflagellate lineages where such a level of redundancy is maintained (). Indeed across plastid containing organisms, be they primary or secondary plastids, the norm is that such redundancy has long since been eliminated. If the chromalveolate hypothesis is correct plastid gain and the acquisition of this redundancy was ancient. In fact it would have to predate not only the divergence of ciliates, but most other major eukaryotic lineages given present placement of “chromalveolate” taxa on eukaryotic phylogenies (). Maintenance of redundant pathways through all of this time is difficult to reconcile with the rapid losses of different elements of this redundancy evident since apicomplexans and dinoflagellates radiated. The alternative explanation is that an independent plastid gain (or gains) occurred much more recently in the lineage(s) leading to extant apicomplexans and dinoflagellates. In this case maintenance of redundancy might only have been short lived, before divergence of these lineages and development of the different combinations of loss that we see today in apicomplexans and dinoflagellates. Perhaps, even, gain of a plastid was a catalyst for the remarkable radiation of apicomplexans and dinoflagellates as algae, symbionts and parasites from their more invariant free-living colponemid-like ancestors.Citation15,16
Figure 2. Schematic of eukaryotic phylogeny of major lineages based on recent published phylogenomic analyses e.g.,Citation26,27 Red algal-derived secondary plastid containing lineages are shown in red. For simplicity, only select aplastidic lineages related to ochrophytes, haptophytes and cryptophytes are shown. Primary plastid-containing lineages are shown in green. Uncertainty in lineage branching order is shown either as polytomies or broken lines. The chromalveolate hypothesis predicts that a common ancestor that gave rise to all red lineages acquired a single red algal-derived plastid. Current eukaryotic phylogenies require this to be very early in eukaryotic evolution, and for multiple cases of plastid loss in descendant aplastidic lineages.
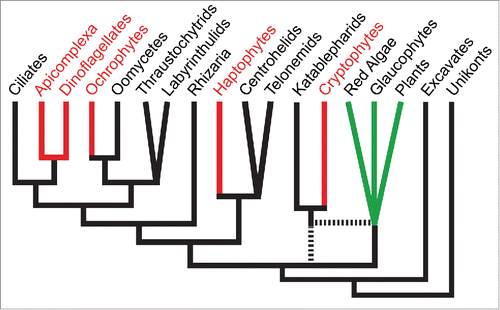
The failure of phylogenies to support the simple radiation of eukaryotes with secondary red algal-derived plastids that the chromalveolate hypothesis predicted has given rise to alternative proposals of multiple lateral endosymbiotic gains and/or exchanges of red algal symbionts.Citation17-23 These include secondary, tertiary and even quaternary endosymbiotic events, the permutations of which remain fluid as molecular phylogenies continue to expand in coverage and vacillate in the host and plastid relationships that they describe. Nevertheless, there is growing consideration of multiple endosymbiotic gains and/or exchanges of red algal-derived plastids across eukaryotes. On the other hand, the argument for a single red plastid gain is sustained by the presence of common derived features of protein-targeting shared by many secondary red algal-derived plastids—the so-called SELMA (symbiont-specific endoplasmic reticulum [ER]-like machinery) that was repurposed from the symbiont ER-associated protein degradation (ERAD) pathway.Citation24,25 This argument hinges on the perceived improbability that either: 1) the ERAD machinery could be repurposed for plastid protein targeting in red algal-derived plastids more than once; or 2) SELMA could be inherited during tertiary or quaternary endosymbioses of algae that already possessed this machinery. But these probabilities are unknown, making the SELMA argument difficult to assess. Metabolic redundancy in lineages that have gained plastid organelles, however, is uncommon and thus the likelihood of its maintenance over very long evolutionary timescales is apparently very low. Evidence of redundancy in the apicomplexan-dinoflagellate common ancestor, therefore, favors a more recent gain of a plastid in this lineage, after ciliates diverged, and perhaps relatively soon before radiation of the remarkably diverse apicomplexan-dinoflagellate group. These data are also consistent with possible further recent plastid endosymbioses that might have either replaced this plastid, or contributed to metabolic capabilities found in extant taxa. In either case, these data argue that the chromalveolate hypothesis, with a single ancient gain of a red complex plastid, is unlikely to be correct.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Chris Howe for comments on this manuscript.
Funding
This work was supported by the MRC (MR/M011690/1). SGG was supported by Science Foundation Ireland Grant 13/SIRG/2125.
References
- Waller RF, Keeling PJ, Donald RG, Striepen B, Handman E, Lang-Unnasch N, Cowman AF, Besra GS, Roos DS, McFadden GI. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci USA 1998; 95:12352-7; PMID:9770490; http://dx.doi.org/10.1073/pnas.95.21.12352
- Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Türbachova I, Eberl M, Zeidler J, Lichtenthaler HK, et al. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 1999; 285:1573-6; PMID:10477522; http://dx.doi.org/10.1126/science.285.5433.1573
- Yu M, Kumar TRS, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, et al. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe 2008; 4:567-78; PMID:19064257; http://dx.doi.org/10.1016/j.chom.2008.11.001
- Yeh E, DeRisi JL. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. Plos Biol 2011; 9:e1001138; PMID:21912516; http://dx.doi.org/10.1371/journal.pbio.1001138
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol 2004; 2:203-16; PMID:15083156; http://dx.doi.org/10.1038/nrmicro843
- Seeber F, Soldati-Favre D. Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol 2010; 281:161-228; PMID:20460186; http://dx.doi.org/10.1016/S1937-6448(10)81005-6
- Gornik SG, Febrimarsa, Cassin AM, Macrae JI, Ramaprasad A, Rchiad Z, McConville MJ, Bacic A, McFadden GI, Pain A, et al. Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc Natl Acad Sci USA 2015; 112:5767-72
- Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, et al. The genome of Cryptosporidium hominis. Nature 2004; 431:1107-12; PMID:15510150; http://dx.doi.org/10.1038/nature02977
- Janouškovec J, Tikhonenkov DV, Burki F, Howe AT, Kolisko M, Mylnikov AP, Keeling PJ. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci USA 2015; 112:10200-7; http://dx.doi.org/10.1073/pnas.1423790112
- Woo YH, Ansari H, Otto TD, Klinger CM, Kolisko M, Michálek J, Saxena A, Shanmugam D, Tayyrov A, Veluchamy A, et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. eLife 2015; 4:e06974; PMID:26175406; http://dx.doi.org/10.7554/eLife.06974
- Bentlage B, Rogers TS, Bachvaroff TR, Delwiche CF. Complex ancestries of isoprenoid synthesis in dinoflagellates. J Euk Micro 2015; 63:123-137
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. Plos Biol 2006; 4:e286; PMID:16933976; http://dx.doi.org/10.1371/journal.pbio.0040286
- Hudson AO, Singh BK, Leustek T, Gilvarg C. An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol 2006; 140:292-301; PMID:16361515; http://dx.doi.org/10.1104/pp.105.072629
- McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, Maurelli AT. L, L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc Natl Acad Sci USA 2006; 103:17909-14; PMID:17093042; http://dx.doi.org/10.1073/pnas.0608643103
- Janouškovec J, Tikhonenkov DV, Mikhailov KV, Simdyanov TG, Aleoshin VV, Mylnikov AP, Keeling PJ. Colponemids represent multiple ancient alveolate lineages. Curr Biol 2013; 23:2546-52; http://dx.doi.org/10.1016/j.cub.2013.10.062
- Tikhonenkov DV, Janouškovec J, Mylnikov AP, Mikhailov KV, Simdyanov TG, Aleoshin VV, Keeling PJ. Description of Colponema vietnamica spn. and Acavomonas peruviana n. gen. n. sp., Two new alveolate phyla (Colponemidia nom. nov. and Acavomonidia nom. nov.) and their contributions to reconstructing the ancestral state of alveolates and eukaryotes. Plos One 2014; 9:e95467; PMID:24740116; http://dx.doi.org/10.1371/journal.pone.0095467
- Puerta MS, Delwiche CF. A hypothesis for plastid evolution in chromalveolates. J Phycol 2008; 44:1097-107; http://dx.doi.org/10.1111/j.1529-8817.2008.00559.x
- Bodył A, Stiller JW, Mackiewicz P. Chromalveolate plastids: direct descent or multiple endosymbioses? Trends Ecol Evol 2009; 24:119-21; PMID:19200617; http://dx.doi.org/10.1016/j.tree.2008.11.003
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol 2010; 27:1698-709; PMID:20194427; http://dx.doi.org/10.1093/molbev/msq059
- Petersen J, Ludewig A-K, Michael V, Bunk B, Jarek M, Baurain D, Brinkmann H. Chromera velia, endosymbioses and the rhodoplex hypothesis—plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Gen Biol Evol 2014; 6:666-84; http://dx.doi.org/10.1093/gbe/evu043
- Stiller JW. Toward an empirical framework for interpreting plastid evolution. J Phycol 2014; 50:462-471.
- Stiller JW, Schreiber J, Yue J, Guo H, Ding Q, Huang J. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat Commun 2014; 5:5764; PMID:25493338; http://dx.doi.org/10.1038/ncomms6764
- Ševčíková T, Horák A, Klimeš V, Zbránková V, Demir-Hilton E, Sudek S, Jenkins J, Schmutz J, Přibyl P, Fousek J, et al. Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci Rep 2015; 5:10134; http://dx.doi.org/10.1038/srep10134
- Gould SB, Maier UG, Martin WF. Protein import and the origin of red complex plastids. Curr Biol 2015; 25:R515-21; PMID:26079086; http://dx.doi.org/10.1016/j.cub.2015.04.033
- Stork S, Moog D, Przyborski JM, Wilhelmi I, Zauner S, Maier UG. Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation. Euk Cell 2012; 11:1472-81; http://dx.doi.org/10.1128/EC.00183-12
- Burki F, Shalchian-Tabrizi K, Minge M, Skjæveland Å, Nikolaev SI, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. Plos One 2007; 2:e790; PMID:17726520; http://dx.doi.org/10.1371/journal.pone.0000790
- Burki F, Okamoto N, Pombert JF, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci 2012; 279:2246-54; PMID:22298847; http://dx.doi.org/10.1098/rspb.2011.2301
